1. Introduction
Recurrent implantation failure (RIF) after in vitro fertilization-embryo transfer (IVF-ET) is diagnosed when high-quality embryos fail to implant in at least three consecutive IVF attempts, and is common challenge encountered by reproductive specialists [
1,
2,
3].
One of the critical factors for implantation failure is considered to be thin endometrium, in which the thickness of the endometrium is less than 7 mm during the “implantation window”. Pathophysiological features of “thin endometrium” include insufficient growth of the glandular epithelium, decreased expression of vascular endothelial growth factor (VEGF), and depletion of blood vessels [
4]. This is especially true for pathological conditions associated with thin endometrium. In the thin endometrium, in contrast to normal, local microenvironment in the proliferation phase characterized by a decreasing of stromal, epithelial, natural killers, T cells and increasing of cellular senescence in perivascular cells [
5]. The thin endometrium observed in patients with RIF remains a challenge in understanding and effectively addressing these conditions.
Studies over the past decade have shed light on structural changes in the endometrium, including the development of pinopode, morphological changes, shifts in the expression of molecular markers of endometrial receptors and the secretion of regulatory factors [
6,
7]. These changes have been associated with gene transcriptional activity, highlighting the intricate link between compromised receptivity due to a thin endometrium and impaired implantation [
8]. The studies using transcriptomic and molecular analysis of the endometrium have shed light on deciphering of the implantation processes and on the identifying potential molecular biomarkers of endometrial receptivity [
9,
10].
A significant number of genes are differentially expressed between the pre-receptive and receptive endometrial phases [
11,
12,
13,
14]. However, differentially expressed genes varied from 107 to 2878, due to the diversity of patients included in different studies [
15]. Due to the significant role of numerous immune pathways in the pathogenesis of unexplained RIF, it is difficult to identify a limited number of genes that predict endometrial receptivity [
16].
The bioinformatics prediction identifies 30 genes associated with endometrial receptivity, meta-signature genes highlight the importance of immune responses, the complement cascade pathway in endometrial mid-secretory functions [
17]. A panel for determining the human endometrial receptivity ER Map®/ER Grade® showed that in healthy women and patients undergoing infertility treatment, the expression of 85 genes significantly changed in the pre-receptive and receptive endometrium [
13].
Various mechanisms have been proposed to explain embryo implantation failure, but most involve disruption of endometrial receptivity due to abnormal activation of the innate immune system [
18,
19,
20]. Gene ontology analysis showed that the most widely represented endometrial receptivity genes were the group responsible for vascular proliferation [
21] and immunological activity [
22,
23]. Therefore, the genes identified in several studies may become potential biomarkers of endometrial receptivity [
17].
In this study, we focused on 10 genes also identified and showed up-regulation in the receptive phase in healthy women and responsible for the pro-inflammatory signaling cascade - C-X-C motif chemokine ligand 8 (CXCL8) and C-X-C motif chemokine ligand 1 (CXCL1), complement cascade - Complement Component 4 Binding Protein Alpha (C4BPA), breakdown of extracellular matrix in the in normal physiological processes, such as embryonic development, reproduction, and tissue remodeling - Matrix metallopeptidase 10 (MMP10), generation of purine nucleotides - Hypoxanthine phosphoribosyltransferase 1 (HPRT1), cell adhesion - Tenascin C (TNC), vascular proliferation pathway - VEGF-B, innate and adaptive immune systems - interferon gamma (INFG), Heart- and neural crest derivatives-expressed transcript 2 (HAND2), interleukin 15 (IL-15). HAND2 regulates IL15, which is required for the activation and survival of uterine natural killer cells [
22,
23].
Another reason for the failure of implantation is associated with a decrease in the activity of the tenascin C (TNC) gene, which is involved in reducing cell adhesion on the surface of the endometrium, facilitating the attachment of the embryo to the endometrium. TNC gene expression level is significantly (p < 0,05) down-regulated in the RIF samples compared to the control group [
19,
24].
Thus, 10 immune response genes were selected (CXCL8, CXCL1, HPRT1, MMP10, INFG, C4BPA, TNC, VEGFB, HAND2 and IL15), the activities of which were increased during the receptive endometrial phase in healthy women. In addition, two of them (C4BPA and TNC) showed down-regulation upon implantation failure in women. It is assumed that a decrease in the expression of these transcriptomes will indicate implantation failure in assisted pregnancies with thin endometrium. The research question whether the selected genes would show downregulation in the case of thin endometrium in patients with recurrent implantation failures was posed in the present study.
