Submitted:
13 August 2024
Posted:
14 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
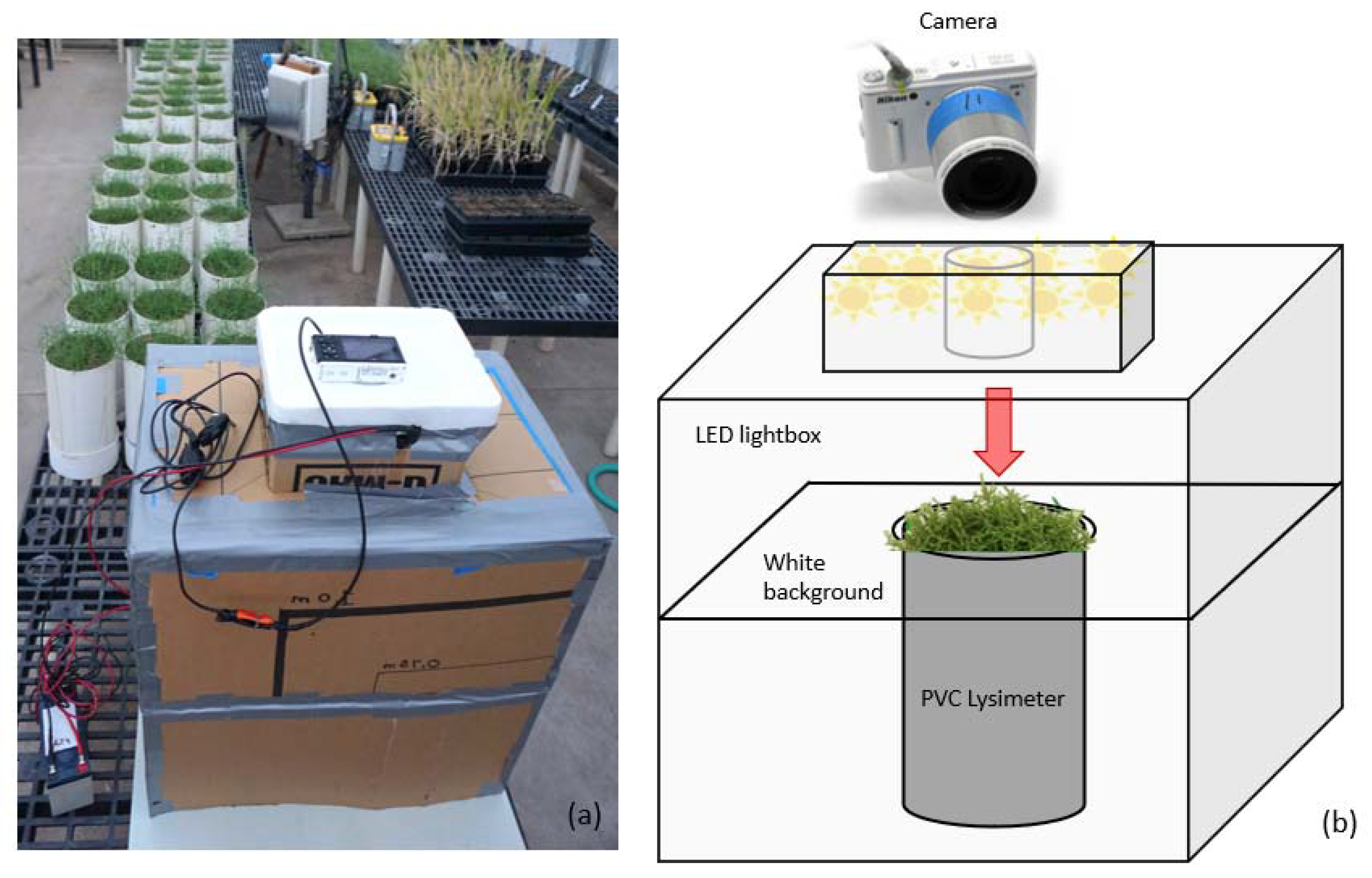
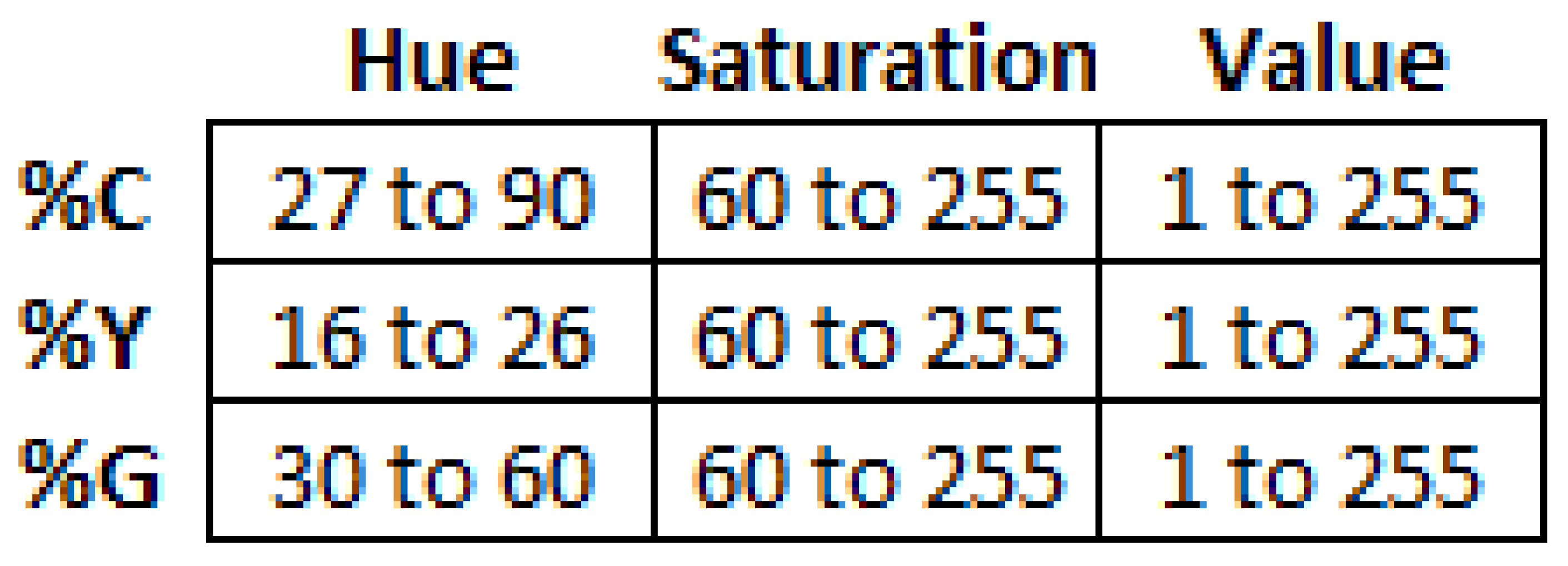
2. Materials and Methods
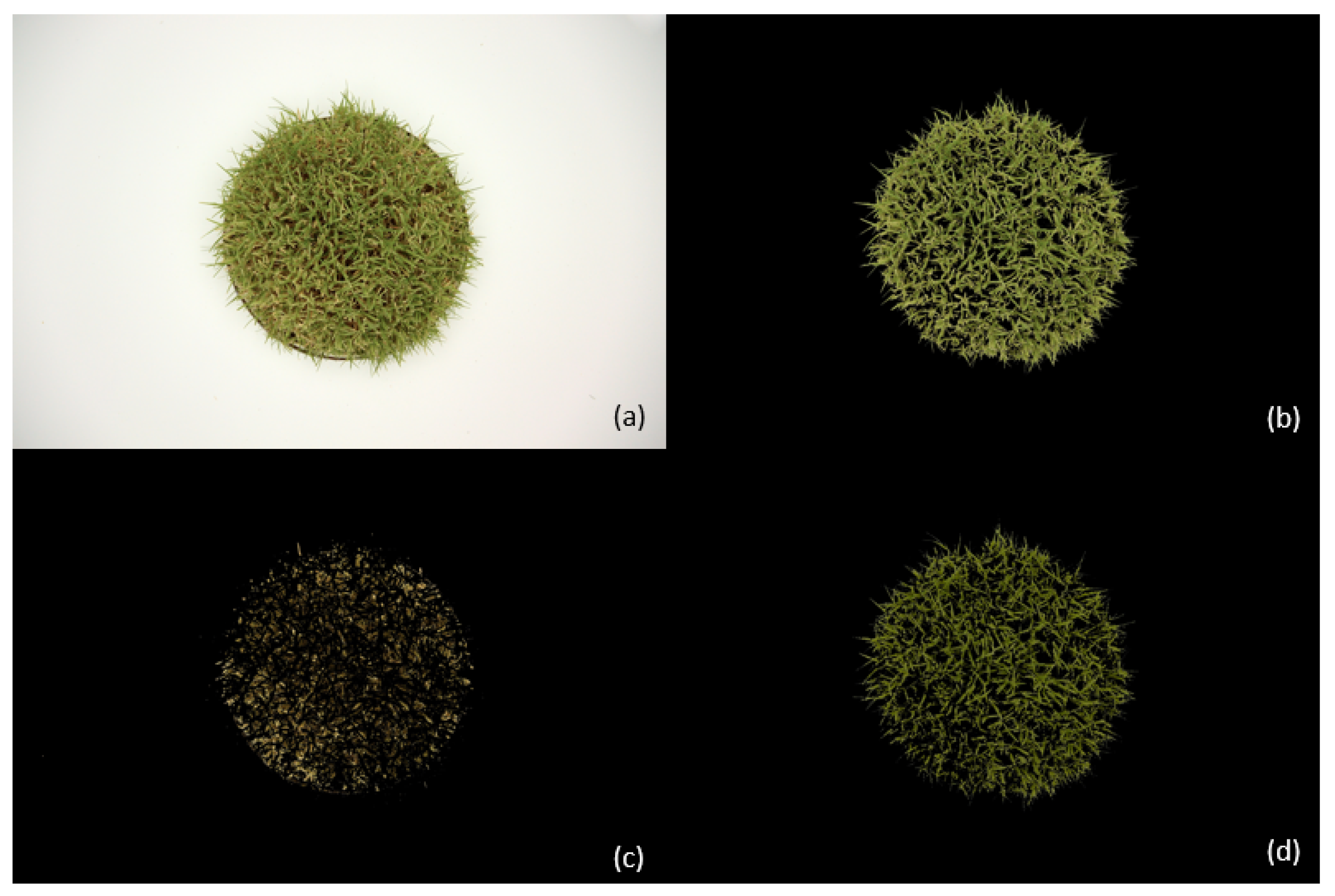
3. Results
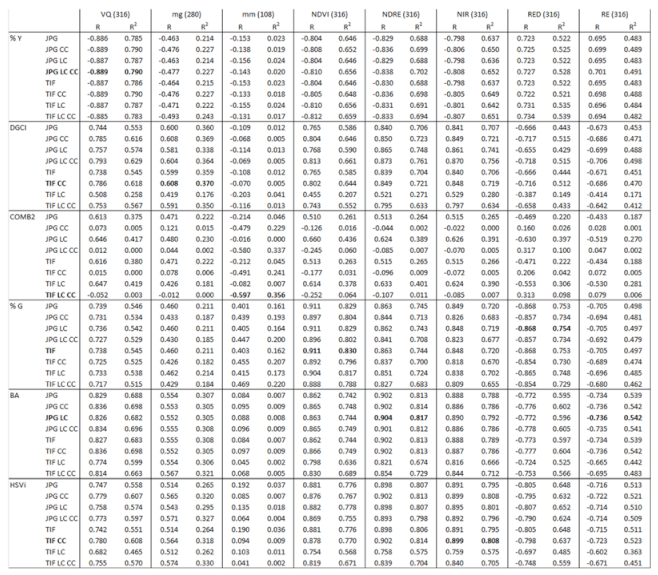
3.1. Linear Correlation between Variables
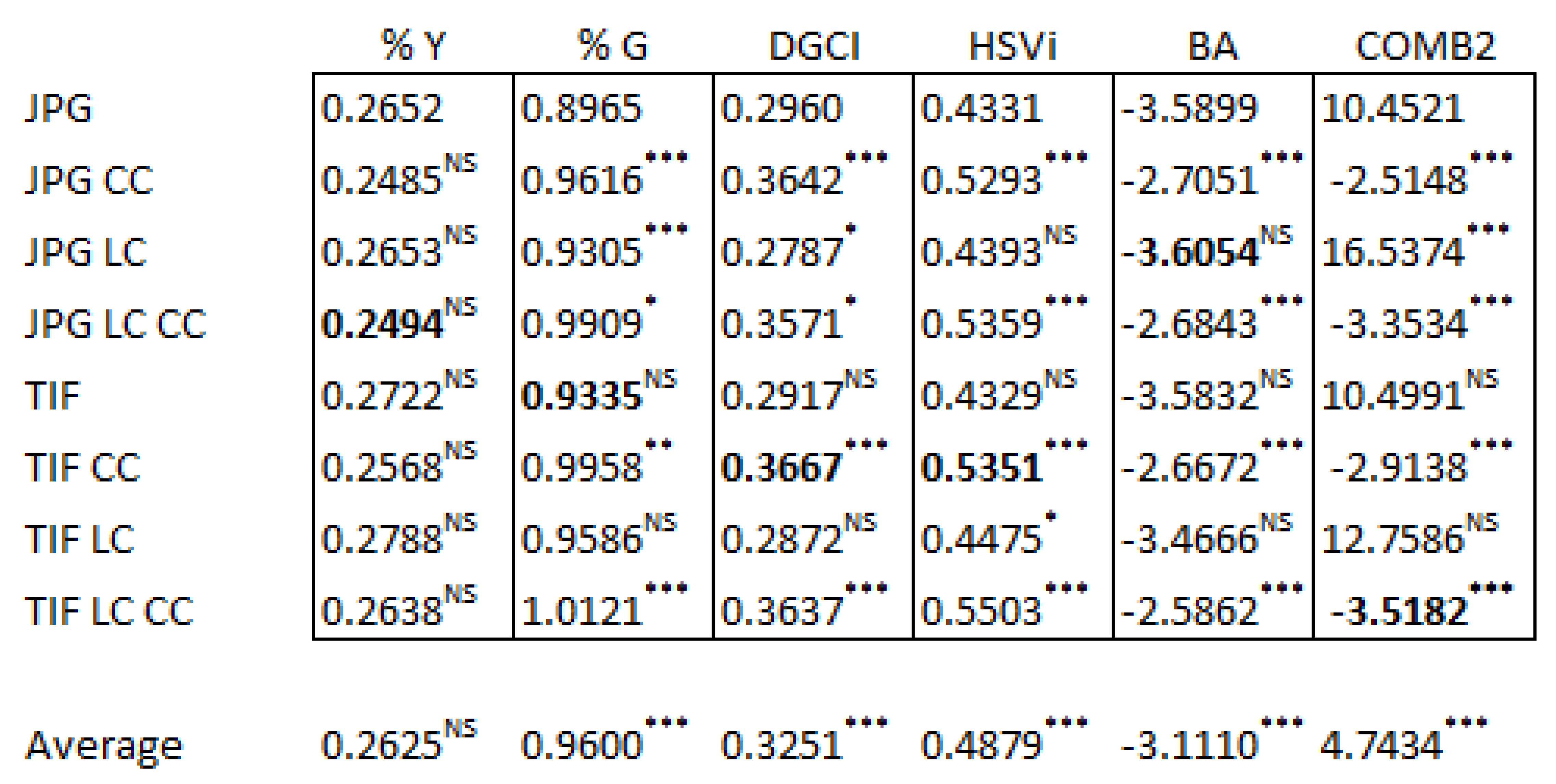
3.2. Effects of Image Corrections on Calculated Metrics
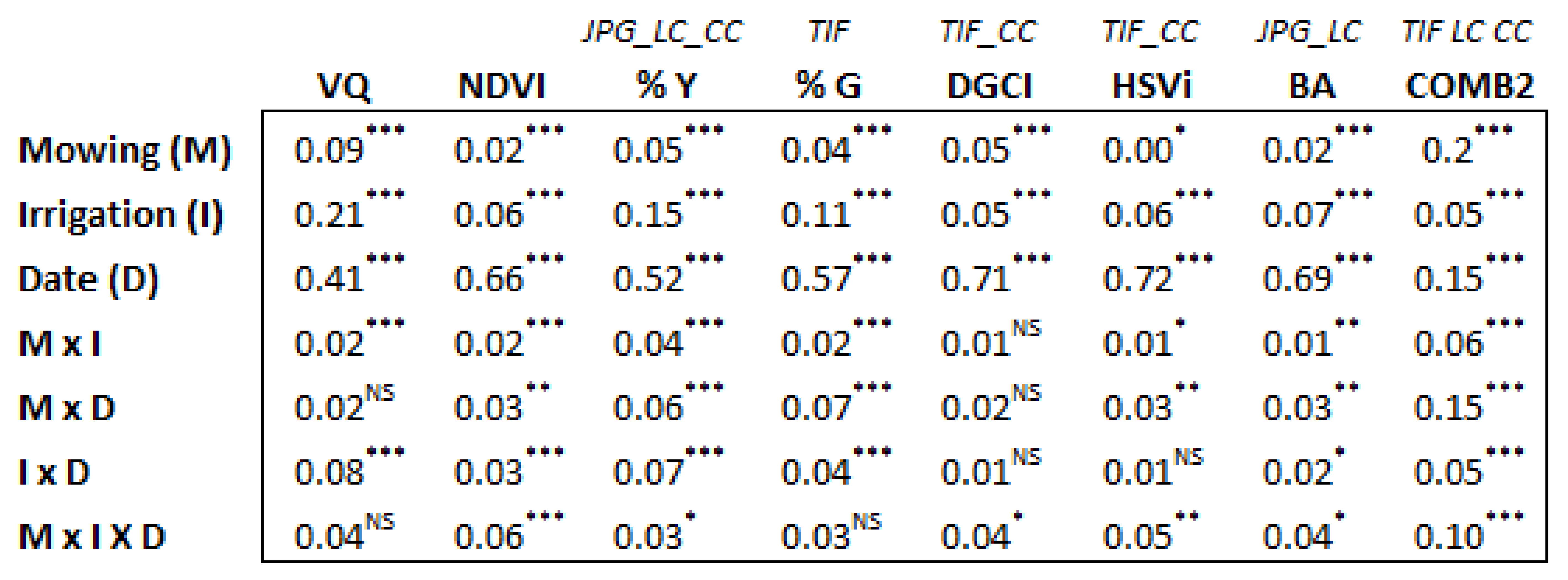
3.3. Detection of Experiment Treatment Effects
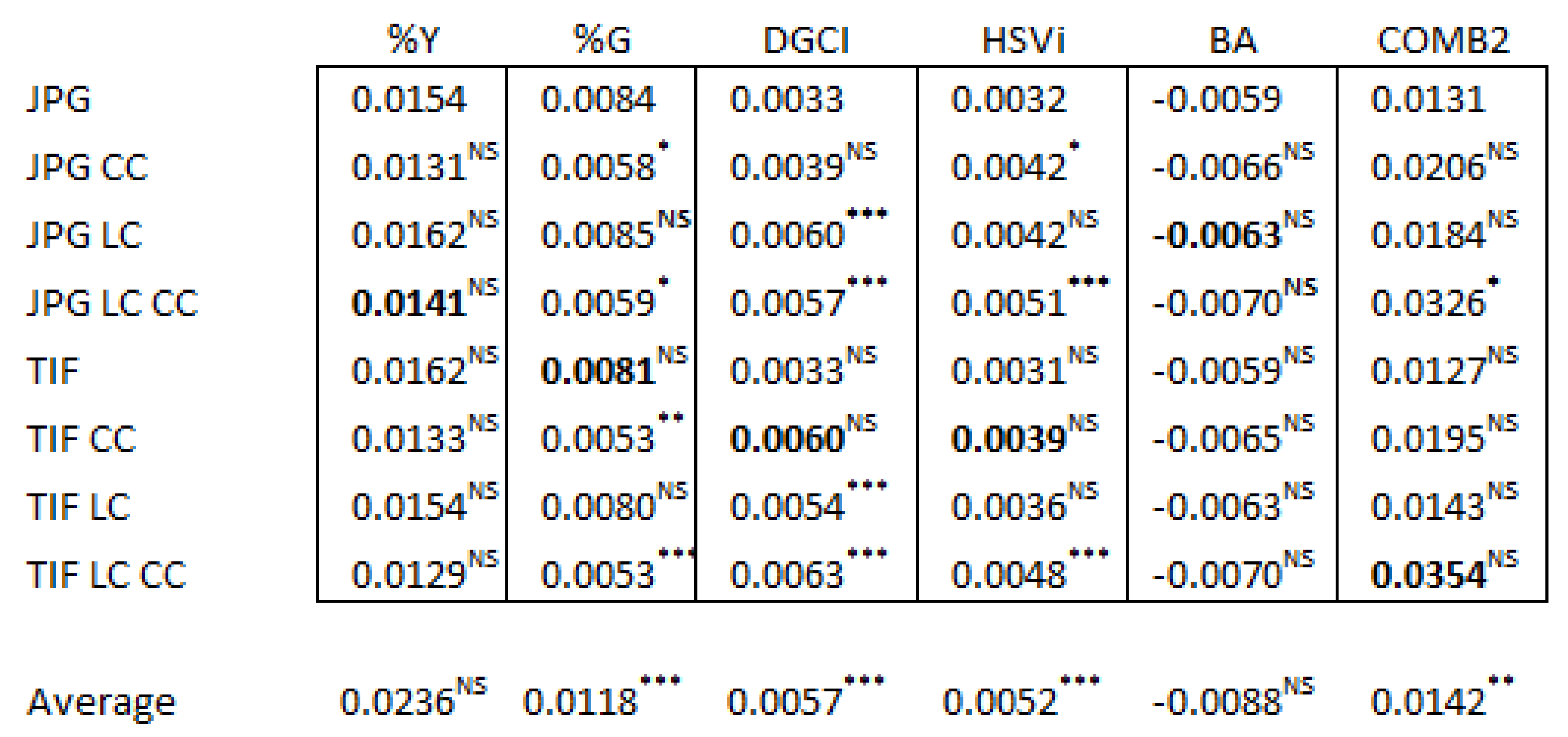
3.4. Image Corrections Effect on Replicates
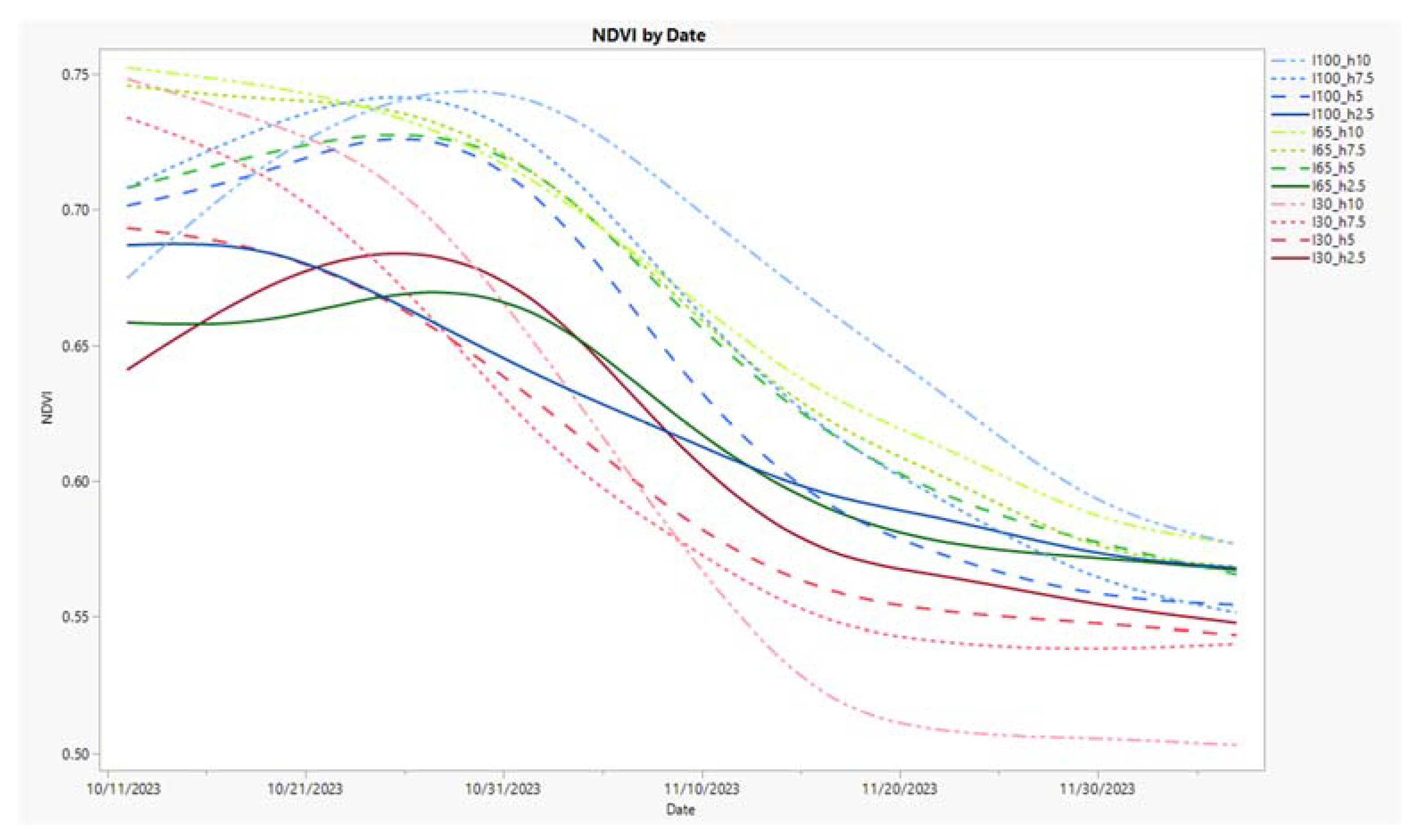
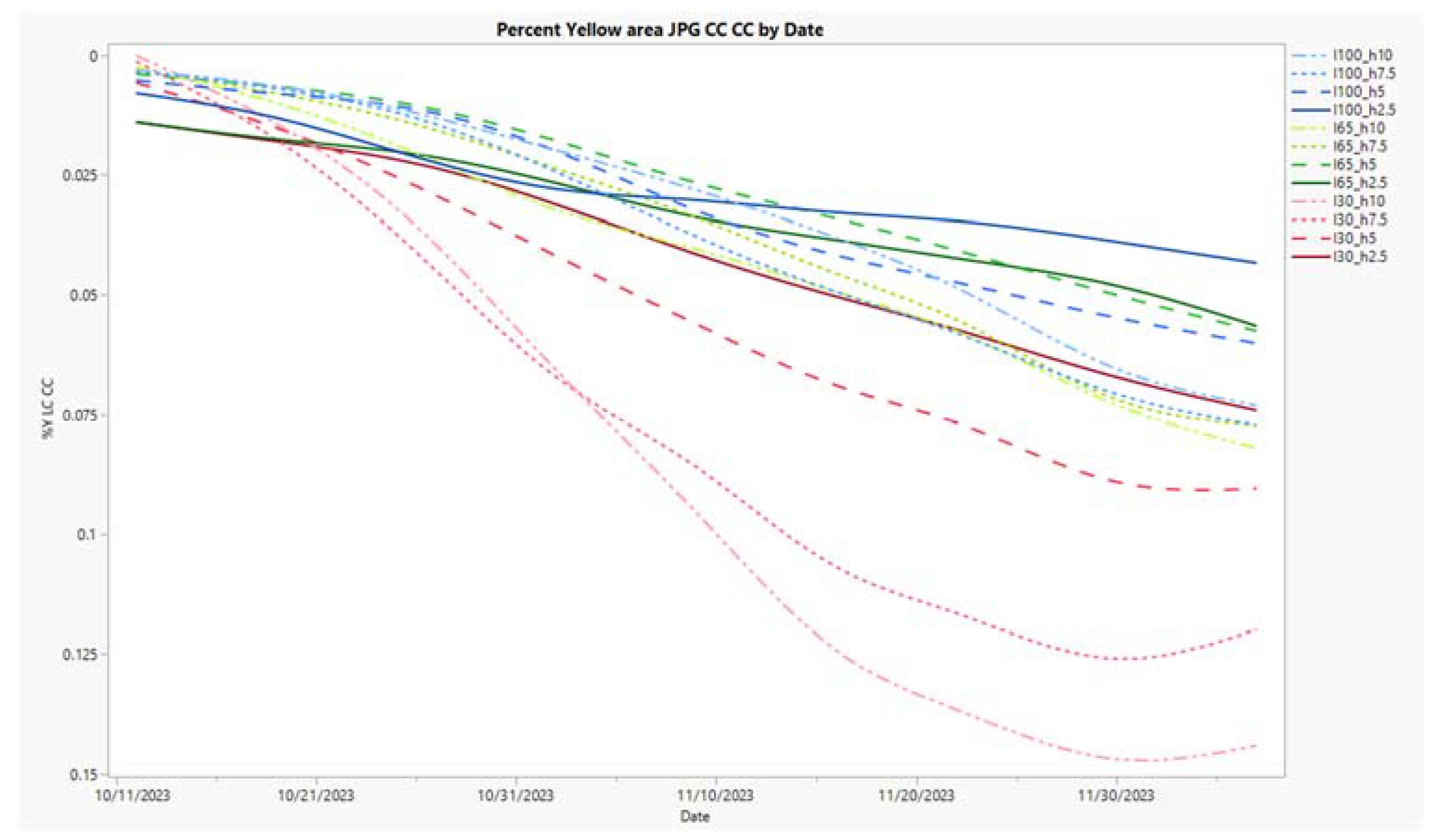
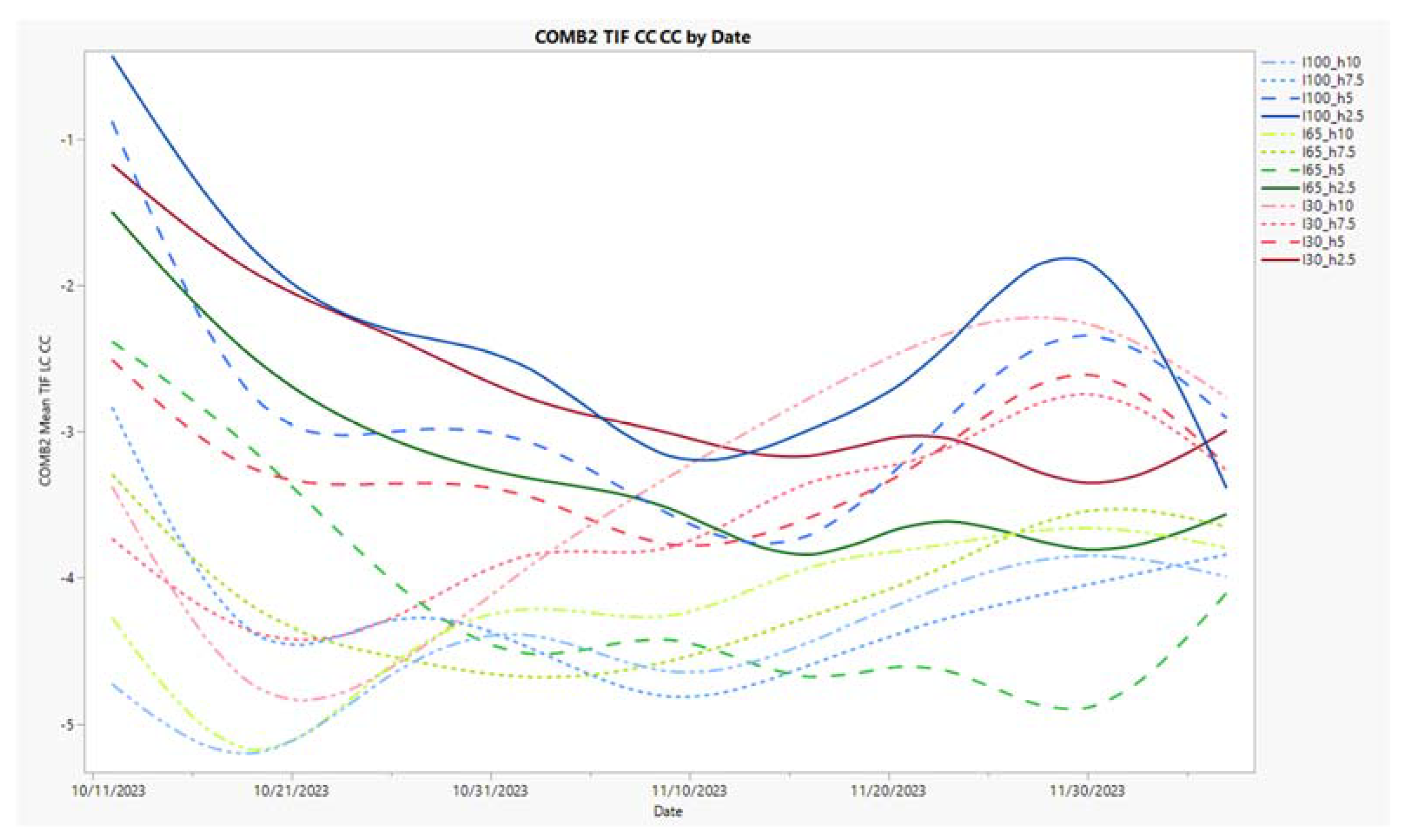
3.5. Time Series Charts for Three Metrics, NDVI, %Y, and COMB2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
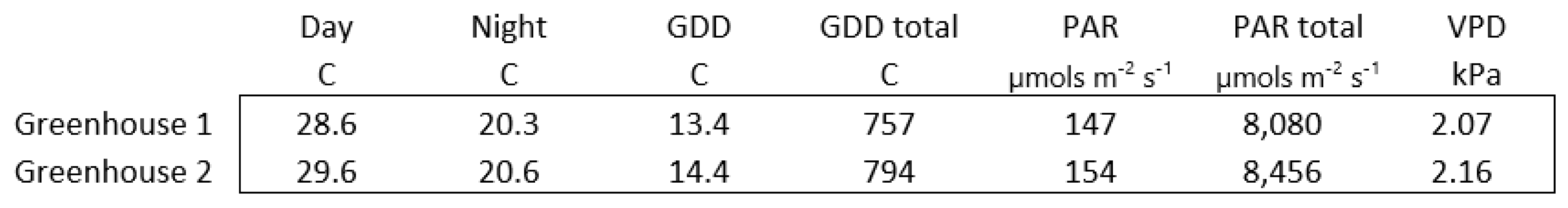
Appendix A. Description of Environmental Measurements
References
- J. B. Beard and R. L. Green, “The Role of Turfgrasses in Environmental Protection and Their Benefits to Humans,” J of Env Quality, vol. 23, no. 3, pp. 452–460, 1994. [CrossRef]
- P. Niazi, O. Alimyar, A. Azizi, A. W. Monib, and H. Ozturk, “People-plant Interaction: Plant Impact on Humans and Environment,” jeas, vol. 4, no. 2, pp. 01–07, May 2023. [CrossRef]
- I. Rehman, B. Hazhirkarzar, and B. C. Patel, “Anatomy, Head and Neck, Eye,” in StatPearls, Treasure Island (FL): StatPearls Publishing, 2024. Accessed: Jun. 14, 2024. [Online]. Available: http://www.ncbi.nlm.nih.gov/books/NBK482428/.
