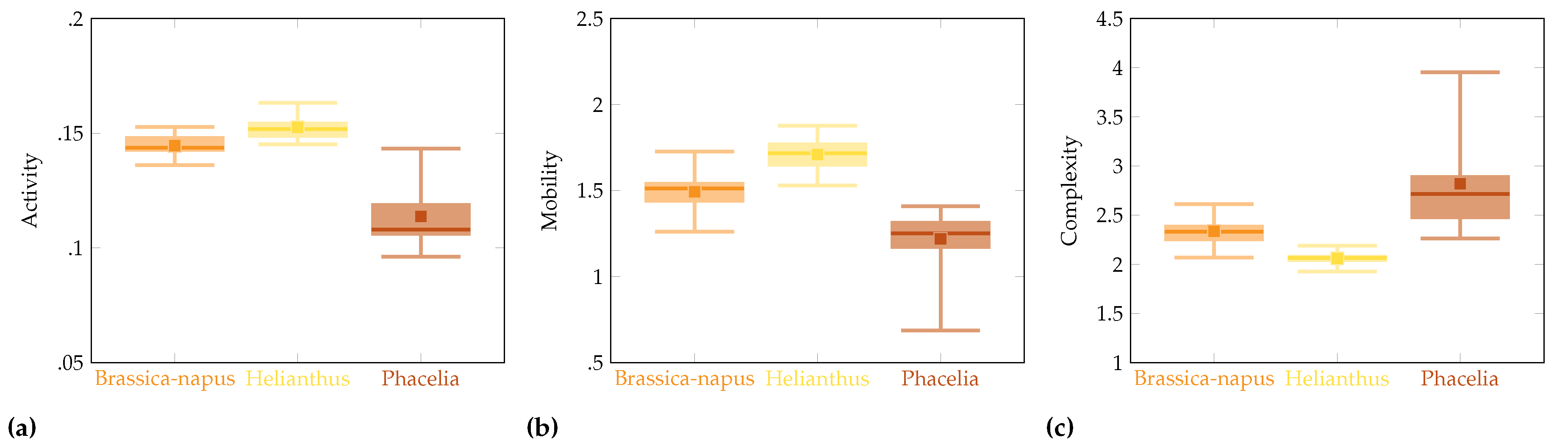1. Introduction
In the modern economy, honey has become subject to improper labeling and the resulting consequences of falsifying its properties [
1,
2]. Due to the prevalence of fake honey products, the authentication of honey has become an active area of technical testing of the quality of honey. Among many countries that play an important role in the production market for honey and products derived from honey, New Zealand and the EU countries are the leaders, taking into account the protection of the interests of consumers and producers [
3,
4]. In the context of the above statement, more and more advanced methods for representing tested products are being sought. Research is usually of an indirect nature; that is, a medium is sought that will well represent the food product while at the same time creating the possibility of using the most advanced information processing technologies regarding the tested feature or the entire product. In the case of analyzing a food product such as honey, properly prepared pollen images or carrying out the so-called pollen analysis regarding pollen images [
5,
6]. There is considerable research on the composition and pharmacological bioactivity of bee pollen (BP), indicating its usefulness and safety [
7]. Numerous studies have shown that BP has a rich and well-balanced composition to serve as a human food and supplement, while its rich bioactive compounds, especially polyphenols, provide a variety of biological and pharmacological activities [
8]. BP-related risks usually come from external factors [
9] or from improper storage and processing conditions [
8].
Allergic reactions are rare, and BP is perceived as a safe product in most physiological situations, including childhood, older age, and disease recovery [
10,
11]. BP from different floral sources has been reported to have anesthetic, antiallergic, anti-androgen, anti-atherosclerotic, anticancer (anticarcinogenic and anti-mutagenic), anti-inflammatory, antimicrobial (antibacterial, antifungal, and antiviral), antioxidant, antiulcer, and immunostimulant activities [
12]. In metabolic pathophysiology, it has been shown to possess anti-obesity, antidiabetic [
13], hypocholesterolemic [
14], and hepatoprotective [
11] effects. In the digestive system, it has been shown to maintain [
10], improve [
11], and regulate [
14] intestinal functions. In the cardiovascular system, it can reduce capillary fragility [
12] and improve overall cardiovascular health [
15]. Some authors reported that BP can contribute to the prevention of and positively impact some degenerative processes such as neurodegeneration [
15], general aging [
13,
14], and cellular apoptosis [
16], and can promote recovery from chronic diseases and possess chemo-preventive properties [
3]. It has also been shown to improve skin health and reparation, including many valuable cosmetic qualities [
13].
Due to its established nutritional use, an ISO Norm (ISO 24382:2023) has been published to standardize the quality of bee pollen as a food product [
17]. This clear interest in BP is evidently based on a solid background of evidence originating from the ethnopharmacological heritage and increasingly collected experimental data. Although BP in its isolated pellet format may be relatively recent and emerged with the elaboration of mechanical pollen traps, bee bread and plant pollen have been used since ancient times. Ancient Egyptians described pollen as a ”life-giving dust” [
18], and Greeks believed that pollen and honey gave youth to kings [
19]. Pollen was used for cosmetic purposes in ancient China [
20]. Ancient Egyptians, Greeks, native Americans, Chinese, and Indians have been reported to have used BP for food and energy on long journeys, as well as for other health benefits [
21]. Based on this nutritional and ethnomedicinal history, BP use has been spreading at a very rapid pace during the last few years due to emerging scientific evidence and the extensive development of the dietary supplement market around the world. BP is widely commercialized as a standalone food and nutritional supplement that benefits from its rich and well-balanced composition. The global market is expected to reach around 670 million Euros in 2024 [
22].