2. Materials and Methods
2.1. Subjects
In accordance with the purpose of the study, two groups were formed from participants who applied to the Scientific Center of Obstetrics, Gynecology and Perinatology (Almaty, Kazakhstan) in 2023. An individual follow-up chart was compiled for each woman, including physical examination findings, complaints, obstetric and gynecological history data, lab tests, as well as diagnostic tests where indicated: pelvic ultrasound, blood hormone level testing, genetic counseling and karyotyping.
The inclusion criterion for RIF group was the presence of 3 or more implantation failures after the IVF procedure, as well as the presence of thin endometrium (less than 7 mm during the implantation window during ultrasound examination). Fertile women had at least one term and healthy newborn and was without any reproductive losses in anamnesis.
Demographic and clinical characteristics of the study groups are presented in
Table 1.
When clinical characteristics were compared between two groups, no statistical differences were found by age and BMI. All participants were under 40 years of age. In gynecological history, the presence of chronic salpingoophoritis and operations on the pelvic organs were more often noted in patients of RIF group compared to fertile group. There were no significant differences between the groups in hemostasis parameters, blood hormone levels.
2.2. Ethics Approval
This study was approved by the Ethical Committee of Al Farabi Kazakh National University, Kazakhstan (Code: IRBA400/IRB 00010790). All participants provided written informed consent for the use of biomaterials in this study.
2.3. Sample Processing
Endometrial samples were collected in the implantation window days (LH+7 – LH+10) of the natural menstrual cycle from RIF patients with thin endometrium (n=20) and fertile women (n=14). Endometrial tissue was collected using Pipelle biopsy with Goldstein catheter (SonoBiopsyTM J-GSBX-072026 size (Fr) 7.2 Cook Incorporated, USA). Tissue samples were transferred from the catheter into a cryotube with 1 ml RNA-later stabilization solution (Thermo Fisher Scientific) to store in a refrigerator 4°C for 12 h. The next day, samples were transferred to freezer −20°C. After sample collection was completed, they were transferred to M. Aitkhozhin Institute of Molecular Biology and Biochemistry (Almaty, Kazakhstan).
2.4. RNA Isolation from Endometrial Samples
Isolation of total RNA from endometrial samples was performed using Dynabeads™ mRNA DIRECT™ Purification Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. The isolated RNA was immediately subjected to further analysis.
2.5. cDNA Synthesis and Quantitative Polymerase Chain Reaction (PCR)
Reverse-transcription and quantitative PCR was performed using primers and probes from TaqMan™ Gene Expression Assay (Applied Biosystems, USA). cDNA was obtained using High-capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. Quantitative PCR was performed in duplicates using TaqMan Fast Advanced Master Mix (Applied Biosystems) under the conditions recommended by the manufacturer on the StepOnePlus Real-Time PCR System (Applied Biosystems). Primary processing was performed using StepOnePlus 2.2.2 and ExpressionSuite v1.3 programs. Relative quantification of gene expression is carried out using the comparative Ct (ΔΔCt) method with modifications as described by Königshoff M. et al. (2009) [
25]. Relative transcript abundance is expressed in ΔCt values (ΔCt = Ctreference − Cttarget). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ) housekeeping genes was used as a reference, in accordance with previous study [
13]. ΔΔCt value (ΔΔCt = ΔCtcase − ΔCtcontrol) was considered as log2 fold change.
2.6. Statistical Analysis
Most of the statistics were performed in the Jamovi program (
https://www.jamovi.org). Statistical significance of the differences in ΔCt between the groups was calculated using the two-tailed Mann–Whitney U test. Spearman's rank correlation method was used to examine the relationship between quantitative variables. P<0.05 was considered statistically significant. For multiple comparisons, the online calculator of FDR correction was used to adjust the p-values (
https://www.sdmproject.com/utilities/?show=FDR). For a comparative visualization of gene expression levels, box plots were constructed using a web tool BoxPlotR (
http://shiny.chemgrid.org/boxplotr). The characteristics of the markers were evaluated by ROC analysis using the web-tool easy ROC [
26], and Jamovi. Youden’s index method was used to calculate optimal cut-off points. Evaluation of classifiers by interpretation of the area under the ROC curve (AUC) was performed as described by [
27].