- S. Delgado, “Dziga Vertov’s ‘Man with a Movie Camera’ and the Phenomenology of Perception,” Film Criticism, vol. 34, no. 1, pp. 1–16, 2009, Accessed: Jun. 14, 2024. [Online]. Available: https://www.jstor.org/stable/24777403.
- M. Wrathall, Skillful Coping: Essays on the Phenomenology of Everyday Perception and Action. Oxford University Press, 2014.
- D. Ekdahl, “Review of Daniel O’Shiel, The Phenomenology of Virtual Technology: Perception and Imagination in a Digital Age, Dublin: Bloomsbury Academic, 2022,” Phenom Cogn Sci, pp. s11097-023-09925-y, Jul. 2023. [CrossRef]
- R. Sesario, Nugroho Djati Satmoko, Errie Margery, Yusi Faizathul Octavia, and Miska Irani Tarigan, “The Comparison Analysis of Brand Association, Brand Awareness, Brand Loyalty and Perceived Quality of Two Top of Mind Camera Products,” jemsi, vol. 9, no. 2, pp. 388–392, Apr. 2023. [CrossRef]
- S. Goma, M. Aleksic, and T. Georgiev, “Camera Technology at the dawn of digital renascence era,” in 2010 Conference Record of the Forty Fourth Asilomar Conference on Signals, Systems and Computers, Nov. 2010, pp. 847–850. [CrossRef]
- M. Esposito, M. Crimaldi, V. Cirillo, F. Sarghini, and A. Maggio, “Drone and sensor technology for sustainable weed management: a review,” Chem. Biol. Technol. Agric., vol. 8, no. 1, p. 18, Mar. 2021. [CrossRef]
- C. Edwards, R. Nilchiani, A. Ganguly, and M. Vierlboeck, “Evaluating the Tipping Point of a Complex System: The Case of Disruptive Technology.” Sep. 27, 2022. [CrossRef]