In this study, we introduce a novel approach to the analysis of bee pollen using Hjorth descriptors [
5,
6] to create a tool for a relatively simple and reliable classification that distinguishes counterfeit products that lack appropriate metrological characteristics.
The structure of the paper is as follows: in
Section 2, we introduce the database of photographed honey samples in (
Section 2.1) and the data processing pipeline consisting of data set preparation, normalization and filtration, feature extraction and selection using Hjort descriptors, and classification (
Section 2.2).
Section 3 presents the results and the summary is presented in
Section 4 and
Section 5.
2. Materials and Methods
2.1. Materials
2.1.1. Image Acquisition Procedure
The photographed honey sample was prepared as twice-centrifuged honey sediment taken from 10 g of honey. The centrifugation process lasted 10 minutes each time at a speed of 3000 rpm. Before the first centrifugation, the sample was topped with distilled water at 20 ml and then after centrifugation, the liquid above the sediment was removed and refilled with distilled water at 20 ml. After the second centrifugation, the liquid was decanted and the remaining sediment was mixed and placed on a glass slide.
The photos were taken using a digital camera with a resolution of 5 megapixels, connected to the tube of an optical microscope, the parameter defining the lens magnification was ×60.
Bee pollen was extracted from the photos using a convolutional neural network (CNN). The pre-trained ResNet50 model [
23] was used as a base. The ResNet50 model was then trained based on the prepared pollen masks, which enabled it to effectively extract pollen images from honey samples.