3. Results
3.1. The level of mRNA in endometrial samples of RIF patients in comparison with the fertile group
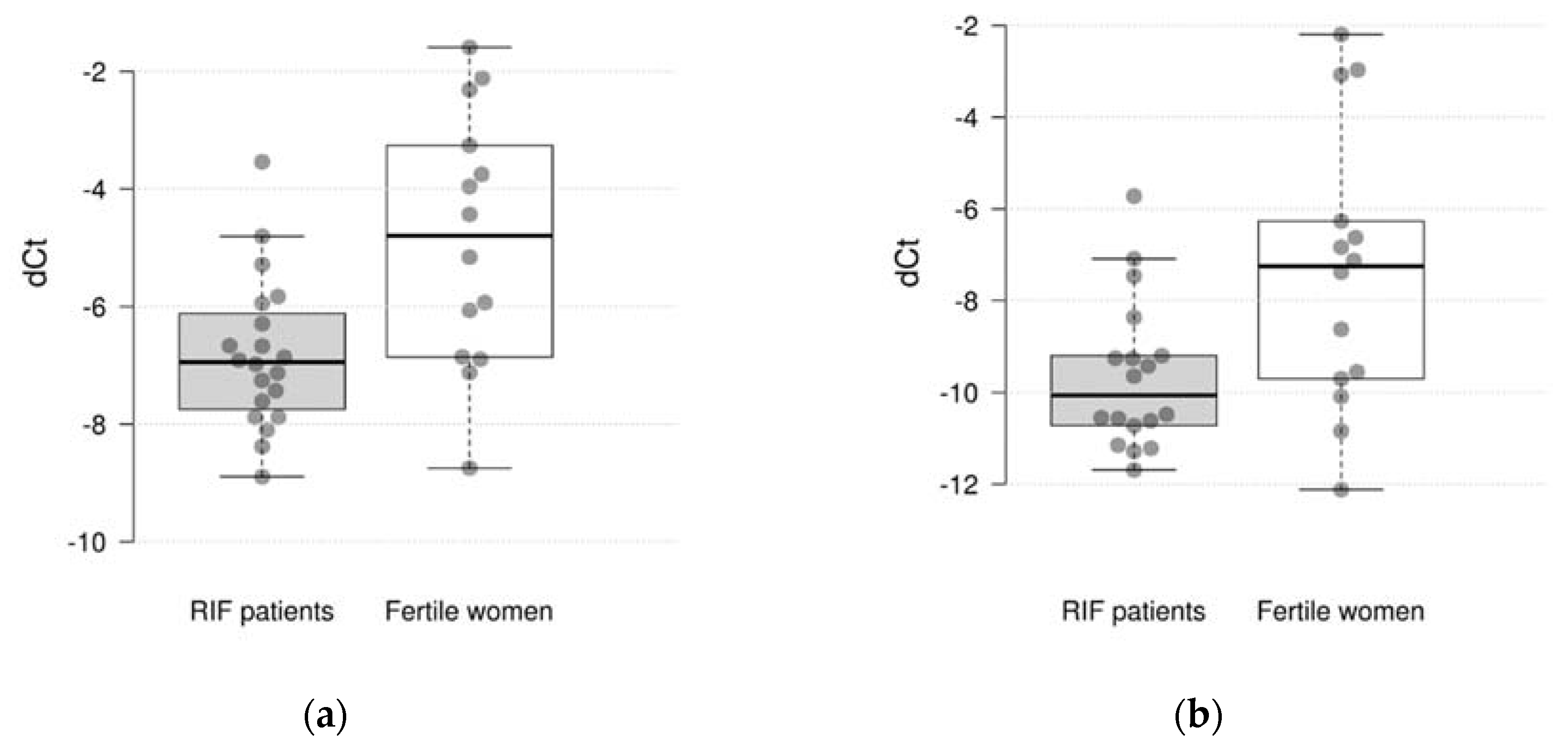
Comparative gene expression statistics between RIF and fertile women are presented in
Table 2. The study showed that the expression of 2 out of 10 genes differs significantly between the studied groups. It was found, that the expression of the CXCL1 and C4BPA were significantly decreased in RIF with thin endometrium patients compared to the fertile women (log
2 fold change=-1.95 and -2.09, p = 0.005 and 0.030, respectively) (
Figure 1). However, after adjustment for multiple comparisons only the difference in CXCL1 expression remained significant (pFDR = 0.050 and 0.150, respectively). There were not significant differences in expression of CXCL8, HPRT1, MMP10, INFG, VEGFB, HAND2, IL15 and TNC genes between groups.
3.2. Associations with Clinical and Laboratory Characteristics
Next, we analyzed differences in gene expression among groups with different clinical characteristics. Among fertile women, C4BPA expression was significantly increased, HAND2 expression was significantly decreased in the endometrium of women with polyps compared to those without them (in both cases, p=0.020). Among RIF women, there were no significant differences in gene expression levels between patients with and without endometriosis.