- X. Yue and E. R. Fossum, “Simulation and design of a burst mode 20Mfps global shutter high conversion gain CMOS image sensor in a standard 180nm CMOS image sensor process using sequential transfer gates,” ei, vol. 35, no. 6, pp. 328-1-328–5, Jan. 2023. [CrossRef]
- M. Riccardi, G. Mele, C. Pulvento, A. Lavini, R. d’Andria, and S.-E. Jacobsen, “Non-destructive evaluation of chlorophyll content in quinoa and amaranth leaves by simple and multiple regression analysis of RGB image components,” Photosynth Res, vol. 120, no. 3, pp. 263–272, 2014. [CrossRef]
- Y. Chang, S. L. Moan, and D. Bailey, “RGB Imaging Based Estimation of Leaf Chlorophyll Content,” in 2019 International Conference on Image and Vision Computing New Zealand (IVCNZ), Dunedin, New Zealand: IEEE, 2019, pp. 1–6. [CrossRef]
- H. Zhang, Y. Ge, X. Xie, A. Atefi, N. K. Wijewardane, and S. Thapa, “High throughput analysis of leaf chlorophyll content in sorghum using RGB, hyperspectral, and fluorescence imaging and sensor fusion,” Plant Methods, vol. 18, no. 1, p. 60, 2022. [CrossRef]
- P. Majer, L. Sass, G. V. Horváth, and É. Hideg, “Leaf hue measurements offer a fast, high-throughput initial screening of photosynthesis in leaves,” Journal of Plant Physiology, vol. 167, no. 1, pp. 74–76, 2010. [CrossRef]
- I. A. T. F. Taj-Eddin et al., “Can we see photosynthesis? Magnifying the tiny color changes of plant green leaves using Eulerian video magnification,” J. Electron. Imaging, vol. 26, no. 06, p. 1, Nov. 2017. [CrossRef]
