Submitted:
12 August 2024
Posted:
13 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Methods
2.1. Microarray Data Sources
2.2. Data and Statistical Analysis
2.3. Identification Key Gene Modules by WGCNA and Functional Enrichment Analysis
2.4. Identification of Hub Genes
3. Results
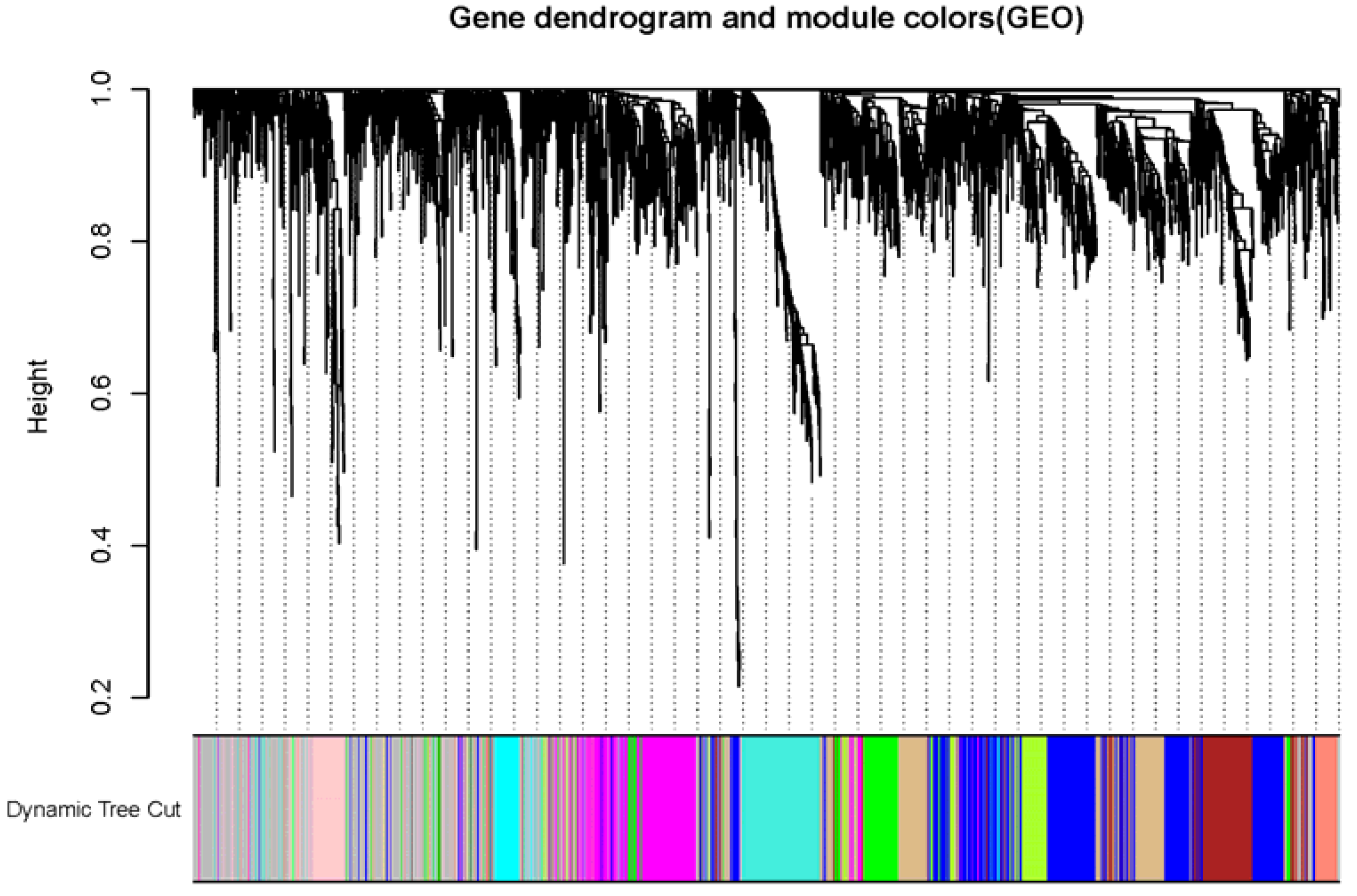
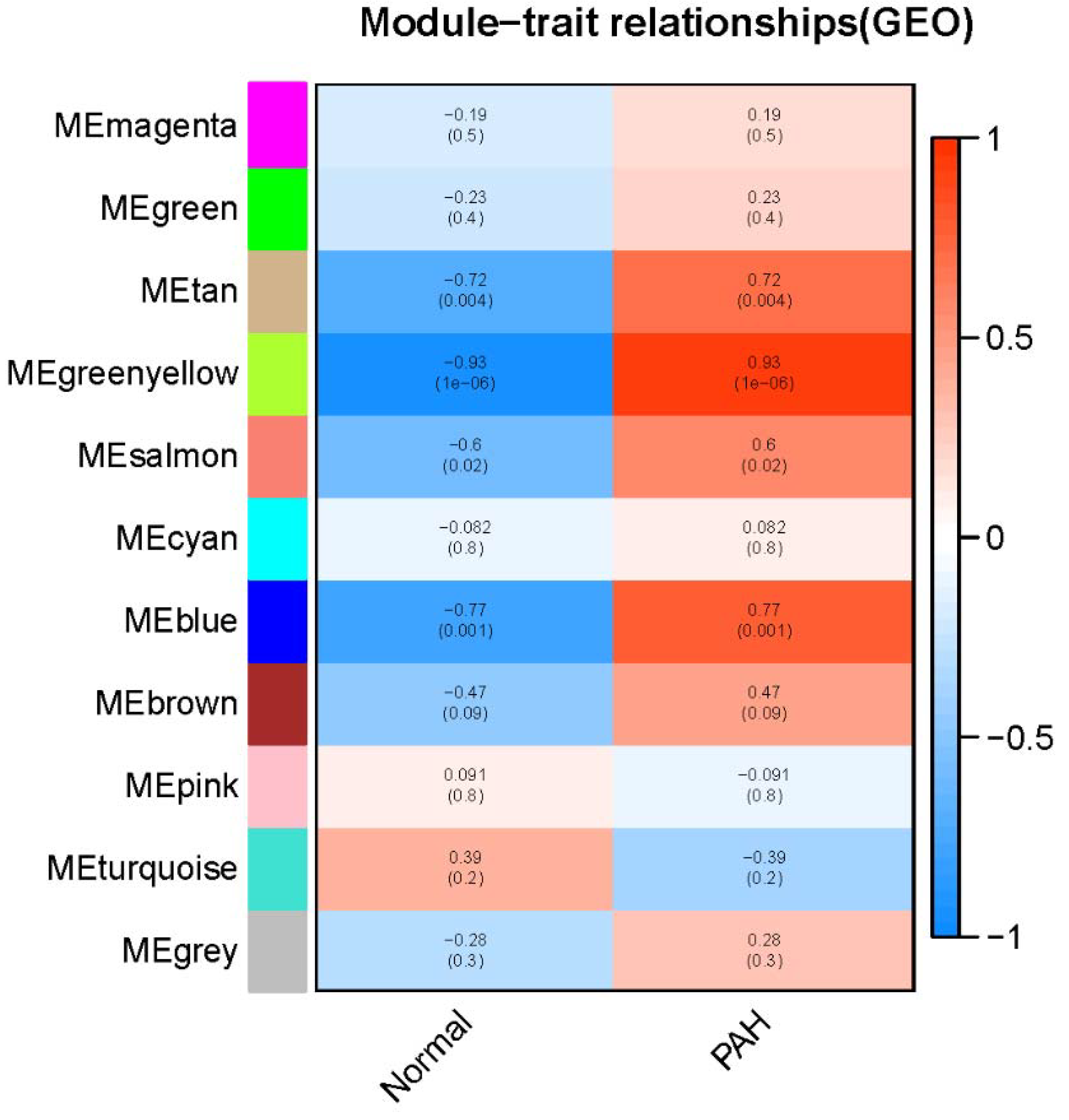
3.1. Identification Key Gene Modules by WGCNA
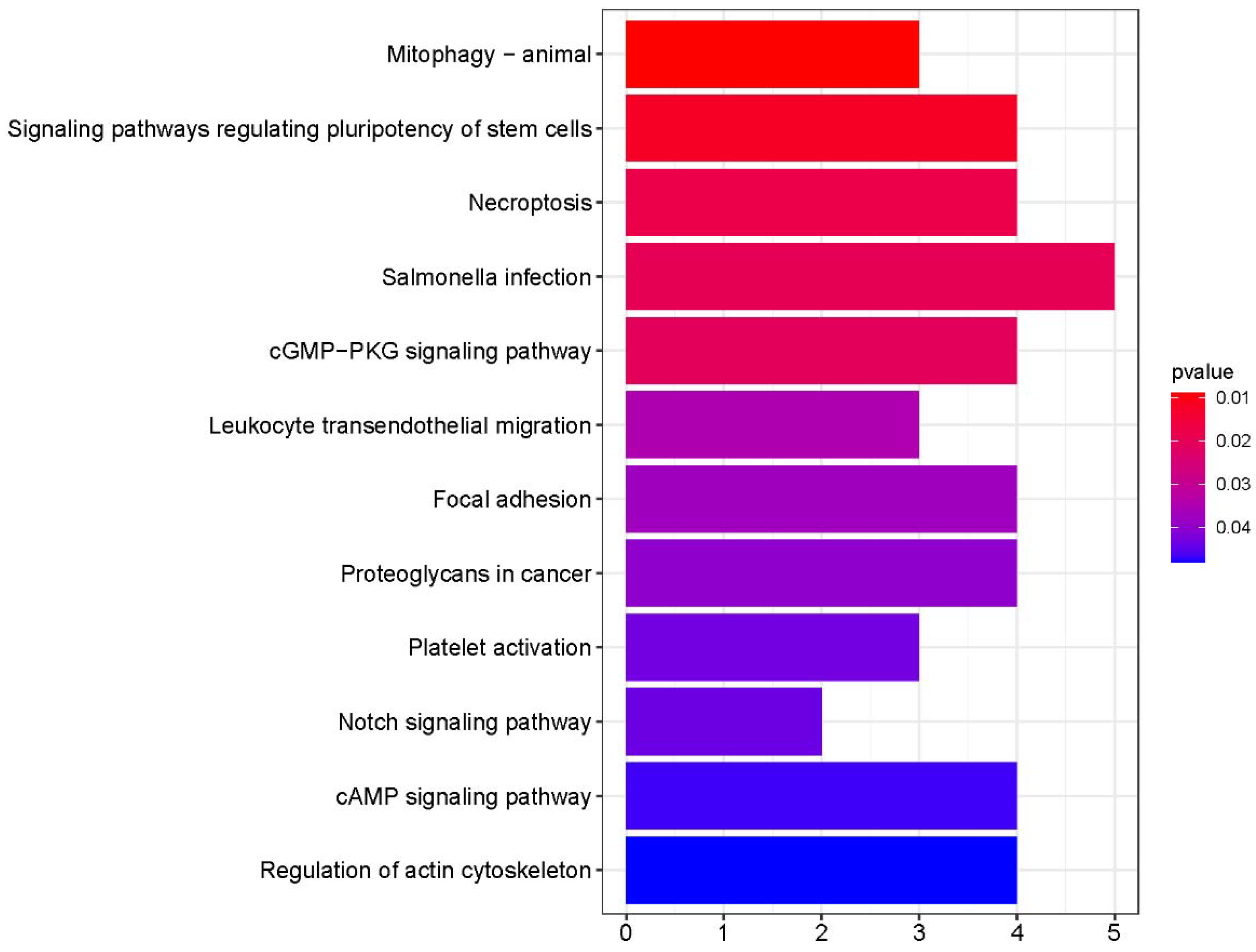
3.2. Functional GO and KEGG Pathway Enrichment Analyses of the Key Module
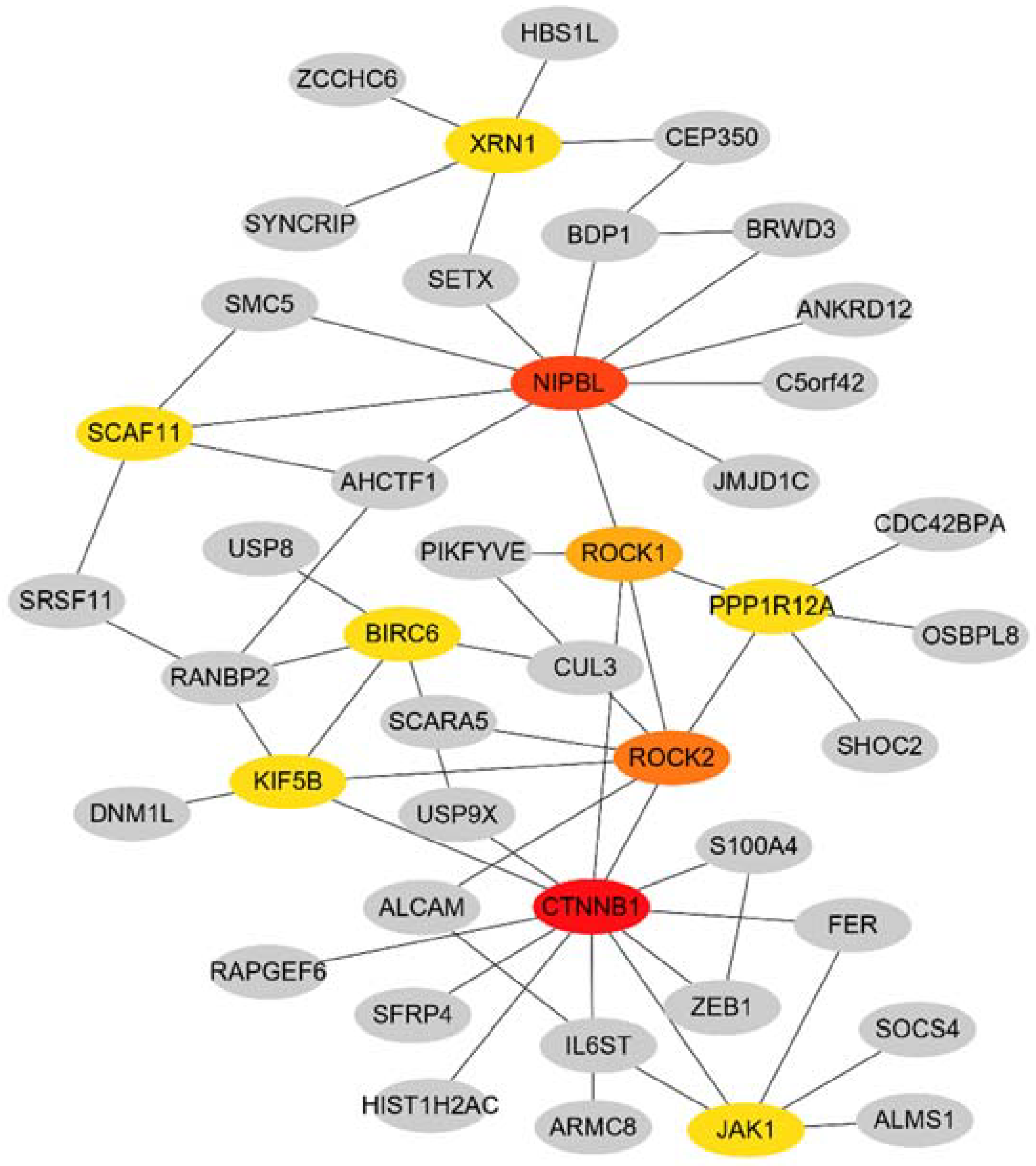
3.3. PPI Network Construction
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PAH | Pulmonary arterial hypertension |
| GEO | Gene Expression Omnibus |
| PPI | protein–protein interaction |
| WHO | World Health Organization |
| mPAP | mean pulmonary artery pressure |
| PCWP | pulmonary capillary wedge pressure |
| PVR | pulmonary vascular resistance |
| RV | right ventricular |
| WGCNA | Weighted gene co-expression network analysis |
| miRs | microRNAs |
| RMA | Robust Multi-array Average |
| MM | module membership |
| GS | gene significance |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MCC | Maximal Clique Centrality |
| BP | biological process |
| CC | cellular component |
| MF | molecular function |
| PAECs | pulmonary artery endothelial cells |
| PASMCs | pulmonary artery smooth muscle cells |
| ROCK | rho-associated coiled-coil–containing protein kinase |