Spearman's rank correlation method was used to examine the relationship between expression of studied genes and quantitative laboratory characteristics in two groups. The correlations are presented in
Table 3. In RIF patients with thin endometrium group significant inverse moderate correlations have been established: between HAND2 and TSH levels (rho = -0.671, p = 0.001), HAND2 and fibrinogen levels (rho = -0.465, p = 0.038), VEGFB and prolactin levels (rho = -0.446, p = 0.049), VEGFB and TSH levels (rho = -0.526, p = 0.017). In the fertile group an inverse moderate relationship between HAND2 and BMI (rho = -0.688, p = 0.008). It should be noted, however, that after adjustment for multiple comparisons none correlations remained significant.
3.3. ROC Analysis
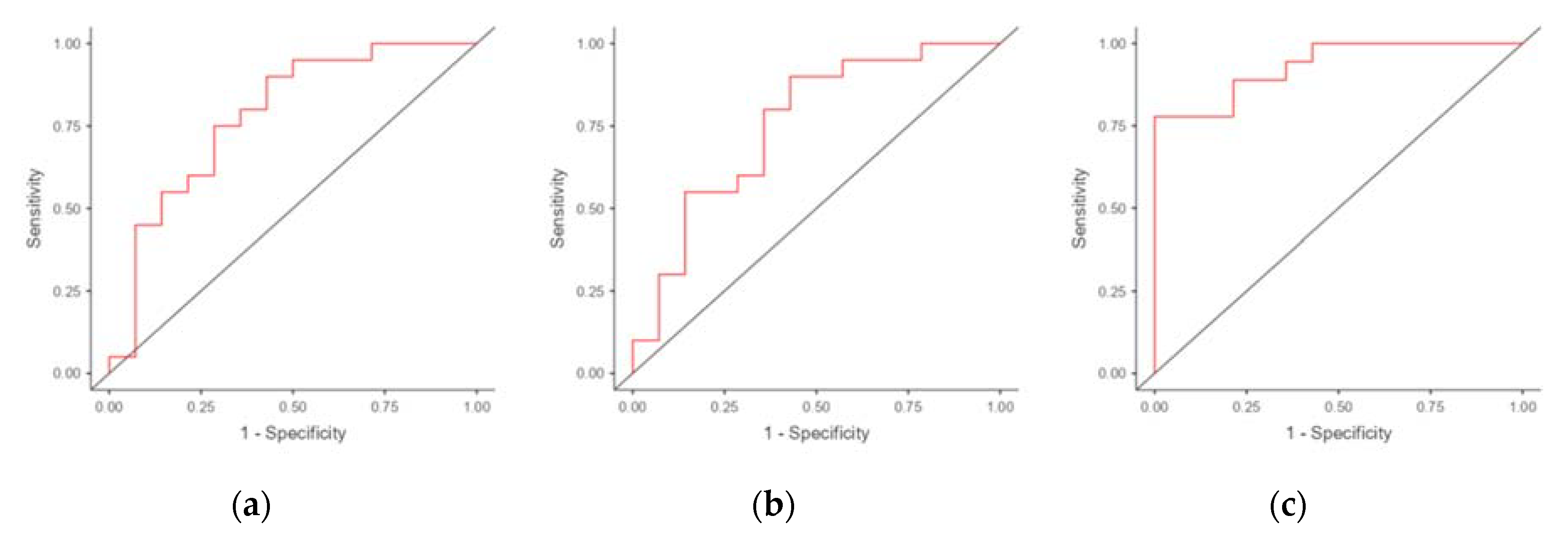
We performed ROC analysis to evaluate the potential of using mRNA of genes differentially expressed in the endometrium of RIF patients as markers for predicting RIF. The results are presented in
Table 4. The areas under the ROC curve (AUC) were obtained for CXCL1 (0.782) and C4BPA (0.726) (
Figure 2). The use of combination of C4BPA and CXCL1 resulted in an increase in AUC to 0.806 (with a specificity of 64.3% and a sensitivity of 83.3%).
4. Discussion
Patients suffering from thin endometrium are more susceptible to fertility impairments [
28,
29]. RIF caused by thin endometrium may be associated with abnormal activation of the inflammatory environment together with an abnormally reduced response to oxidative stress [
30]. Therefore, studying the cellular and molecular mechanisms of the thin endometrium with the elucidation of dysfunctional signaling pathways is important.