ResNet-50 is CNN architecture that belongs to the ResNet (Residual Networks) family, a series of models designed to address the challenges associated with training deep neural networks. Developed by researchers at Microsoft Research Asia, ResNet-50 is renowned for its depth and efficiency in image classification tasks. ResNet architectures come in various depths, such as ResNet-18, ResNet-32, etc., with ResNet-50 being a midsized variant. ResNet-50 was released in 2015 but remains a notable model in the history of image classification.
The primary problem ResNet solved was the degradation problem in deep neural networks. As networks become deeper, their accuracy saturates and then degrades rapidly. This degradation is not caused by overfitting, but rather by the difficulty of optimizing the training process.
ResNet solved this problem using Residual Blocks that allow for the direct flow of information through the skip connections, mitigating the vanishing gradient problem.
2.1.2. Database
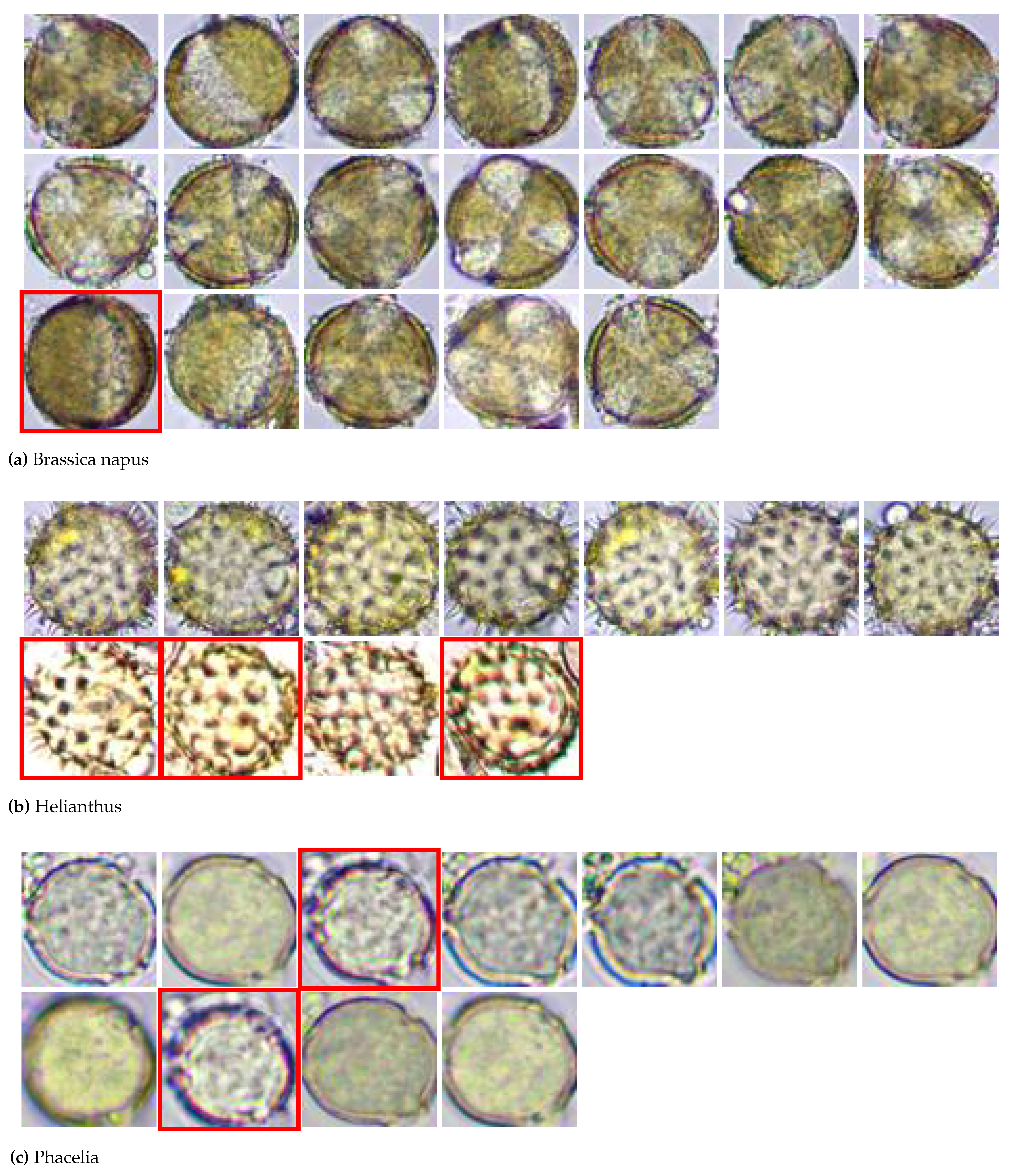
The testing database contained 41 color images divided into three groups: Brassica napus (19 images), Helianthus (11 images) and Phacelia (11 images), each with a resolution of 256×256 pixels and an 8-bit depth. The whole database is shown in
Figure 1. In each of the three groups, some images were excluded (images marked in red boxes: Brassica napus 1 image, Helianthus 3 images, and Phacelia 2 images) due to their high mismatch with the group.
Finally, 35 images (18 images of Brassica napus, 8 images of Helianthus, and 9 images of Phacelia) were used for further analysis.
2.2. Methods
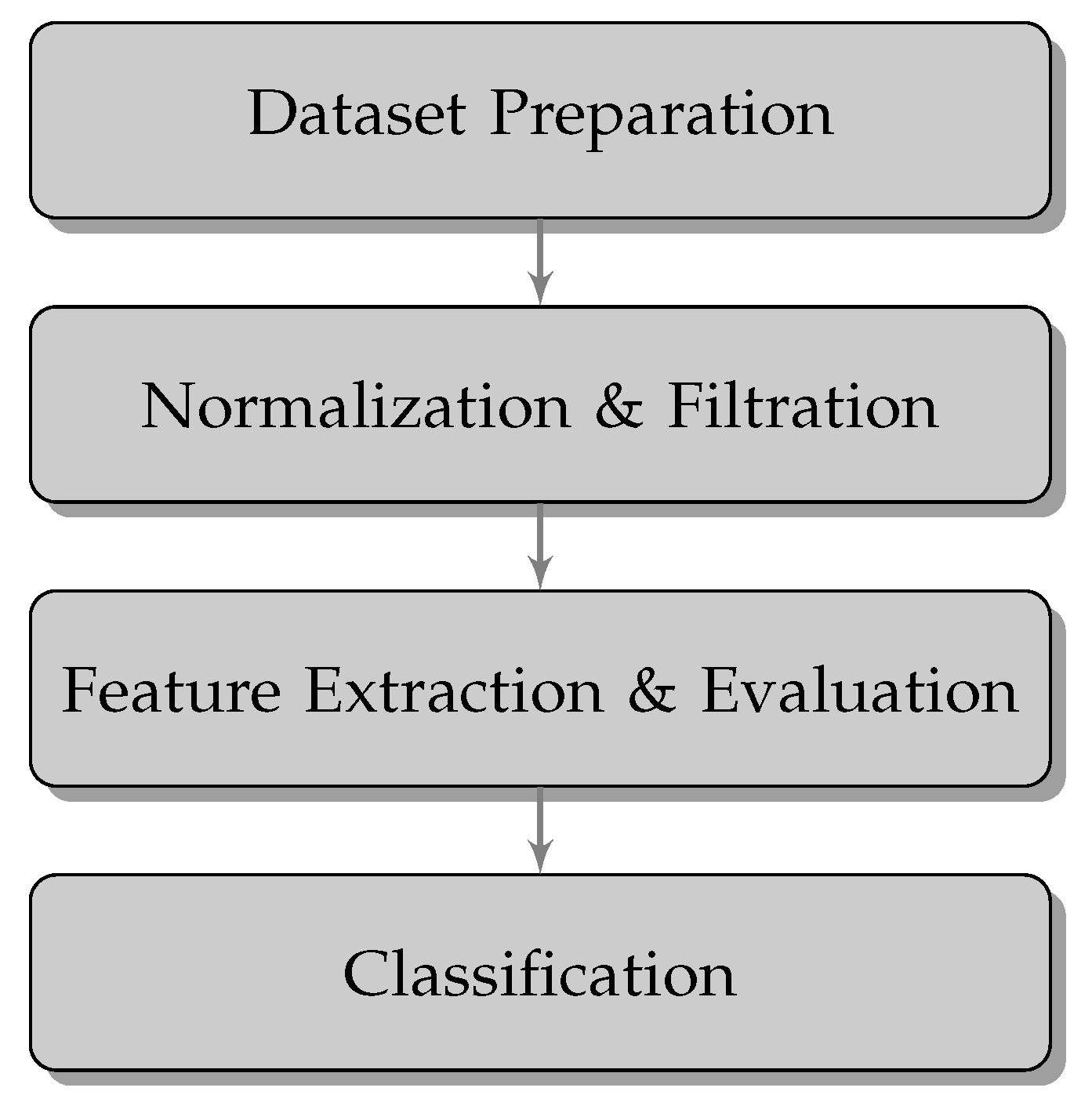
The proposed fully automatic Bee Pollen classification algorithm consists of several steps presented in
Figure 2. After the
preparation of the database procedure (described in
Section 2.1 images are subjected to a
Normalization & Filtration stage. Then,
Feature Extraction & Evaluation procedure is implemented. Finally, based on extracted features, image
Classification stage is performed.