- M. Vasilev, V. Stoykova, P. Veleva, and Z. Zlatev, “Non-Destructive Determination of Plant Pigments Based on Mobile Phone Data,” TEM Journal, pp. 1430–1442, Aug. 2023. [CrossRef]
- S. Kandel et al., “Research directions in data wrangling: Visualizations and transformations for usable and credible data,” Information Visualization, vol. 10, no. 4, pp. 271–288, 2011. [CrossRef]
- E. P. White, E. Baldridge, Z. T. Brym, K. J. Locey, D. J. McGlinn, and S. R. Supp, “Nine simple ways to make it easier to (re)use your data,” Ideas in Ecology and Evolution, vol. 6, no. 2, Aug. 2013, Accessed: Jun. 14, 2024. [Online]. Available: https://ojs.library.queensu.ca/index.php/IEE/article/view/4608.
- A. Goodman et al., “Ten Simple Rules for the Care and Feeding of Scientific Data,” PLoS Comput Biol, vol. 10, no. 4, p. e1003542, Apr. 2014. [CrossRef]
- T. U. Wall, E. McNie, and G. M. Garfin, “Use-inspired science: making science usable by and useful to decision makers,” Frontiers in Ecol & Environ, vol. 15, no. 10, pp. 551–559, 2017. [CrossRef]
- D. Gašparovič, J. Žarnovský, H. Beloev, and P. Kangalov, “Evaluation of the Quality of the Photographic Process at the Components Dimensions Measurement,” Agricultural, Forest and Transport Machinery and Technologies, vol. Volume II – Issue 1, no. ISSN: 2367– 5888, 2015, [Online]. Available: https://aftmt.uni-ruse.bg/images/vol.2.1/AFTMT_V_II-1-2015-3.pdf.