| DEGs | differentially expressed genes |
References
- "2015 ESC/ERS Guidelines for the diagnosis and treatment of pulmonary hypertension. The Joint Task Force for the Diagnosis and Treatment of Pulmonary Hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society (ERS)." Nazzareno Galiè, Marc Humbert, Jean-Luc Vachiery, Simon Gibbs, Irene Lang, Adam Torbicki, Gérald Simonneau, Andrew Peacock, Anton Vonk Noordegraaf, Maurice Beghetti, Ardeschir Ghofrani, Miguel Angel Gomez Sanchez, Georg Hansmann, Walter Klepetko, Patrizio Lancellotti, Marco Matucci, Theresa McDonagh, Luc A. Pierard, Pedro T. Trindade, Maurizio Zompatori and Marius Hoeper. Eur Respir J 2015; 46: 903-975. Eur. Respir. J. 2015, 46, 1855–1856. [CrossRef]
- Ruopp, N.F.; Cockrill, B.A. Diagnosis and Treatment of Pulmonary Arterial Hypertension: A Review. Jama 2022, 327, 1379–1391. [Google Scholar] [CrossRef]
- Hashimoto, R.; Lanier, G.M.; Dhagia, V.; et al. , Pluripotent hematopoietic stem cells augment α-adrenergic receptor-mediated contraction of pulmonary artery and contribute to the pathogenesis of pulmonary hypertension. Am. J. Physiol.. Lung Cell. Mol. Physiol. 2020, 318, L386–l401. [Google Scholar] [CrossRef]
- Steppan, J.; Wang, H.; Nandakumar, K.; et al. LOXL2 inhibition ameliorates pulmonary artery remodeling in pulmonary hypertension. Am. J. Physiol.. Lung Cell. Mol. Physiol. 2024. [Google Scholar] [CrossRef]