2.2.1. Data Preparation
Since the numerical analysis uses monochromatic images, the first step is to convert the color RGB images (such as shown in
Figure 1) to a grayscale. A grayscale image may be defined as a two-dimensional function
, where
x and
y are spatial (plane) coordinates, and the amplitude of
I in any pair of coordinates
is called the gray level of the image at that point. For simplicity, such an image will be denoted as
I.
2.2.2. Normalization & Filtration
The next stage after data preparation is intensity normalization and image filtration. The commonly used normalization approach, where the image intensity range gets transformed into the range from 0–1, is a Min-Max normalization or rescaling. It works according to the formula:
where
denotes original image. This transformation does not affect the image itself.
Because the derivatives used later are very sensitive to noise, before they are determined, the normalized image is filtered using the average filter. Filter coefficients are calculated according to the average and standard deviation of the image.
2.2.3. Feature Extraction and Evaluation
Further stages of the analysis will use image derivatives. As is commonly known, in the case of 2D signals, the derivative may be directional. However, the directionality of the derivatives in the developed approach is unimportant. For this reason, the gradient modules were determined using combined filters. In the case of the first derivative, the following formula was used:
while for the second derivative:
For simplicity, the first and the second image derivatives in any pair of coordinates will be denoted as and , respectively.
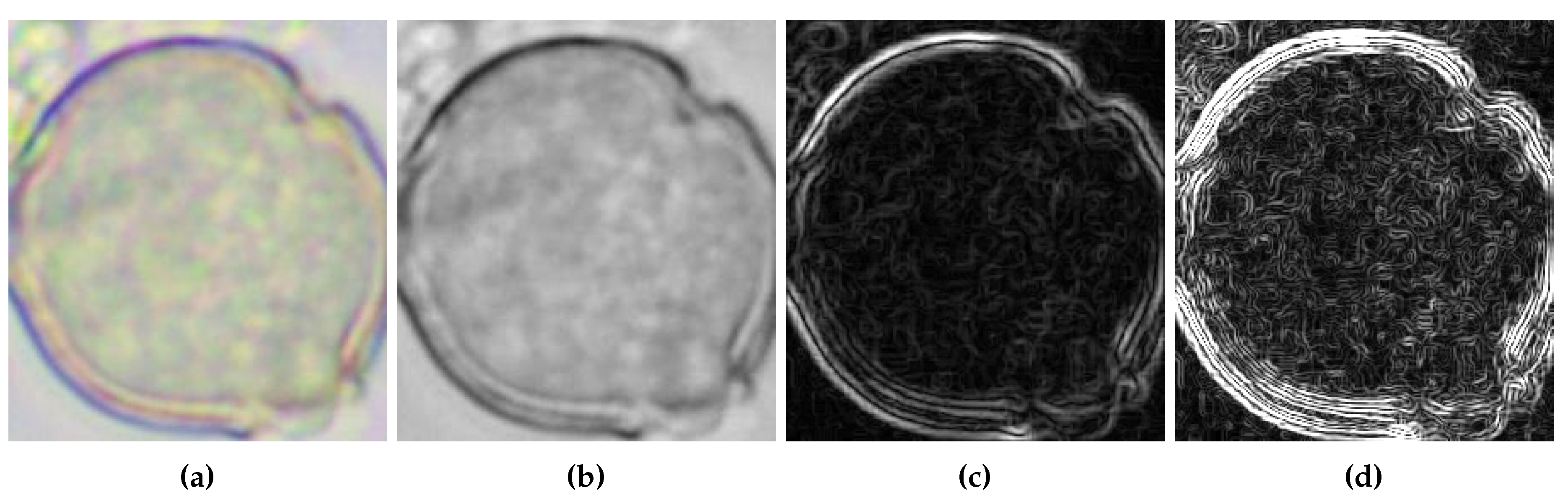
The above stages of image preparation and derivative calculation are illustrated in
Figure 3. For further analysis, the images presented in
Figure 3b–d will be used.