- G. Liu, S. Tian, Y. Mo, R. Chen, and Q. Zhao, “On the Acquisition of High-Quality Digital Images and Extraction of Effective Color Information for Soil Water Content Testing,” Sensors, vol. 22, no. 9, p. 3130, Apr. 2022. [CrossRef]
- D. Chen et al., “Dissecting the Phenotypic Components of Crop Plant Growth and Drought Responses Based on High-Throughput Image Analysis,” The Plant Cell, vol. 26, no. 12, pp. 4636–4655, Jan. 2015. [CrossRef]
- N. Honsdorf, T. J. March, B. Berger, M. Tester, and K. Pillen, “High-Throughput Phenotyping to Detect Drought Tolerance QTL in Wild Barley Introgression Lines,” PLoS ONE, vol. 9, no. 5, p. e97047, May 2014. [CrossRef]
- Y. Wang, D. Wang, P. Shi, and K. Omasa, “Estimating rice chlorophyll content and leaf nitrogen concentration with a digital still color camera under natural light,” Plant Methods, vol. 10, no. 1, p. 36, 2014. [CrossRef]
- K. M. Veley, J. C. Berry, S. J. Fentress, D. P. Schachtman, I. Baxter, and R. Bart, “High-throughput profiling and analysis of plant responses over time to abiotic stress,” Plant Direct, vol. 1, no. 4, p. e00023, 2017. [CrossRef]
- Z. Liang et al., “Conventional and hyperspectral time-series imaging of maize lines widely used in field trials,” GigaScience, vol. 7, no. 2, Feb. 2018. [CrossRef]
- B. Horvath and J. Vargas Jr, “Analysis of Dollar Spot Disease Severity Using Digital Image Analysis,” International Turfgrass Society Research Journal, vol. Volume 10, pp. 196–201, 2005, [Online]. Available: https://www.researchgate.net/publication/268359230_ANALYSIS_OF_DOLLAR_SPOT_DISEASE_SEVERITY_USING_DIGITAL_IMAGE_ANALYSIS.
- F. G. Horgan, A. Jauregui, A. Peñalver Cruz, E. Crisol Martínez, and C. C. Bernal, “Changes in reflectance of rice seedlings during planthopper feeding as detected by digital camera: Potential applications for high-throughput phenotyping,” PLoS ONE, vol. 15, no. 8, p. e0238173, Aug. 2020. [CrossRef]
- F. J. Adamsen et al., “Measuring Wheat Senescence with a Digital Camera,” Crop Sci., vol. 39, no. 3, pp. 719–724, 1999. [CrossRef]
- J. Friell, E. Watkins, and B. Horgan, “Salt Tolerance of 74 Turfgrass Cultivars in Nutrient Solution Culture,” Crop Science, vol. 53, no. 4, pp. 1743–1749, 2013. [CrossRef]
- J. Zhang et al., “Drought responses of above-ground and below-ground characteristics in warm-season turfgrass,” J Agronomy Crop Science, vol. 205, no. 1, pp. 1–12, 2019. [CrossRef]
- E. V. Lukina, M. L. Stone, and W. R. Raun, “Estimating vegetation coverage in wheat using digital images,” Journal of Plant Nutrition, vol. 22, no. 2, pp. 341–350, 1999. [CrossRef]
- D. E. Karcher and M. D. Richardson, “Batch Analysis of Digital Images to Evaluate Turfgrass Characteristics,” Crop Science, vol. 45, no. 4, pp. 1536–1539, 2005. [CrossRef]
- D. J. Bremer, H. Lee, K. Su, and S. J. Keeley, “Relationships between Normalized Difference Vegetation Index and Visual Quality in Cool-Season Turfgrass: II. Factors Affecting NDVI and its Component Reflectances,” Crop Science, vol. 51, no. 5, pp. 2219–2227, 2011. [CrossRef]
- B. S. Bushman, B. L. Waldron, J. G. Robins, K. Bhattarai, and P. G. Johnson, “Summer Percent Green Cover among Kentucky Bluegrass Cultivars, Accessions, and Other Poa Species Managed under Deficit Irrigation,” Crop Science, vol. 52, no. 1, pp. 400–407, 2012. [CrossRef]
- A. Patrignani and T.E. Ochsner, “Canopeo: A Powerful New Tool for Measuring Fractional Green Canopy Cover,” Agronomy Journal, vol. 107, no. 6, pp. 2312–2320, 2015. [CrossRef]
- S. O. Chung, M. S. N. Kabir, and Y. J. Kim, “Variable Fertilizer Recommendation by Image-based Grass Growth Status,” IFAC-PapersOnLine, vol. 51, no. 17, pp. 10–13, 2018. [CrossRef]
- K. R. Ball et al., “High-throughput, image-based phenotyping reveals nutrient-dependent growth facilitation in a grass-legume mixture,” PLoS ONE, vol. 15, no. 10, p. e0239673, Oct. 2020. [CrossRef]
- H. C. Wright, F. A. Lawrence, A. J. Ryan, and D. D. Cameron, “Free and open-source software for object detection, size, and colour determination for use in plant phenotyping,” Plant Methods, vol. 19, no. 1, p. 126, Nov. 2023. [CrossRef]
- J. N. Cobb, G. DeClerck, A. Greenberg, R. Clark, and S. McCouch, “Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement,” Theor Appl Genet, vol. 126, no. 4, pp. 867–887, 2013. [CrossRef]
- B.T. Gouveia et al., “Multispecies genotype × environment interaction for turfgrass quality in five turfgrass breeding programs in the southeastern United States,” Crop Science, vol. 61, no. 5, pp. 3080–3096, 2021. [CrossRef]
- M. F. McCabe and M. Tester, “Digital insights: bridging the phenotype-to-genotype divide,” Journal of Experimental Botany, vol. 72, no. 8, pp. 2807–2810, Apr. 2021. [CrossRef]
- M. F. Danilevicz et al., “Plant Genotype to Phenotype Prediction Using Machine Learning,” Front. Genet., vol. 13, p. 822173, May 2022. [CrossRef]
- S. A. Tsaftaris, M. Minervini, and H. Scharr, “Machine Learning for Plant Phenotyping Needs Image Processing,” Trends in Plant Science, vol. 21, no. 12, pp. 989–991, 2016. [CrossRef]
- U. Lee, S. Chang, G. A. Putra, H. Kim, and D. H. Kim, “An automated, high-throughput plant phenotyping system using machine learning-based plant segmentation and image analysis,” PLoS ONE, vol. 13, no. 4, p. e0196615, Apr. 2018. [CrossRef]
- J. C. O. Koh, G. Spangenberg, and S. Kant, “Automated Machine Learning for High-Throughput Image-Based Plant Phenotyping,” Remote Sensing, vol. 13, no. 5, p. 858, Feb. 2021. [CrossRef]
- Z. Li, R. Guo, M. Li, Y. Chen, and G. Li, “A review of computer vision technologies for plant phenotyping,” Computers and Electronics in Agriculture, vol. 176, p. 105672, 2020. [CrossRef]
- D.T. Smith, A. B. Potgieter, and S. C. Chapman, “Scaling up high-throughput phenotyping for abiotic stress selection in the field,” Theor Appl Genet, vol. 134, no. 6, pp. 1845–1866, 2021. [CrossRef]
- L. Li, Q. Zhang, and D. Huang, “A Review of Imaging Techniques for Plant Phenotyping,” Sensors, vol. 14, no. 11, pp. 20078–20111, Oct. 2014. [CrossRef]
- Rousseau, H. D. Dee, and T. Pridmore, “Imaging Methods for Phenotyping of Plant Traits,” in Phenomics in Crop Plants: Trends, Options and Limitations, J. Kumar, A. Pratap, and S. Kumar, Eds., New Delhi: Springer India, 2015, pp. 61–74. [CrossRef]
- M. Tariq, M. Ahmed, P. Iqbal, Z. Fatima, and S. Ahmad, “Crop Phenotyping,” in Systems Modeling, M. Ahmed, Ed., Singapore: Springer Singapore, 2020, pp. 45–60. [CrossRef]
- Cardellicchio; et al. , “Detection of tomato plant phenotyping traits using YOLOv5-based single stage detectors,” Computers and Electronics in Agriculture, vol. 207, p. 107757, 2023. [CrossRef]
- N. Harandi, B. Vandenberghe, J. Vankerschaver, S. Depuydt, and A. Van Messem, “How to make sense of 3D representations for plant phenotyping: a compendium of processing and analysis techniques,” Plant Methods, vol. 19, no. 1, p. 60, Jun. 2023. [CrossRef]
- C. Zhang, J. Kong, D. Wu, Z. Guan, B. Ding, and F. Chen, “Wearable Sensor: An Emerging Data Collection Tool for Plant Phenotyping,” Plant Phenomics, vol. 5, p. 0051, 2023. [CrossRef]
- Y. Zhang and N. Zhang, “Imaging technologies for plant high-throughput phenotyping: a review,” Front. Agr. Sci. Eng., vol. 0, no. 0, p. 0, 2018. [CrossRef]
- S. Das Choudhury, A. Samal, and T. Awada, “Leveraging Image Analysis for High-Throughput Plant Phenotyping,” Front. Plant Sci., vol. 10, p. 508, Apr. 2019. [CrossRef]
- Department of Computer Science and IT, N.M.S. Sermathai Vasan College for Women, Madurai, India. and K. Rani, “Image Analysis Techniqueson Phenotype for Plant System,” IJEAT, vol. 9, no. 1s4, pp. 565–568, Dec. 2019. [CrossRef]
- M. Kamran Omari, J. Lee, M. Akbar Faqeerzada, R. Joshi, E. Park, and B.-K. Cho, “Digital image-based plant phenotyping: a review,” Korean Journal of Agricultural Science, vol. 47(1), pp. 119–130, 2020, [Online]. Available: https://www.researchgate.net/publication/340032840_Digital_image-based_plant_phenotyping_a_review.
- H. L. Shantz, “The Place of Grasslands in the Earth’s Cover,” Ecology, vol. 35, no. 2, pp. 143–145, 1954. [CrossRef]
- B. F. Jacobs, J. D. Kingston, and L. L. Jacobs, “The Origin of Grass-Dominated Ecosystems,” Annals of the Missouri Botanical Garden, vol. 86, no. 2, pp. 590–643, 1999. [CrossRef]
- A. R. Watkinson and S. J. Ormerod, “Grasslands, Grazing and Biodiversity: Editors’ Introduction,” Journal of Applied Ecology, vol. 38, no. 2, pp. 233–237, 2001, Accessed: Jun. 14, 2024. [Online]. Available: https://www.jstor.org/stable/2655793.
- C. A. E. Strömberg, “Evolution of Grasses and Grassland Ecosystems,” Annu. Rev. Earth Planet. Sci., vol. 39, no. 1, pp. 517–544, May 2011. https://doi.org/10.1146/annurev-earth-040809-152402. [CrossRef]
- S. L. Chawla, M. A. Roshni Agnihotri, P. Sudha, and H. P. Shah, “Turfgrass: A Billion Dollar Industry,” in National Conference on Floriculture for Rural and Urban Prosperity in the Scenario of Climate Change, 2018. [Online]. Available: https://www.researchgate.net/publication/324483293_Turfgrass_A_Billion_Dollar_Industry.