We focused on 10 genes involved in immune pathways of endometrial receptivity, which have shown up-regulations in receptive endometrial phase of healthy women.
In this study, gene expression of RIF patients with thin endometrium along with the matched endometrial tissues from fertile women were explored, and we revealed the abnormal activation of the inflammatory environment in thin endometrium. It was found, that the expression C4BPA and CXCL1 genes were significantly decreased in RIF with thin endometrium patients compared to the fertile women (p=0.030 and p=0.005, respectively).
Complement Component 4 Binding Protein Alpha (C4BPA) gene codes an inhibitor of complement system activation, which reduce of a misdirected complement attack to the embryo [
31]. Abnormally decreased levels of C4BPA have been detected in the mid-secretory endometrium of women with implantation failure [
18] and endometriosis [
32]. Moreover, in the thin endometrium, the ligand-receptor pair genes in CD45 and complement signals were upregulated [
30]. In this study also highlighted that the mis-activation of immune cells and disorder of cell dialogue to other cell types may contribute to the pathology to thin endometrium. Through activation of the immune system and enhancement of signaling pathways associated with lymphoid cells, the expression of chemokine family genes can be examined for the risk of thin endometrium [
30].
In our study, the down-regulation of C-X-C motif chemokine ligand 1 (CXCL1) gene indicates a proinflammatory signaling cascade activation in RIF patients with thin endometrium. At the same time, there were not significant differences in concentrations for CXCL8, HPRT1, MMP10, INFG, VEGFB, HAND2, IL15 and TNC genes expression between groups. Our findings indirectly indicate “silent endometrium” is observed in thin endometrium. Pathologies that directly affect the uterine mucosa, including the thin endometrium, can provoke a refractory endometrium, characterized by the absence of proliferation and abnormal thickness [
33]. When examining intercellular communication in the thin endometrium using CellChat, it was shown that the number and strength of interactions between stromal cells and proliferating stromal cells were reduced [
30]. In the late-proliferative phase, suppressed cell proliferation was detected in women with thin endometrium and reduced expression of PDZ-binding kinase in endometrial stromal cells, which was facilitated by the presence of an aberrantly proinflammatory environment [
34].
In our study, in RIF patients with thin endometrium significant (p<0.05) inverse moderate correlations were shown. It was noted that the decreasing of HAND2 gene level was associated to increasing of TSH and fibrinogen levels, and in fertile women the decreasing of HAND2 gene level was associated with increasing of BMI. Heart- and neural crest derivatives-expressed transcript 2 (HAND2) regulates interleukin 15 (IL-15), a key immune factor required for the activation and survival of uterine natural killers, briefly induces immunotolerance. During the secretory phase, levels of HAND2 and IL-15 are significantly increased in the healthy human endometrium [
23]. The same trends were observed in VEGFB with TSH and prolactin levels. VEGF-B has been shown to be a survival factor for vascular endothelial cells [
21]. These finding confirmed that RIF population is highly heterogenous, and numerous risk factors for RIF have been identified lifestyle factors, including BMI, uterine and ovarian factors (endometriosis, polyps, adenomyosis, polycystic syndrome), thrombophilia [
35,
36]. The following studies should be performed taking these factors into account, depending on the clinical forms. The endometrial receptivity is characterized by complex dynamics of interactive networks and regulatory functions in different aspects of the implantation process, and high phenotype variability between individuals implicate them as central gatekeepers of receptivity for embryo implantation [
37]. However, our studies showed the prognostic potential of a combination of C4BPA + CXCL1 genes to predict RIF.
Our studies had limitations that need to be addressed in future studies. It was a small sample sizes, it is recommended to increase the observational cohorts according to clinical forms. Transcriptional analysis of thin endometrial tissue should be validated using multi-omics data, including proteomics (cell immunophenotyping) or metabolomics.
5. Conclusions
Our study revealed differential expression of C4BPA and CXCL1 genes in RIF patients with thin endometrium compared with the endometrium of fertile women. These data allowed us to evaluate the predictive value of the combined model for predicting implantation failure depending on endometrial thickness. The data also provide potential therapeutic targets for the treatment of thin endometrium in patients undergoing ART programs.