Hjorth Descriptors
The next step after image preparation, normalization, and filtering is feature extraction and evaluation. In the current submission, Hjorth descriptors were used to extract image features.
The information processing method that uses Hjorth descriptors is a little over 50 years old because, in 1970 [
5], Swedish researcher Bo Hjorth proposed the construction of three descriptors: Activity, Mobility, and Complexity to extract classification features and effectively evaluate the EEG signal.
The research using Hjorth descriptors is based on a specific procedure that is relatively typical in terms of recognition and classification issues (see
Figure 2).
The EEG signal is, as we know, a one-dimensional signal. Hence, an important novelty in the developed analysis methodology is an attempt to modify the above-mentioned descriptors in order to create the possibility of their application to two-dimensional signals, such as the images of bee pollen being analyzed.
The definition of Hjorth descriptors is derived from the frequency domain. The spectral moment of order
n is defined as:
where
is the power spectrum function. This function is computed as the product of the Fourier transform of the signal
described as
and its complex conjugate
. The spectral moment of order 0 (zero)
represents the total power of the signal in the frequency domain:
According to Parseval’s theorem [
24], the mean power in the time domain is equal to the total power of the signal in the frequency domain, that is:
This spectral moment of order 0 is the first Hjorth’s descriptors called Activity [
25]. The Activity of a signal is defined as the variance of the signal amplitude:
The dimension of the Activity parameter is the square value of the considered signal. It is worth noting that the variance of a zero-mean signal is equal to its mean power in the time domain:
By analogy, the spectral moment of order 2 represents the average frequency of the signal:
is known the second Hjorth’s descriptors called Mobility:
And finally, the spectral moment of order 4 represents the bandwidth of the signal:
is the third Hjorth’s descriptors called Complexity:
3. Results
The considerations carried out allowed for the obtaining of the following results, which will be presented in several aspects. First, a collective analysis of the Hjorth descriptors proposed for bee pollen image research was presented to calculate them in an appropriate manner for the three groups of bee pollen and the three descriptors described above (
Figure 4).
This first approach seems to create a reasonable hope of achieving the intended goal, which is a general and independent classifier of bee pollen for their general distinguishability and, subsequently, for checking possible frauds in the production of the final product.
3.1. Statistical Analysis
A statistical analysis has been performed to check whether Hjorth descriptors can distinguish the various bee pollen groups; in our case, the groups are Brassica-napus, Helianthus, and Phacelia. In the first step of analysis, the assumptions of a simple ANOVA test have been verified. Due to the lack of normal distribution and homogeneity of variance, confirmed by the Shapiro-Wilk test and the Levene test (
Table 1 and
Table 2), respectively, and additionally due to the small size of the samples, the nonparametric Kruskal-Wallis ANOVA test, based on ranks instead of measurements, has been applied for the Hjorth descriptors (i.e., Activity, Mobility, and Complexity) representing the bee pollen groups.
Kruskal-Wallis ANOVA test assesses whether n independent samples come from the same population (distribution) or the population with the same median (null hypothesis). According to the results summarized in
Table 3, there is a significant difference between the bee pollen groups under consideration regarding Activity p<0.00001, Mobility p<0.0001, and Complexity p<0.00001. Based on the results presented, it can be stated that Activity, Mobility, and Complexity have a significant statistical impact on the affiliation of bee pollen to a specific group (Brassica-napus, Helianthus, and Phacelia).
Multiple comparison post hoc tests have been performed to answer the question of which of the compared groups is responsible for rejecting the null hypothesis, that is, to indicate which two pollen groups differ significantly. In line with the result obtained (
Table 4,
Figure 4) for Activity, significant differences have been pointed out between groups Brassica-napus & Phacelia, Helianthus & Phacelia, for Mobility between groups Brassica-napus & Helianthus, Helianthus & Phacelia, and Complexity between groups Brassica-napus & Helianthus, Helianthus & Phacelia.
The separation of individual image groups based on the Hjorth descriptors is also visible in
Figure 5.
4. Discussion
This paper presents a novel approach to using the Hjorth descriptor method, known for many years, originally developed for the analysis of one-dimensional electroencephalographic (EEG) signals in the field of analysis of two-dimensional signals, such as bee pollen images. A detailed procedure for acquiring the images mentioned above, which became the basis for the analysis of their discrimination, was described. Therefore, a successful attempt was made to use image parameterization using Hjorth descriptors to construct an initial classifier of bee pollen images.
It should be clearly emphasized here that the primary goal for which such an attempt was made is to introduce some order into the honey production and distribution market, as there are rather chaotic and sporadic attempts to use modern digital tools to make honey production an important element of food product management.
In the paper, intensive statistical analyzes were performed after a detailed description of the tool and the subject of the study, leading to interesting conclusions. Using the descriptors Activity, Mobility and Complexity, it was possible to effectively distinguish the three groups of bee pollen studied. Therefore, appropriate tests of normality of the distribution of the parameters studied (Shapiro-Wilk test) and homogeneity of variance tests (Levene test) were performed. In turn, ANOVA rank (Kruskal-Wallis test) clearly indicated that the listed descriptors in the post hock test clearly allow for distinguishing the groups of bee pollen described above.