- J. Wu and M. E. Bauer, “Estimating Net Primary Production of Turfgrass in an Urban-Suburban Landscape with QuickBird Imagery,” Remote Sensing, vol. 4, no. 4, pp. 849–866, Mar. 2012. [CrossRef]
- Milesi, S. W. Running, C. D. Elvidge, J. B. Dietz, B. T. Tuttle, and R. R. Nemani, “Mapping and Modeling the Biogeochemical Cycling of Turf Grasses in the United States,” Environmental Management, vol. 36, no. 3, pp. 426–438, 2005. [CrossRef]
- C. A. Blanco-Montero, T. B. Bennett, P. Neville, C. S. Crawford, B. T. Milne, and C. R. Ward, “Potential environmental and economic impacts of turfgrass in Albuquerque, New Mexico (USA),” Landscape Ecol, vol. 10, no. 2, pp. 121–128, 1995. [CrossRef]
- J. V. Krans and K. Morris, “Determining a Profile of Protocols and Standards used in the Visual Field Assessment of Turfgrasses: A Survey of National Turfgrass Evaluation Program-Sponsored University Scientists,” Applied Turfgrass Science, vol. 4, no. 1, pp. 1–6, 2007. [CrossRef]
- J. B. Beard, Turfgrass: Science and culture. Englewood Cliffs, NJ: Prentice Hall, 1973. [Online]. Available: https://catalogue.nla.gov.au/catalog/2595129.
- A. Calera, C. Martínez, and J. Melia, “A procedure for obtaining green plant cover: Relation to NDVI in a case study for barley,” International Journal of Remote Sensing, vol. 22, no. 17, pp. 3357–3362, 2001. [CrossRef]
- L. Cabrera-Bosquet, G. Molero, A. Stellacci, J. Bort, S. Nogués, and J. Araus, “NDVI as a potential tool for predicting biomass, plant nitrogen content and growth in wheat genotypes subjected to different water and nitrogen conditions,” Cereal Research Communications, vol. 39, no. 1, pp. 147–159, 2011. [CrossRef]
- R. L. Rorie et al., “Association of ‘Greenness’ in Corn with Yield and Leaf Nitrogen Concentration,” Agronomy Journal, vol. 103, no. 2, pp. 529–535, 2011. [CrossRef]
- M. OZYAVUZ, B. C. BILGILI, and S. SALICI, “Determination of Vegetation Changes with NDVI Method,” Journal of Environmental Protection and Ecology, vol. 16, no. 1, pp. 264–273, 2015, [Online]. Available: https://www.researchgate.net/publication/284981527_Determination_of_vegetation_changes_with_NDVI_method.
- S. Huang, L. Tang, J. P. Hupy, Y. Wang, and G. Shao, “A commentary review on the use of normalized difference vegetation index (NDVI) in the era of popular remote sensing,” J. For. Res., vol. 32, no. 1, pp. 1–6, 2021. [CrossRef]
- Y. Xu, Y. Yang, X. Chen, and Y. Liu, “Bibliometric Analysis of Global NDVI Research Trends from 1985 to 2021,” Remote Sensing, vol. 14, no. 16, p. 3967, Aug. 2022. [CrossRef]
- T. N. Carlson and D. A. Ripley, “On the relation between NDVI, fractional vegetation cover, and leaf area index,” Remote Sensing of Environment, vol. 62, no. 3, pp. 241–252, 1997. [CrossRef]
- T. R. Tenreiro, M. García-Vila, J. A. Gómez, J. A. Jiménez-Berni, and E. Fereres, “Using NDVI for the assessment of canopy cover in agricultural crops within modelling research,” Computers and Electronics in Agriculture, vol. 182, p. 106038, 2021. [CrossRef]
- P. V. Lykhovyd, R. A. Vozhehova, S. O. Lavrenko, and N. M. Lavrenko, “The Study on the Relationship between Normalized Difference Vegetation Index and Fractional Green Canopy Cover in Five Selected Crops,” The Scientific World Journal, vol. 2022, pp. 1–6, Mar. 2022. [CrossRef]
- M. Pagola et al., “New method to assess barley nitrogen nutrition status based on image colour analysis,” Computers and Electronics in Agriculture, vol. 65, no. 2, pp. 213–218, 2009. [CrossRef]
- C. M. Straw and G. M. Henry, “Spatiotemporal variation of site-specific management units on natural turfgrass sports fields during dry down,” Precision Agric, vol. 19, no. 3, pp. 395–420, 2018. [CrossRef]
- R. Hejl, C. Straw, B. Wherley, R. Bowling, and K. McInnes, “Factors leading to spatiotemporal variability of soil moisture and turfgrass quality within sand-capped golf course fairways,” Precision Agric, vol. 23, no. 5, pp. 1908–1917, 2022. [CrossRef]
- S. Kawashima, “An Algorithm for Estimating Chlorophyll Content in Leaves Using a Video Camera,” Annals of Botany, vol. 81, no. 1, pp. 49–54, 1998. [CrossRef]
- E. R. Hunt, P. C. Doraiswamy, J. E. McMurtrey, C. S. T. Daughtry, E. M. Perry, and B. Akhmedov, “A visible band index for remote sensing leaf chlorophyll content at the canopy scale,” International Journal of Applied Earth Observation and Geoinformation, vol. 21, pp. 103–112, 2013. [CrossRef]
- M. Schiavon, B. Leinauer, E. Sevastionova, M. Serena, and B. Maier, “Warm-season Turfgrass Quality, Spring Green-up, and Fall Color Retention under Drip Irrigation,” Applied Turfgrass Science, vol. 8, no. 1, pp. 1–9, 2011. [CrossRef]
- J. Marín, S. Yousfi, P. V. Mauri, L. Parra, J. Lloret, and A. Masaguer, “RGB Vegetation Indices, NDVI, and Biomass as Indicators to Evaluate C3 and C4 Turfgrass under Different Water Conditions,” Sustainability, vol. 12, no. 6, p. 2160, Mar. 2020. [CrossRef]
- G. E. Bell et al., “Vehicle-Mounted Optical Sensing: An Objective Means for Evaluating Turf Quality,” Crop Science, vol. 42, no. 1, pp. 197–201, 2002. [CrossRef]
- G. E. Bell, D. L. Martin, K. Koh, and H. R. Han, “Comparison of Turfgrass Visual Quality Ratings with Ratings Determined Using a Handheld Optical Sensor,” hortte, vol. 19, no. 2, pp. 309–316, 2009. [CrossRef]
- D. E. Karcher and M. D. Richardson, “Digital Image Analysis in Turfgrass Research,” in Turfgrass: Biology, Use, and Management, J. C. Stier, B. P. Horgan, and S. A. Bonos, Eds., Madison, WI, USA: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America, 2015, pp. 1133-1149–2. [CrossRef]
- B. D. S. Barbosa et al., “RGB vegetation indices applied to grass monitoring: a qualitative analysis,” p. 675.9Kb, 2019. [CrossRef]
- B. Whitman, B. V. Iannone, J. K. Kruse, J. B. Unruh, and A. G. Dale, “Cultivar blends: A strategy for creating more resilient warm season turfgrass lawns,” Urban Ecosyst, vol. 25, no. 3, pp. 797–810, 2022. [CrossRef]
- D. Hahn, A. Morales, C. Velasco-Cruz, and B. Leinauer, “Assessing Competitiveness of Fine Fescues (Festuca L. spp.) and Tall Fescue (Schedonorus arundinaceous (Schreb.) Dumort) Established with White Clover (Trifolium repens L., WC), Daisy (Bellis perennis L.) and Yarrow (Achillea millefolium L.),” Agronomy, vol. 11, no. 11, p. 2226, Nov. 2021. [CrossRef]
- B. Schwartz, J. Zhang, J. Fox, and J. Peake, “Turf Performance of Shaded ‘TifGrand’ and ‘TifSport’ Hybrid Bermudagrass as Affected by Mowing Height and Trinexapac-ethyl,” hortte, vol. 30, no. 3, pp. 391–397, 2020. [CrossRef]
- M. D. Richardson, D. E. Karcher, A. J. Patton, and J. H. McCalla, “Measurement of Golf Ball Lie in Various Turfgrasses Using Digital Image Analysis,” Crop Science, vol. 50, no. 2, pp. 730–736, 2010. [CrossRef]
- A. Walter, B. Studer, and R. Kölliker, “Advanced phenotyping offers opportunities for improved breeding of forage and turf species,” Annals of Botany, vol. 110, no. 6, pp. 1271–1279, 2012. [CrossRef]
- J. B. Hu, M. X. Dai, and S. T. Peng, “An automated (novel) algorithm for estimating green vegetation cover fraction from digital image: UIP-MGMEP,” Environ Monit Assess, vol. 190, no. 11, p. 687, 2018. [CrossRef]
- P. Xu, N. Wang, X. Zheng, G. Qiu, and B. Luo, “A New Turfgrass Coverage Evaluation Method Based on Two-Stage k-means Color Classifier,” in 2019 Boston, Massachusetts July 7- July 10, 2019, American Society of Agricultural and Biological Engineers, 2019. [CrossRef]
- S. Xie, C. Hu, M. Bagavathiannan, and D. Song, “Toward Robotic Weed Control: Detection of Nutsedge Weed in Bermudagrass Turf Using Inaccurate and Insufficient Training Data,” IEEE Robot. Autom. Lett., vol. 6, no. 4, pp. 7365–7372, 2021. [CrossRef]
- J. B. Ortiz, C. N. Hirsch, N. J. Ehlke, and E. Watkins, “SpykProps: an imaging pipeline to quantify architecture in unilateral grass inflorescences,” Plant Methods, vol. 19, no. 1, p. 125, Nov. 2023. [CrossRef]
- D. J. Bremer, H. Lee, K. Su, and S. J. Keeley, “Relationships between Normalized Difference Vegetation Index and Visual Quality in Cool-Season Turfgrass: I. Variation among Species and Cultivars,” Crop Science, vol. 51, no. 5, pp. 2212–2218, 2011. [CrossRef]
- H.Lee, D. J. Bremer, K. Su, and S. J. Keeley, “Relationships between Normalized Difference Vegetation Index and Visual Quality in Turfgrasses: Effects of Mowing Height,” Crop Science, vol. 51, no. 1, pp. 323–332, 2011. [CrossRef]
- B.Leinauer, D. M. VanLeeuwen, M. Serena, M. Schiavon, and E. Sevostianova, “Digital Image Analysis and Spectral Reflectance to Determine Turfgrass Quality,” Agronomy Journal, vol. 106, no. 5, pp. 1787–1794, 2014. [CrossRef]
- A.Haghverdi, A. Sapkota, A. Singh, S. Ghodsi, and M. Reiter, “Developing Turfgrass Water Response Function and Assessing Visual Quality, Soil Moisture and NDVI Dynamics of Tall Fescue Under Varying Irrigation Scenarios in Inland Southern California,” Journal of the ASABE, vol. 66, no. 6, pp. 1497–1512, 2023. [CrossRef]
- R. W. Hejl, M. M. Conley, D. D. Serba, and C. F. Williams, “Mowing Height Effects on ‘TifTuf’ Bermudagrass during Deficit Irrigation,” Agronomy, vol. 14, no. 3, p. 628, Mar. 2024. [CrossRef]
- B.Boiarskii, “Comparison of NDVI and NDRE Indices to Detect Differences in Vegetation and Chlorophyll Content,” JMCMS, vol. spl1, no. 4, Nov. 2019. [CrossRef]
- F.Pallottino, F. Antonucci, C. Costa, C. Bisaglia, S. Figorilli, and P. Menesatti, “Optoelectronic proximal sensing vehicle-mounted technologies in precision agriculture: A review,” Computers and Electronics in Agriculture, vol. 162, pp. 859–873, 2019. [CrossRef]
- R. K. Kurbanov and N. I. Zakharova, “Application of Vegetation Indexes to Assess the Condition of Crops,” S-h. maš. tehnol., vol. 14, no. 4, pp. 4–11, Dec. 2020. [CrossRef]
- J.Frossard and, O. Renaud, “Permutation Tests for Regression, ANOVA, and Comparison of Signals: The permuco Package,” J. Stat. Soft., vol. 99, no. 15, 2021. [CrossRef]
- M. D. Richardson, D. E. Karcher, and L. C. Purcell, “Quantifying Turfgrass Cover Using Digital Image Analysis,” Crop Science, vol. 41, no. 6, pp. 1884–1888, 2001. [CrossRef]
- C.Zhang, G. D. Pinnix, Z. Zhang, G. L. Miller, and T. W. Rufty, “Evaluation of Key Methodology for Digital Image Analysis of Turfgrass Color Using Open-Source Software,” Crop Science, vol. 57, no. 2, pp. 550–558, 2017. [CrossRef]
- M. S. Woolf, L. M. Dignan, A. T. Scott, and J. P. Landers, “Digital postprocessing and image segmentation for objective analysis of colorimetric reactions,” Nat Protoc, vol. 16, no. 1, pp. 218–238, 2021. [CrossRef]
- D.E. Karcher and M. D. Richardson, “Quantifying Turfgrass Color Using Digital Image Analysis,” Crop Science, vol. 43, no. 3, pp. 943–951, 2003. [CrossRef]
- M. Anderson, R. Motta, S. Chandrasekar, and M. Stokes, “Proposal for a Standard Default Color Space for the Internet—sRGB,” in The Fourth Color Imaging Conference, Society of Imaging Science and Technology, 1996, pp. 239–245. [Online]. Available: https://library.imaging.org/admin/apis/public/api/ist/website/downloadArticle/cic/4/1/art00061.