Author Contributions
Conceptualization, G.K., Y.A., A.K. and N.M.; Funding acquisition, G.M., G.A. and A.T.; Investigation, Y.A.; Methodology, G.K., Y.A., A.K., N.M., D.S.; Resources, A.K. and G.M.; Supervision, N.M.; Visualization, A.K. and G.M.; Writing – original draft, Y.A. and A.K.; Writing – review & editing, G.K., Y.A., A.K., N.M., G.M., G.A., D.S. and A.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science Committee of the Ministry of Science and High Education of the Republic of Kazakhstan (Grant No. AP 14870089).
Institutional Review Board Statement
This study was approved by the Ethical Committee of Al Farabi Kazakh National University, Kazakhstan (Code: IRBA400/IRB 00010790).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Experimental and clinical-pathological data is available.
Acknowledgments
The authors are appreciative to all clinicians of Scientific Center of Obstetrics, Gynecology and Perinatology, and scientists of M. Aitkhozhin Institute of Molecular Biology and Biochemistry.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Simon, A.; Laufer, N. Assessment and treatment of repeated implantation failure (RIF). J Assist Reprod Genet. 2012, 11, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Kasius, A.; Smit, J.G.; Torrance, H.L.; Eijkemans, M.J.; Mol, B.W.; Opmeer, B.C.; Broekmans, F.J. Endometrial thickness and pregnancy rates after IVF: a systematic review and meta-analysis. Hum Reprod Update. 2014, 20, 530–541. [Google Scholar] [CrossRef]
- Lessey, B.A.; Young, S.L. What exactly is endometrial receptivity? Fertil Steril. 2019, 111, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, R.; Taketani, T.; Mihara, Y.; Sato, S.; Okada, M.; Tamura, I.; Jozaki, K.; Kajimura, T.; Asada, H.; Tamura, H.; Takasaki, A.; Sugino, N. Thin endometrium transcriptome analysis reveals a potential mechanism of implantation failure. Reprod Med Biol. 2017, 16, 206–227. [Google Scholar] [CrossRef]
- Lv, H.; Zhao, G.; Jiang, P.; Wang, H.; Wang, Z.; Yao, S.; Zhou, Z.; Wang, L.; Liu, D.; Deng, W.; Dai, J.; Hu, Y. Deciphering the endometrial niche of human thin endometrium at single-cell resolution. Proc Natl Acad Sci USA. 2022, 119, e2115912119. [Google Scholar] [CrossRef]
- Boeing, S.; Williamson, L.; Encheva, V.; Gori, I.; Saunders, R.E.; Instrell, R.; Aygün, O.; Rodriguez-Martinez, M.; Weems, J.C.; Kelly, G.P.; Conaway, J.W.; Conaway, R.C.; Stewart, A.; Howell, M.; Snijders, A.P.; Svejstrup, J.Q. Multiomic Analysis of the UV-Induced DNA Damage Response. Cell Rep. 2016, 15, 1597–1610. [Google Scholar] [CrossRef] [PubMed]
- Bassil, R.; Casper, R.; Samara, N.; Hsieh, T.B.; Barzilay, E.; Orvieto, R.; Haas, J. Does the endometrial receptivity array really provide personalized embryo transfer? J Assist Reprod Genet. 2018, 35, 1301–1305. [Google Scholar] [CrossRef]
- Saha, S.; Matthews, D. A.; Bessant, C. High Throughput Discovery of Protein Variants Using Proteomics Informed by Transcriptomics. Nucleic Acids Res 2018, 46, 4893–4902. [Google Scholar] [CrossRef]
- Hu, S.; Yao, G.; Wang, Y.; Xu, H.; Ji, X.; He, Y.; Zhu, Q.; Chen, Z.; Sun, Y. Transcriptomic changes during the pre-receptive to receptive transition in human endometrium detected by RNA-seq. J Clin Endocrinol Metab. 2014, 99, E2744–E2753. [Google Scholar] [CrossRef]
- Wang, X.; Yu, Q. An update on the progress of transcriptomic profiles of human endometrial receptivity. Biol Reprod. 2018, 98, 440–448. [Google Scholar] [CrossRef]