- Xiao, L.; Tong, X. [Advances in molecular mechanism of vascular remodeling in pulmonary arterial hypertension]. Zhejiang Da Xue Xue Bao. Yi Xue Ban = J. Zhejiang University. Med. Sci. 2019, 48, 102–110. [Google Scholar] [CrossRef]
- Benza, R.L.; Miller, D.P.; Barst, R.J.; et al. An evaluation of long-term survival from time of diagnosis in pulmonary arterial hypertension from the REVEAL Registry. Chest 2012, 142, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Albulushi, A.; Kashoub, M.; Al-Saidi, K.; et al. Iron Deficiency in Pulmonary Hypertension. Int. Heart J. 2024. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, Y. Pathophysiology and Treatment of Pulmonary Arterial Hypertension. Int. J. Mol. Sci. 2024, 25. [Google Scholar] [CrossRef] [PubMed]
- Wołowiec, Ł.; Mędlewska, M.; Osiak, J.; et al. MicroRNA and lncRNA as the Future of Pulmonary Arterial Hypertension Treatment. Int. J. Mol. Sci. 2023, 24. [Google Scholar] [CrossRef]
- Southgate, L.; Machado, R.D.; Gräf, S.; et al. Molecular genetic framework underlying pulmonary arterial hypertension. Nat. Rev.. Cardiol. 2020, 17, 85–95. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Barabási, A.L.; Oltvai, Z.N. Network biology: understanding the cell's functional organization. Nat. Rev.. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Mura, M.; Cecchini, M.J.; Joseph, M.; et al. Osteopontin lung gene expression is a marker of disease severity in pulmonary arterial hypertension. Respirology 2019, 24, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics : A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–d368. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl 4), S11. [Google Scholar] [CrossRef] [PubMed]