- J. Casadesús and, D. Villegas, “Conventional digital cameras as a tool for assessing leaf area index and biomass for cereal breeding,” JIPB, vol. 56, no. 1, pp. 7–14, 2014. [CrossRef]
- D.Karcher, C. Purcell, and K. Hignight, “Devices, systems and methods for digital image analysis,” US20220270206A1, Aug. 25, 2022 Accessed: Jun. 14, 2024. [Online]. Available: https://patents.google.com/patent/US20220270206A1/en.
- M. Schiavon, B. Leinauer, M. Serena, R. Sallenave, and B. Maier, “Establishing Tall Fescue and Kentucky Bluegrass Using Subsurface Irrigation and Saline Water,” Agronomy Journal, vol. 105, no. 1, pp. 183–190, 2013. [CrossRef]
- M. Giolo, C. Pornaro, A. Onofri, and S. Macolino, “Seeding Time Affects Bermudagrass Establishment in the Transition Zone Environment,” Agronomy, vol. 10, no. 8, p. 1151, Aug. 2020. [CrossRef]
- M. Schiavon, C. Pornaro, and S. Macolino, “Tall Fescue (Schedonorus arundinaceus (Schreb.) Dumort.) Turfgrass Cultivars Performance under Reduced N Fertilization,” Agronomy, vol. 11, no. 2, p. 193, Jan. 2021. [CrossRef]
- B.Zhao et al., “Estimating the Growth Indices and Nitrogen Status Based on Color Digital Image Analysis During Early Growth Period of Winter Wheat,” Front. Plant Sci., vol. 12, p. 619522, Apr. 2021. [CrossRef]
- A. Sierra Augustinus, P. H. McLoughlin, A. F. Arevalo Alvarenga, J. B. Unruh, and M. Schiavon, “Evaluation of Different Aerification Methods for Ultradwarf Hybrid Bermudagrass Putting Greens,” hortte, vol. 33, no. 4, pp. 333–341, 2023. [CrossRef]
- S. Singh et al., “Genetic Variability of Traffic Tolerance and Surface Playability of Bermudagrass (Cynodon spp.) under Fall Simulated Traffic Stress,” horts, vol. 59, no. 1, pp. 73–83, 2024. [CrossRef]
- T. Glab, W. Szewczyk, and K. Gondek, “Response of Kentucky Bluegrass Turfgrass to Plant Growth Regulators,” Agronomy, vol. 13, no. 3, p. 799, Mar. 2023. [CrossRef]
- M. Guijarro, G. Pajares, I. Riomoros, P. J. Herrera, X. P. Burgos-Artizzu, and A. Ribeiro, “Automatic segmentation of relevant textures in agricultural images,” Computers and Electronics in Agriculture, vol. 75, no. 1, pp. 75–83, Jan. 2011. [CrossRef]
- D.M. Woebbecke, G. E. Meyer, K. Von Bargen, and D. A. Mortensen, “Color Indices for Weed Identification Under Various Soil, Residue, and Lighting Conditions,” Transactions of the ASAE, vol. 38, no. 1, pp. 259–269, 1995. [CrossRef]
- T. Kataoka, T. Kaneko, H. Okamoto, and S. Hata, “Crop growth estimation system using machine vision,” in Proceedings 2003 IEEE/ASME International Conference on Advanced Intelligent Mechatronics (AIM 2003), Kobe, Japan: IEEE, 2003, pp. b1079–b1083. [CrossRef]
- J.A. Marchant and C. M. Onyango, “Shadow-invariant classification for scenes illuminated by daylight,” J. Opt. Soc. Am. A, JOSAA, vol. 17, no. 11, pp. 1952–1961, Nov. 2000. [CrossRef]
- T. Hague, N. D. Tillett, and H. Wheeler, “Automated Crop and Weed Monitoring in Widely Spaced Cereals,” Precision Agric, vol. 7, no. 1, pp. 21–32, Mar. 2006. [CrossRef]
- R. Robertson, “The CIE 1976 Color-Difference Formulae,” Color Research & Application, vol. 2, no. 1, pp. 7–11, 1977. [CrossRef]
- J. Schwiegerling, Field Guide to Visual and Ophthalmic Optics, vol. FG04. Bellingham, Washington, USA: SPIE, 2004. [Online]. Available: https://spie.org/publications/spie-publication-resources/optipedia-free-optics-information/fg04_p12_phoscoresponse?
- A. Koschan and M., A. Abidi, Digital Color Image Processing, 1st ed. Wiley, 2008. [CrossRef]
- J. Chopin, P. Kumar, and S. J. Miklavcic, “Land-based crop phenotyping by image analysis: consistent canopy characterization from inconsistent field illumination,” Plant Methods, vol. 14, no. 1, p. 39, 2018. [CrossRef]
- J.M. Pape and C. Klukas, “Utilizing machine learning approaches to improve the prediction of leaf counts and individual leaf segmentation of rosette plant images,” Department of Molecular Genetics, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), 06466 Gatersleben, Germany, 2015. [Online]. Available: https://openimageanalysisgroup.github.io/MCCCS/publications/Pape_Klukas_LSC_2015.pdf.
- S. C. Kefauver et al., “Comparative UAV and Field Phenotyping to Assess Yield and Nitrogen Use Efficiency in Hybrid and Conventional Barley,” Front. Plant Sci., vol. 8, p. 1733, Oct. 2017. [CrossRef]
- J. Du, B. Li, X. Lu, X. Yang, X. Guo, and C. Zhao, “Quantitative phenotyping and evaluation for lettuce leaves of multiple semantic components,” Plant Methods, vol. 18, no. 1, p. 54, 2022. [CrossRef]
- X. Xie et al., “A Novel Feature Selection Strategy Based on Salp Swarm Algorithm for Plant Disease Detection,” Plant Phenomics, vol. 5, p. 0039, 2023. [CrossRef]
- W. Wahono, D. Indradewa, B. H. Sunarminto, E. Haryono, and D. Prajitno, “CIE L*a*b* Color Space Based Vegetation Indices Derived from Unmanned Aerial Vehicle Captured Images for Chlorophyll and Nitrogen Content Estimation of Tea (Camellia sinensis L. Kuntze) Leaves,” Ilmu Pertanian (Agricultural Science), vol. 4, no. 1, pp. 46–51, May 2019. [CrossRef]
- R. R. De Casas, P. Vargas, E. Pérez-Corona, E. Manrique, C. García-Verdugo, and L. Balaguer, “Sun and shade leaves of Olea europaea respond differently to plant size, light availability and genetic variation: Canopy plasticity in Olea europea,” Functional Ecology, vol. 25, no. 4, pp. 802–812, 2011. [CrossRef]
- S. Rasmann, E. Chassin, J. Bilat, G. Glauser, and P. Reymond, “Trade-off between constitutive and inducible resistance against herbivores is only partially explained by gene expression and glucosinolate production,” Journal of Experimental Botany, vol. 66, no. 9, pp. 2527–2534, 2015. [CrossRef]
- J. C. Berry et al., “Increased signal-to-noise ratios within experimental field trials by regressing spatially distributed soil properties as principal components,” eLife, vol. 11, p. e70056, Jul. 2022. [CrossRef]
- E. Fitz–Rodríguez and C., Y. Choi, “Monitoring Turfgrass Quality Using Multispectral Radiometry,” Transactions of the ASAE, vol. 45, no. 3, 2002. [CrossRef]
- H.M. Aynalem, T. L. Righetti, and B. M. Reed, “Non-destructive evaluation of in vitro-stored plants: A comparison of visual and image analysis,” In Vitro Cell. Dev. Biol. - Plant, vol. 42, no. 6, pp. 562–567, 2006. [CrossRef]
- A.S. Kaler et al., “Genome-Wide Association Mapping of Dark Green Color Index using a Diverse Panel of Soybean Accessions,” Sci Rep, vol. 10, no. 1, p. 5166, Mar. 2020. [CrossRef]
- M.S. Brown, “Color Processing for Digital Cameras,” in Fundamentals and Applications of Colour Engineering, 1st ed., P. Green, Ed., Wiley, 2023, pp. 81–98. [CrossRef]
- J. Casadesús et al., “Using vegetation indices derived from conventional digital cameras as selection criteria for wheat breeding in water-limited environments,” Annals of Applied Biology, vol. 150, no. 2, pp. 227–236, 2007. [CrossRef]
- K. W. Houser, M. Wei, A. David, M. R. Krames, and X. S. Shen, “Review of measures for light-source color rendition and considerations for a two-measure system for characterizing color rendition,” Opt. Express, vol. 21, no. 8, p. 10393, Apr. 2013. [CrossRef]
- W. L. Wu et al., “High Color Rendering Index of Rb 2 GeF 6 :Mn 4+ for Light-Emitting Diodes,” Chem. Mater., vol. 29, no. 3, pp. 935–939, Feb. 2017. [CrossRef]
- J. Schewe, The Digital Negative: Raw Image Processing in Lightroom, Camera Raw, and Photoshop. Peachpit Pr, 2012.