- Tapia, A.; Gangi, L.M.; Zegers-Hochschild, F.; Balmaceda, J.; Pommer, R.; Trejo, L.; Pacheco, I.M.; Salvatierra, A.M.; Henrıquez, S.; Quezada, M.; Vargas, M.; Ríos, M.; Munroe, D.J.; Croxatto, H.B.; Velasquez, L. Differences in the endometrial transcript profile during the receptive period between women who were refractory to implantation and those who achieved pregnancy. Hum Reprod. 2008, 23, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Sigurgeirsson, B.; Åmark, H.; Jemt, A.; Ujvari, D.; Westgren, M.; Lundeberg, J.; Gidlöf, S. Comprehensive RNA sequencing of healthy human endometrium at two time points of the menstrual cycle. Biol Reprod. 2017, 96, 24–33. [Google Scholar] [CrossRef]
- Enciso, M.; Carrascosa, J.P.; Sarasa, J.; Martínez-Ortiz, P.A.; Munné, S.; Horcajadas, JA.; Aizpurua, J. Development of a new comprehensive and reliable endometrial receptivity map (ER Map/ER Grade) based on RT-qPCR gene expression analysis. Hum Reprod. 2018, 33, 220–228. [Google Scholar] [CrossRef]
- Messaoudi, S.; El Kasmi, I.; Bourdiec, A.; Crespo, K.; Bissonnette, L.; Le Saint, C.; Bissonnette, F.; Kadoch, I.-J. 15 years of transcriptomic analysis on endometrial receptivity: What have we learnt? Fertil. Res. Pract. 2019, 5, 9. [Google Scholar] [CrossRef]
- Maziotis, E.; Kalampokas, T.; Giannelou, P.; Grigoriadis, S.; Rapani, A.; Anifantakis, M.; Kotsifaki, A.; Pantou, A.; Triantafyllidou, O.; Tzanakaki, D.; Neofytou, S.; Vogiatzi, P.; Bakas, P.; Simopoulou, M.; Vlahos, N. Commercially Available Molecular Approaches to Evaluate Endometrial Receptivity: A Systematic Review and Critical Analysis of the Literature. Diagnostics (Basel). 2022, 12, 2611. [Google Scholar] [CrossRef]
- Li, F.; Gao, W.; Li, Y.; Wang, Y.; Liu, L.; Zhang, X. Potential Biomarkers and Endometrial Immune Microenvironment in Recurrent Implantation Failure. Biomolecules. 2023, 2023 13, 406. [Google Scholar] [CrossRef]
- Altmäe, S.; Koel, M.; Võsa, U.; Adler, P.; Suhorutšenko, M.; Laisk-Podar, T.; Kukushkina, V.; Saare, M.; Velthut-Meikas, A.; Krjutškov, K.; Aghajanova, L.; Lalitkumar, P.G.; Gemzell-Danielsson, K.; Giudice, L.; Simón, C.; Salumets, A. Meta-signature of human endometrial receptivity: a meta-analysis and validation study of transcriptomic biomarkers. Sci Rep. 1007; ;7. [Google Scholar] [CrossRef]
- Herington, J.L.; Guo, Y.; Reese, J.; Paria, B.C. Gene profiling the window of implantation: microarray analyses from human and rodent models. J Reprod Health Med. 2016, 2, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Basatvat, S.; Russell, J.M.; Saare, M.; Thurston, L.M.; Salumets, A.; Fazeli, A. Potential innate immunity-related markers of endometrial receptivity and recurrent implantation failure (RIF). Reprod Biol. 2021, 21, 100569. [Google Scholar] [CrossRef] [PubMed]
- Albayrak, İ.G.; Azhari, F.; Çolak, E.N.; Balcı, B.K.; Ülgen, E.; Sezerman, U.; Baştu, E.; Günel, T. Endometrial gene expression profiling of recurrent implantation failure after in vitro fertilization. Mol Biol Rep. 2021, 48, 5075–5082. [Google Scholar] [CrossRef]
- Li, X.; Kumar, A.; Zhang, F.; Lee, C.; Tang, Z. Complicated life, complicated VEGF-B. Trends Mol Med. 2012, 18, 119–127. [Google Scholar] [CrossRef]
- Lédée, N.; Petitbarat, M.; Rahmati, M.; Dubanchet, S.; Chaouat, G.; Sandra, O.; Perrier-d'Hauterive, S.; Munaut, C.; Foidart, J.M. New pre-conception immune biomarkers for clinical practice: interleukin-18, interleukin-15 and TWEAK on the endometrial side, G-CSF on the follicular side. J Reprod Immunol. 2011, 88, 118–123. [Google Scholar] [CrossRef]
- Murata, H.; Tanaka, S.; Tsuzuki-Nakao, T.; Kido, T.; Kakita-Kobayashi, M.; Kida, N.; Hisamatsu, Y.; Tsubokura, H.; Hashimoto, Y.; Kitada, M.; Okada, H. The transcription factor HAND2 up-regulates transcription of the IL15 gene in human endometrial stromal cells. J Biol Chem. 2020, 295, 9596–9605. [Google Scholar] [CrossRef]