- Zolty, R. Pulmonary arterial hypertension specific therapy: The old and the new. Pharmacol. Ther. 2020, 214, 107576. [Google Scholar] [CrossRef] [PubMed]
- Tuder, R.M.; Archer, S.L.; Dorfmüller, P.; et al. Relevant issues in the pathology and pathobiology of pulmonary hypertension. J. Am. Coll. Cardiol. 2013, 62, D4–D12. [Google Scholar] [CrossRef]
- Gomez-Puerto, M.C.; van Zuijen, I.; Huang, C.J.; et al. Autophagy contributes to BMP type 2 receptor degradation and development of pulmonary arterial hypertension. J. Pathol. 2019, 249, 356–367. [Google Scholar] [CrossRef]
- Chichger, H.; Rounds, S.; Harrington, E.O. Endosomes and Autophagy: Regulators of Pulmonary Endothelial Cell Homeostasis in Health and Disease. Antioxid. Redox Signal. 2019, 31, 994–1008. [Google Scholar] [CrossRef]
- Ornatowski, W.; Lu, Q.; Yegambaram, M.; et al. Complex interplay between autophagy and oxidative stress in the development of pulmonary disease. Redox Biol. 2020, 36, 101679. [Google Scholar] [CrossRef]
- Xiao, G.; Zhuang, W.; Wang, T.; et al. Transcriptomic analysis identifies Toll-like and Nod-like pathways and necroptosis in pulmonary arterial hypertension. J. Cell. Mol. Med. 2020, 24, 11409–11421. [Google Scholar] [CrossRef]