- S. H. Lee and J. S. Choi, “Design and implementation of color correction system for images captured by digital camera,” IEEE Trans. Consumer Electron., vol. 54, no. 2, pp. 268–276, 2008. [CrossRef]
- G. D. Finlayson, M. Mackiewicz, and A. Hurlbert, “Color Correction Using Root-Polynomial Regression,” IEEE Trans. on Image Process., vol. 24, no. 5, pp. 1460–1470, 2015. [CrossRef]
- V. Senthilkumaran, “Color Correction Using Color Checkers,” presented at the Proceedings of the First International Conference on Combinatorial and Optimization, ICCAP 2021, December 7-8 2021, Chennai, India, Dec. 2021. Accessed: Jun. 14, 2024. [Online]. Available: https://eudl.eu/doi/10.4108/eai.7-12-2021.2314537.
- D. Okkalides, “Assessment of commercial compression algorithms, of the lossy DCT and lossless types, applied to diagnostic digital image files,” Computerized Medical Imaging and Graphics, vol. 22, no. 1, pp. 25–30, 1998. [CrossRef]
- V. Lebourgeois, A. Bégué, S. Labbé, B. Mallavan, L. Prévot, and B. Roux, “Can Commercial Digital Cameras Be Used as Multispectral Sensors? A Crop Monitoring Test,” Sensors, vol. 8, no. 11, pp. 7300–7322, Nov. 2008. [CrossRef]
- A. Zabala and, X. Pons, “Effects of lossy compression on remote sensing image classification of forest areas,” International Journal of Applied Earth Observation and Geoinformation, vol. 13, no. 1, pp. 43–51, 2011. [CrossRef]
- A. Zabala and, X. Pons, “Impact of lossy compression on mapping crop areas from remote sensing,” International Journal of Remote Sensing, vol. 34, no. 8, pp. 2796–2813, Apr. 2013. [CrossRef]
- J. Casadesús, C. Biel, and R. Savé, “Turf color measurement with conventional digital cameras,” in Vila Real, Universidade de Trás-os-Montes e Alto Douro, 2005.
- Ku, S. Mansoor, G. D. Han, Y. S. Chung, and T. T. Tuan, “Identification of new cold tolerant Zoysia grass species using high-resolution RGB and multi-spectral imaging,” Sci Rep, vol. 13, no. 1, p. 13209, Aug. 2023. [CrossRef]
- S. S. Mutlu, N. K. Sönmez, M. Çoşlu, H. R. Türkkan, and D. Zorlu, “UAV-based imaging for selection of turfgrass drought resistant cultivars in breeding trials,” Euphytica, vol. 219, no. 8, p. 83, 2023. [CrossRef]
- R. Matsuoka, K. Asonuma, G. Takahashi, T. Danjo, and K. Hirana, “Evaluation of Correction Methods of Chromatic Aberration in Digital Camera Images,” ISPRS Ann. Photogramm. Remote Sens. Spatial Inf. Sci., vol. I–3, pp. 49–55, Jul. 2012. [CrossRef]
- M. Mackiewicz, C. F. Andersen, and G. Finlayson, “Method for hue plane preserving color correction,” J. Opt. Soc. Am. A, vol. 33, no. 11, p. 2166, Nov. 2016. [CrossRef]
- J. C. Berry, N. Fahlgren, A. A. Pokorny, R. S. Bart, and K. M. Veley, “An automated, high-throughput method for standardizing image color profiles to improve image-based plant phenotyping,” PeerJ, vol. 6, p. e5727, Oct. 2018. [CrossRef]
- O. Burggraaff et al., “Standardized spectral and radiometric calibration of consumer cameras,” Opt. Express, vol. 27, no. 14, p. 19075, Jul. 2019. [CrossRef]
- L. Tu, Q. Peng, C. Li, and A. Zhang, “2D In Situ Method for Measuring Plant Leaf Area with Camera Correction and Background Color Calibration,” Scientific Programming, vol. 2021, pp. 1–11, Mar. 2021. [CrossRef]
- D. Lozano-Claros, E. Custovic, G. Deng, J. Whelan, and M. G. Lewsey, “ColorBayes: Improved color correction of high-throughput plant phenotyping images to account for local illumination differences.” Mar. 02, 2022. [CrossRef]
- J. Chopin, P. Kumar, and S. J. Miklavcic, “Land-based crop phenotyping by image analysis: consistent canopy characterization from inconsistent field illumination,” Plant Methods, vol. 14, no. 1, p. 39, May 2018. [CrossRef]
- Y. Liang, D. Urano, K.-L. Liao, T. L. Hedrick, Y. Gao, and A. M. Jones, “A nondestructive method to estimate the chlorophyll content of Arabidopsis seedlings,” Plant Methods, vol. 13, no. 1, p. 26, 2017. [CrossRef]
- J. C. O. Koh, M. Hayden, H. Daetwyler, and S. Kant, “Estimation of crop plant density at early mixed growth stages using UAV imagery,” Plant Methods, vol. 15, no. 1, p. 64, 2019. [CrossRef]
- W. H. Ryan et al., “The Use of Photographic Color Information for High-Throughput Phenotyping of Pigment Composition in Agarophyton vermiculophyllum (Ohmi) Gurgel, J.N.Norris & Fredericq,” Cryptogamie, Algologie, vol. 40, no. 7, p. 73, Sep. 2019. [CrossRef]
- I. Borra-Serrano, A. Kemeltaeva, K. Van Laere, P. Lootens, and L. Leus, “A view from above: can drones be used for image-based phenotyping in garden rose breeding?,” Acta Hortic., no. 1368, pp. 271–280, 2023. [CrossRef]
- W. Yin, X. Zang, L. Wu, X. Zhang, and J. Zhao, “A Distortion Correction Method Based on Actual Camera Imaging Principles,” Sensors, vol. 24, no. 8, p. 2406, Apr. 2024. [CrossRef]
- J. F. Paril and A. J. Fournier-Level, “instaGraminoid, a Novel Colorimetric Method to Assess Herbicide Resistance, Identifies Patterns of Cross-Resistance in Annual Ryegrass,” Plant Phenomics, vol. 2019, p. 2019/7937156, 2019. [CrossRef]
- P. L. Vines, J. Zhang, P. L. Vines, and J. Zhang, “High-throughput plant phenotyping for improved turfgrass breeding applications,” G, vol. 2, no. 1, pp. 1–13, Jan. 2022. [CrossRef]
- T. Wang, A. Chandra, J. Jung, and A. Chang, “UAV remote sensing based estimation of green cover during turfgrass establishment,” Computers and Electronics in Agriculture, vol. 194, p. 106721, 2022. [CrossRef]
- A. Atefi, Y. Ge, S. Pitla, and J. Schnable, “In vivo human-like robotic phenotyping of leaf traits in maize and sorghum in greenhouse,” Computers and Electronics in Agriculture, vol. 163, p. 104854, 2019. [CrossRef]
- A. Atefi, Y. Ge, S. Pitla, and J. Schnable, “Robotic Technologies for High-Throughput Plant Phenotyping: Contemporary Reviews and Future Perspectives,” Front. Plant Sci., vol. 12, p. 611940, Jun. 2021. [CrossRef]
- A. Arunachalam and, H. Andreasson, “Real-time plant phenomics under robotic farming setup: A vision-based platform for complex plant phenotyping tasks,” Computers & Electrical Engineering, vol. 92, p. 107098, 2021. [CrossRef]
- H. Fonteijn et al., “Automatic Phenotyping of Tomatoes in Production Greenhouses Using Robotics and Computer Vision: From Theory to Practice,” Agronomy, vol. 11, no. 8, p. 1599, Aug. 2021. [CrossRef]
- L. Yao, R. Van De Zedde, and G. Kowalchuk, “Recent developments and potential of robotics in plant eco-phenotyping,” Emerging Topics in Life Sciences, vol. 5, no. 2, pp. 289–300, May 2021. [CrossRef]
- S. Pongpiyapaiboon et al., “Development of a digital phenotyping system using 3D model reconstruction for zoysiagrass,” The Plant Phenome Journal, vol. 6, no. 1, p. e20076, 2023. [CrossRef]
- H. Bethge, T. Winkelmann, P. Lüdeke, and T. Rath, “Low-cost and automated phenotyping system ‘Phenomenon’ for multi-sensor in situ monitoring in plant in vitro culture,” Plant Methods, vol. 19, no. 1, p. 42, May 2023. [CrossRef]
- K. Ma, X. Cui, G. Huang, and D. Yuan, “Effect of Light Intensity on Close-Range Photographic Imaging Quality and Measurement Precision,” IJMUE, vol. 11, no. 2, pp. 69–78, Feb. 2016. [CrossRef]
- J. Bendig, D. Gautam, Z. Malenovsky, and A. Lucieer, “Influence of Cosine Corrector and Uas Platform Dynamics on Airborne Spectral Irradiance Measurements,” in IGARSS 2018 - 2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia: IEEE, 2018, pp. 8822–8825. [CrossRef]
- Z. Feng, Q. Liang, Z. Zhang, and W. Ji, “Camera Calibration Method Based on Sine Cosine Algorithm,” in 2021 IEEE International Conference on Progress in Informatics and Computing (PIC), Shanghai, China: IEEE, Dec. 2021, pp. 174–178. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