- Bastu, E.; Demiral, I.; Gunel, T.; Ulgen, E.; Gumusoglu, E.; Hosseini, M.K.; Sezerman, U.; Buyru, F.; Yeh, J. Potential Marker Pathways in the Endometrium That May Cause Recurrent Implantation Failure. Reprod Sci. 2019, 26, 879–890. [Google Scholar] [CrossRef]
- Königshoff, M.; Kramer, M.; Balsara, N.; Wilhelm, J.; Amarie, O.V.; Jahn, A.; Rose, F.; Fink, L.; Seeger, W.; Schaefer, L.; Günther, A.; Eickelberg, O. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. Journal of Clinical Investigation. 2009, 119, 772–787. [Google Scholar] [CrossRef]
- Goksuluk, D.; Korkmaz, S.; Zararsiz, G.; Karaagaoglu, A.E. Easy ROC: an interactive web-tool for ROC curve analysis using R language environment. The R Journal. 2016, 8, 213–230. [Google Scholar] [CrossRef]
- Muller, M.P.; Tomlinson, G.; Marrie, T.J.; Tang, P.; McGeer, A.; Low, D.E.; Detsky, A.S.; Gold, W.L. Can routine laboratory tests discriminate between severe acute respiratory syndrome and other causes of community-acquired pneumonia? Clin Infect Dis. 2005, 40, 1079–1086. [Google Scholar] [CrossRef]
- Du, J.; Lu, H.; Yu, X.; Dong, L.; Mi, L.; Wang, J.; Zheng, X.; Feng, K. The effect of icariin for infertile women with thin endometrium: A protocol for systematic review. Medicine (Baltimore). 2020, 99, e19111. [Google Scholar] [CrossRef]
- Xu, L.; Fan, Y.; Wang, J.; Shi, R. Dysfunctional intercellular communication and metabolic signaling pathways in thin endometrium. Front. Physiol. 2022, 13, 1050690. [Google Scholar] [CrossRef]
- Zong, L.; Zheng, S.; Meng, Y.; Tang, W.; Li, D.; Wang, Z.; Tong, X.; Xu, B. Integrated Transcriptomic Analysis of the miRNA–mRNA Interaction Network in Thin Endometrium. Front Genet. 2021, 12, 589408. [Google Scholar] [CrossRef]
- Tapia, A.; Vilos, C.; Marın, J.C.; Croxatto, H.B.; Devoto, L. Bioinformatic detection of E47, E2F1 and SREBP1 transcription factors as potential regulators of genes associated to acquisition of endometrial receptivity. Reprod Biol Endocrinol. 2011, 9, 14. [Google Scholar] [CrossRef]
- Vargas, E.; García-Moreno, E.; Aghajanova, L.; Salumets, A.; Horcajadas, J.A.; Esteban, F.J.; Altmäe, S. The mid-secretory endometrial transcriptomic landscape in endometriosis: a meta-analysis. Hum Reprod Open. 2022, 4, hoac016. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Eguren, A.; Bueno-Fernandez, C.; Gómez-Álvarez, M.; Francés-Herrero, E.; Pellicer, A.; Bellver, J.; Seli, E.; Cervelló, I. Evolution of biotechnological advances and regenerative therapies for endometrial disorders: a systematic review. Hum Reprod Update. 2024, dmae013. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Yao, S.; Dong, Y.; Liu, D.; Wang, H.; Jiang, P.; Dai, C.; Lv, H.; Cao, C.; Zhou, Z.; Wang, L.; Gou, W.; Zhang, X.; Zhao, G.; Hu, Y. Down-regulation of PBK inhibits proliferation of human endometrial stromal cells in thin endometrium. Reprod Biol Endocrinol. 2022, 20, 25. [Google Scholar] [CrossRef] [PubMed]
- Bashiri, A.; Halper, K.I.; Orvieto, R. Recurrent Implantation Failure-update overview on etiology, diagnosis, treatment and future directions. Reprod. Biol. Endocrinol. 2018, 16, 121. [Google Scholar] [CrossRef]
- Bai, X.; Zheng, L.; Li, D.; Xu, Y. Research progress of endometrial receptivity in patients with polycystic ovary syndrome: a systematic review. Reprod Biol Endocrinol. 2021, 19, 122. [Google Scholar] [CrossRef]
- Robertson, S.A.; Moldenhauer, L.M.; Green, E.S.; Care, A.S.; Hull, M.L. Immune determinants of endometrial receptivity: a biological perspective. Fertil Steril. 2022, 117, 1107–1120. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).