- Frantz, R.P.; McLaughlin, V.V.; Sahay, S.; et al. Seralutinib in adults with pulmonary arterial hypertension (TORREY): a randomised, double-blind, placebo-controlled phase 2 trial. Lancet. Respir. Med. 2024, 12, 523–534. [Google Scholar] [CrossRef]
- Vrigkou, E.; Tsantes, A.E.; Kopterides, P.; et al. Coagulation Profiles of Pulmonary Arterial Hypertension Patients, Assessed by Non-Conventional Hemostatic Tests and Markers of Platelet Activation and Endothelial Dysfunction. Diagnostics 2020, 10. [Google Scholar] [CrossRef]
- Luo, J.; Li, H.; Liu, Z.; et al. Integrative analyses of gene expression profile reveal potential crucial roles of mitotic cell cycle and microtubule cytoskeleton in pulmonary artery hypertension. BMC Med. Genom. 2020, 13, 86. [Google Scholar] [CrossRef]
- Li, Q.; Meng, L.; Liu, D. Screening and Identification of Therapeutic Targets for Pulmonary Arterial Hypertension Through Microarray Technology. Front. Genet. 2020, 11, 782. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, S.S.; Feng, Y.Y.; et al. Identification of novel biomarkers involved in pulmonary arterial hypertension based on multiple-microarray analysis. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Jing, T.; Wang, T.; et al. Molecular Characterization and Elucidation of Pathways to Identify Novel Therapeutic Targets in Pulmonary Arterial Hypertension. Front. Physiol. 2021, 12, 694702. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Li, N.; Zheng, Z.; et al. Screening of Hub Genes Associated with Pulmonary Arterial Hypertension by Integrated Bioinformatic Analysis. BioMed Res. Int. 2021, 2021, 6626094. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.H.; Wang, L.M.; Hu, X.H. MiR-135a inhibitor alleviates pulmonary arterial hypertension through β-Catenin/GSK-3β signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9574–9581. [Google Scholar] [CrossRef]
- Karolak, J.A.; Szafranski, P.; Kilner, D.; et al. Heterozygous CTNNB1 and TBX4 variants in a patient with abnormal lung growth, pulmonary hypertension, microcephaly, and spasticity. Clin. Genet. 2019, 96, 366–370. [Google Scholar] [CrossRef]
- Alonso-Gil, D.; Losada, A. NIPBL and cohesin: new take on a classic tale. Trends Cell Biol. 2023, 33, 860–871. [Google Scholar] [CrossRef]
- Zheng, L.; Zhou, H.; Guo, L.; et al. Inhibition of NIPBL enhances the chemosensitivity of non-small-cell lung cancer cells via the DNA damage response and autophagy pathway. OncoTargets Ther. 2018, 11, 1941–1948. [Google Scholar] [CrossRef]
- Shi, J.; Surma, M.; Yang, Y.; et al. Disruption of both ROCK1 and ROCK2 genes in cardiomyocytes promotes autophagy and reduces cardiac fibrosis during aging. FASEB J. : Off. Publ. Fed. Am. Soc. Exp. Biol. 2019, 33, 7348–7362. [Google Scholar] [CrossRef]
- Knipe, R.S.; Probst, C.K.; Lagares, D.; et al. The Rho Kinase Isoforms ROCK1 and ROCK2 Each Contribute to the Development of Experimental Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2018, 58, 471–481. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