Therefore, to summarize this discussion, it can be stated that a successful attempt was made to parameterize bee pollen images using Hjorth descriptors: Activity, Mobility, and Complexity.
5. Conclusions
The conclusions that emerge from this work can be summarized in two directions. First, in the opinion of the authors, a successful attempt was made to use a somewhat forgotten tool, which seems to be Hjorth descriptors, written for the first time over 50 years ago, to parameterize two-dimensional signals, such as images of bee pollen obtained in the manner described in the methods
Section 3.1. Second, the use of the aforementioned descriptors, Activity, Mobility, and Complexity, to develop a methodology for distinguishing selected groups of bee pollen images effectively leads to the development of an effective classifier necessary for conducting qualitative analyses of food products such as honey.
The presented approach, although quite effective, has a few limitations. The basic one is the relatively small number of images analyzed in the three groups: Brassica-napus, Helianthus, and Phacelia. Images of these selected bee pollens were obtained through a rather complicated technological process described in the paper. Work is underway to quantitatively enrich the database used for the author’s analyses. Then, as can be assumed, performing the analyses described in the article will definitely gain credibility and greater representativeness. The authors of this paper set this as another goal aimed at developing an effective classification tool.
Author Contributions
E.T.: conceptualization, writing and editing; P.R.: image database preparation; W.W.: processing methodology, software implementation, validation, writing and editing; B.L.: image database preparation; B.M.: statistical analysis, results verification, writing and editing, S.S.: writing and editing; All authors have read and agreed to the published version of the manuscript.
Funding
This article was supported by the project program NUTRITECH1/0004A/2022 entitled ”Development and implementation of a globally innovative digital honey pollen analysis service using technologies based on automation and artificial intelligence for use in the functional food production sector” selected as part of the 1st NUTRITECH competition – nutrition in the light of the challenges of improving the well-being of society and climate change.
Data Availability Statement
The image data could be provided on request after contact with P.R.
Acknowledgments
The authors would like to express their gratitude to the beneficiary of the competition, that is AI Technika Sp. z o.o., for providing the database of bee pollen images used in the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Puścion-Jakubik, A.; Borawska, M. Odmianowe miody pszczele – pyłki główne i towarzyszące jako podstawa ich zaklasyfikowania. Problemy Higieny i Epidemiologii, 2015, 97, 275–278.
- Criteria for Identifying Manuka Honey. New Zealand Ministry for Primary Industries, 2017.
- Alshallash, K. S.; Abolaban, G.; Elhamamsy, S. M.; Zaghlool, A.; Nasr, A.; Nagib, A.; El-Hakim, A. F. A.; Zahra, A. A.; Hamdy, A. E.; Taha, I. Bee Pollen as a Functional ProductâChemical Constituents and Nutritional Properties. Journal of Ecological Engineering, 2023, 24, 173–183. [CrossRef]
- Coordinated Control Plan to Establish the Prevalence of Fraudulent Practices in the Marketing of Honey, 2015.
- Hjorth, B. EEG analysis based on time domain properties. Electroencephalogr Clin Neurophysiol, 1970, 29, 306. [CrossRef]
- Feradov, F.; Markova, V.; Ganchev, T. Automated Detection of Improper Sitting Postures in Computer Users Based on Motion Capture Sensors. Computers, 2022, 11, 116. [CrossRef]
- Kacemi, R.; Campos, M. G. Translational Research on Bee Pollen as a Source of Nutrients: A Scoping Review from Bench to Real World. Nutrients, 2023, 15, 2413. [CrossRef]
- Campos, M. G.; Frigerio, C.; Bobis, O.; Urcan, A. C.; Gomes, N.G.M. Infrared irradiation drying impact on bee pollen: Case study on the phenolic composition of Eucalyptus globulus labill and Salix atrocinerea Brot. pollens. Processes, 2021, 9, 890. [CrossRef]
- Campos, M. G.; Anjos, O.; Chica, M.; Campoy, P.; Nozkova, J.; Almaraz-Abarca, N.; Barreto, L. M. R. C.; Nordi, J. C.; Estevinho, L. M.; Pascoal, A.; et al. Standard methods for pollen research. J. Apic. Res., 2021, 60, 1–109thods for pollen research. [CrossRef]
- Matuszewska, E.; Plewa, S.; Pietkiewicz, D.; Kossakowski, K.; Matysiak, J.; Rosinski, G.; Matysiak, J. Mass Spectrometry-Based Identification of Bioactive Bee Pollen Proteins: Evaluation of Allergy Risk after Bee Pollen Supplementation. Molecules, 2022, 27, 7733. [CrossRef]
- Kostic, A. Ž.; Milinčić, D.D.; Barać, M.B.; Shariati, M.A.; Tešić, L.; Pešić, M.B. The Application of Pollen as a Functional Food and Feed Ingredient-The Present and Perspectives. Biomolecules, 2020, 10, 84. [CrossRef]
- Baky, M. H.; Abouelela, M. B.; Wang, K.; Farag, M. A. Bee Pollen and Bread as a Super-Food: A Comparative Review of Their Metabolome Composition and Quality Assessment in the Context of Best Recovery Conditions. Molecules, 2023, 28, 715. [CrossRef]
- Chelucci, E.; Chiellini, C.; Cavallero, A.; Gabriele, M. Bio-Functional Activities of Tuscan Bee Pollen. Antioxidants, 2023, 12, 115. [CrossRef]
- Qiao, J.; Feng, Z.; Zhang, Y.; Xiao, X.; Dong, J.; Haubruge, E.; Zhang, H. Phenolamide and flavonoid glycoside profiles of 20 types of monofloral bee pollen. Food Chem., 2023, 405, 134800. [CrossRef]
- Xue, F.; Li, C. Effects of ultrasound assisted cell wall disruption on physicochemical properties of camellia bee pollen protein isolates. Ultrasonics Sonochemistry, 2023, 92, 106249. [CrossRef]
- Han, S.; Chen, L.; Zhang, Y.; Xie, S.; Yang, J.; Su, S.; Yao, H.; Shi, P. Lotus Bee Pollen Extract Inhibits Isoproterenol-Induced Hypertrophy via JAK2/STAT3 Signaling Pathway in Rat H9c2 Cells. Antioxidants, 2022, 12, 88. [CrossRef]
- ISO 24382:2023: Bee pollen, Edition 1. International Organization for Standardization, Geneva, Switzerland. https://www.iso.org/standard/78544.html?browse=tc.
- El-Seedi, H. R.; Khalifa, S. A. M.; El-Wahed, A. A.; Gao, R.; Guo, Z.; Tahir, H. E.; Zhao, C.; Du, M.; Farag, M. A.; Musharraf, S. G.; et al. Honeybee products: An updated review of neurological actions. Trends in Food Science & Technology, 2020, 101, 17–27. [CrossRef]
- Camacho-Bernal, G. I.; Cruz-Cansino, N. d. S.; Ramírez-Moreno, E.; Delgado-Olivares, L.; Zafra-Rojas, Q. Y.; Castañeda-Ovando, A.; Suárez-Jacobo, Á. Addition of Bee Products in Diverse Food Sources: Functional and Physicochemical Properties. Applied Sciences, 2021, 11, 8156. [CrossRef]
- Xi, X.; Li, J.; Guo, S.; Li, Y.; Xu, F.; Zheng, M.; Cao, H.; Cui, X.; Guo, H.; Han, C. The Potential of Using Bee Pollen in Cosmetics: A Review. Journal of Oleo Science, 2018, 67, 1071–1082. [CrossRef]
- Lu, P.; Takiguchi, S.; Honda, Y.; Lu, Y.; Mitsui, T.; Kato, S.; Kodera, R.; Furihata, K.; Zhang, M.; Okamoto, K.; et al. NMR and HPLC profiling of bee pollen products from different countries. Food Chemistry: Molecular Sciences, 2022, 5, 100119. [CrossRef]
- Tao, Y.; Zhou, E.; Li, F.; Meng, L.; Li, Q.; Wu, L. Allergenicity Alleviation of Bee Pollen by Enzymatic Hydrolysis: Regulation in Mice Allergic Mediators, Metabolism, and Gut Microbiota. Foods, 2022, 11, 3454. [CrossRef]
- Potrimba, P. What is ResNet-50? https://blog.roboflow.com/what-is-resnet-50/ [Access: 2024-07-16]et-50? https://blog.roboflow.
- Smith, S. W. The scientist and engineer’s guide to digital signal processing, 1997.
- Cocconcelli, M.; Strozzi, M.; Cavalaglio Camargo Molano J.;Rubini, R. Detectivity: A combination of Hjorth’s parameters for condition monitoring of ball bearings. Mechanical Systems and Signal Processing, 2022, 164, 108247. [CrossRef]
Figure 1.
Bee Pollen images divided into three groups (images marked in a red box were excluded from further analysis)
Figure 1.
Bee Pollen images divided into three groups (images marked in a red box were excluded from further analysis)
Figure 2.
Workflow of the proposed methodology
Figure 2.
Workflow of the proposed methodology
Figure 3.
Single data set: (a) original color image (from Phacelia group), (b) original grayscale image I, (c) first derivative of the image, (d) second derivative of the image
Figure 3.
Single data set: (a) original color image (from Phacelia group), (b) original grayscale image I, (c) first derivative of the image, (d) second derivative of the image
Figure 4.
Box plots for Hjorth descriptors: (a) Activity, (b) Mobility, (c) Complexity determined for three bee pollen groups: Brassica-napus, Helianthus, and Phacelia; the box is drawn from the first to the third quartiles, with a horizontal line drawn inside to denote the median and a dot to present the mean; the whiskers indicate the minimum and maximum descriptor’s value for the group
Figure 4.
Box plots for Hjorth descriptors: (a) Activity, (b) Mobility, (c) Complexity determined for three bee pollen groups: Brassica-napus, Helianthus, and Phacelia; the box is drawn from the first to the third quartiles, with a horizontal line drawn inside to denote the median and a dot to present the mean; the whiskers indicate the minimum and maximum descriptor’s value for the group
Figure 5.
Three-dimensional visualization of analyzed image database of bee pollen three groups represented by points with coordinates of Hjorth descriptors (Activity, Mobility, Complexity)
Figure 5.
Three-dimensional visualization of analyzed image database of bee pollen three groups represented by points with coordinates of Hjorth descriptors (Activity, Mobility, Complexity)
Table 1.
The smallest level of significance (p-value) for which the calculated value of testing statistic leads to rejection of the null hypothesis of normality distribution and homogeneity of variance for Shapiro-Wilk and Levene tests, respectively for Hjorth descriptors: Activity, Mobility, and Complexity
Table 1.
The smallest level of significance (p-value) for which the calculated value of testing statistic leads to rejection of the null hypothesis of normality distribution and homogeneity of variance for Shapiro-Wilk and Levene tests, respectively for Hjorth descriptors: Activity, Mobility, and Complexity
| Hjorth |
Normal distribution |
Homogeneity of variance |
| descriptors |
Shapiro-Wilk test |
Levene test |
| variable |
p – value |
p – value |
| Activity |
0.000177 |
0.001101 |
| Mobility |
0.017085 |
0.031298 |
| Complexity |
0.000026 |
0.000900 |
Table 2.
The exact results for the Levene test performed for the variance homogeneity of Hjorth descriptors (i.e., Activity, Mobility, and Complexity) to check the assumption of simple ANOVA method of variance analysis
Table 2.
The exact results for the Levene test performed for the variance homogeneity of Hjorth descriptors (i.e., Activity, Mobility, and Complexity) to check the assumption of simple ANOVA method of variance analysis
| Statistical |
Hjorth |
Effect |
Error |
F |
p |
| test |
descriptors |
SS |
df |
MS |
SS* |
df* |
MS* |
| Levene |
Activity |
0.000395 |
2 |
0.000197 |
0.000744 |
32 |
0.000023 |
8.491422 |
0.001101 |
| Mobility |
0.079244 |
2 |
0.039622 |
0.327807 |
32 |
0.010244 |
3.867815 |
0.031298 |
| Complexity |
0.563294 |
2 |
0.281647 |
1.023957 |
32 |
0.031999 |
8.801832 |
0.000900 |
Table 3.
The results for the ANOVA rank Kruskal-Wallis test performed for Hjorth descriptors (i.e., Activity, Mobility, and Complexity) obtained for three bee pollen groups Brassica-napus, Helianthus, and Phacelia
Table 3.
The results for the ANOVA rank Kruskal-Wallis test performed for Hjorth descriptors (i.e., Activity, Mobility, and Complexity) obtained for three bee pollen groups Brassica-napus, Helianthus, and Phacelia
| Hjorth |
Group |
Group |
Rank |
Rank |
Kruskal-Wallis |
| descriptors |
|
size |
sum |
average |
Test value |
p–value |
| |
Brassica-napus |
18 |
348 |
19.333 |
|
|
| Activity |
Helianthus |
8 |
229 |
28.625 |
21.478 |
0.00001 |
| |
Phacelia |
9 |
53 |
5.888 |
|
|
| |
Brassica-napus |
18 |
310 |
17.222 |
|
|
| Mobility |
Helianthus |
8 |
242 |
30.250 |
19.004 |
0.0001 |
| |
Phacelia |
9 |
78 |
8.666 |
|
|
| |
Brassica-napus |
18 |
339 |
18.833 |
|
|
| Complexity |
Helianthus |
8 |
41 |
5.125 |
20.944 |
0.00001 |
| |
Phacelia |
9 |
250 |
27.777 |
|
|
Table 4.
The results (p-values) of multiple comparisons post hoc tests performed for Hjorth descriptors: (a) Activity, (b) Mobility, and (c) Complexity determined for three groups of bee pollen Brassica-napus, Helianthus, and Phacelia; statistical significant differences between analyzed groups are marked with color.
Table 4.
The results (p-values) of multiple comparisons post hoc tests performed for Hjorth descriptors: (a) Activity, (b) Mobility, and (c) Complexity determined for three groups of bee pollen Brassica-napus, Helianthus, and Phacelia; statistical significant differences between analyzed groups are marked with color.
| (a) Activity |
|
|
|
| Group |
Brassica-napus |
Helianthus |
Phacelia |
| RA=19.333 |
RA=28.625 |
RA=5.888 |
| Brassica-napus |
|
0.098529 |
0.003929 |
| Helianthus |
0.098529 |
|
0.000015 |
| Phacelia |
0.003929 |
0.000015 |
|
| (b) Mobility |
|
|
|
| Group |
Brassica-napus |
Helianthus |
Phacelia |
| RA=17.222 |
RA=30.250 |
RA=8.666 |
| Brassica-napus |
|
0.008313 |
0.122515 |
| Helianthus |
0.008313 |
|
0.000044 |
| Phacelia |
0.122515 |
0.000044 |
|
| (c) Complexity |
|
|
|
| Group |
Brassica-napus |
Helianthus |
Phacelia |
| RA=18.833 |
RA=5.125 |
RA=27.777 |
| Brassica-napus |
|
0.004926 |
0.097518 |
| Helianthus |
0.004926 |
|
0.000016 |
| Phacelia |
0.097518 |
0.000016 |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).