1. Introduction
Breast cancer continues to be a highly common and lethal type of cancer that affects women globally [
1,
2]. A mass of aberrant cells that develops within the breast tissue is called a breast tumor, which can be benign or malignant. Because they can spread (metastasize) to other parts of the body and cause major health issues as well as, frequently, death, malignant tumors are especially dangerous [
3]. Effective treatment and higher survival rates depend on early discovery and precise diagnosis. The heterogeneity in tumor size, form, and appearance on medical imaging, however, presents substantial barriers to the process of identifying and recognizing breast cancers [
4,
5].
Many imaging modalities, such as mammography, ultrasound, and magnetic resonance imaging (MRI), can identify and detect breast cancers. The most popular screening method is mammography, which produces finely detailed images of the breast using low-dose X-rays. Using high-frequency sound waves to create images, ultrasound is especially helpful in differentiating between cysts filled with fluid and solid tumors. Conversely, magnetic resonance imaging (MRI) yields high-contrast images and is frequently employed for high-risk patients or to delve deeper into unclear results from other imaging modalities. Although each technique has advantages and disadvantages, correctly interpreting the images they generate is essential to a successful diagnosis [
1,
6].
One of the most important steps in the diagnostic workflow is the segmentation of breast tumors, which is the act of defining the boundaries of the tumor inside an image. This has always been done manually by radiologists, which takes a lot of time and is prone to differences in observation quality [
7,
8]. The process of manual segmentation necessitates a high degree of expertise and can be error-prone because it includes delineating the tumor on each image slice. Computational algorithms-driven automated segmentation techniques seek to address these issues by delivering repeatable and consistent outcomes [
9,
10].
Automated breast tumor segmentation, however, provides numerous benefits. It offers reliable and replicable outcomes, lessens the burden on healthcare practitioners, and enables quick analysis of extensive information. In addition, automated techniques can identify intricate patterns and characteristics that may go unnoticed by human observation, hence potentially resulting in timelier and more precise diagnoses [
11,
12]. Conventional image processing methods frequently encounter difficulties when dealing with the diversity and intricacy of medical images. Deep learning (DL) approaches have become effective tools for analyzing medical images in order to tackle these difficulties [
7,
13].
Convolutional neural networks (CNNs), in particular, are DL models that have shown a remarkable ability to learn intricate patterns and features from vast datasets [
4,
14,
15]. DL models have the ability to automatically extract hierarchical features from raw images, in contrast to typical machine learning techniques that necessitate manual feature extraction. Because of this flexibility, they are especially well-suited for the complex task of segmenting breast tumors, where there can be a significant degree of diversity in tumor appearance [
13,
16,
17,
18,
19].
The consistency and scalability of the outcomes are two main ways that human and automatic segmentation differ from one another. Because of the radiologist’s experience, manual segmentation may be more accurate in specific circumstances, but it is impractical for handling massive amounts of data. On the other hand, automated segmentation with DL methods can process large volumes of imaging data quickly and reliably, lessening the effort required of medical personnel and lowering the possibility of human error. Furthermore, when more annotated data becomes available, DL models can adjust to new imaging modalities and changing clinical practices, enhancing their performance over time [
15,
20,
21,
22].
There are numerous advantages to using DL for breast tumor segmentation. These models help with early and accurate tumor border delineation, which is crucial for effective treatment outcomes. They also improve the precision of tumor boundary delineation. Moreover, multi-modal imaging data can be included in DL models, offering a thorough analysis that incorporates the advantages of several imaging modalities [
21,
23]. Personalized treatment regimens and better clinical decision-making may result from this all-encompassing strategy. Deep learning’s potential to revolutionize breast cancer diagnosis and treatment could be realized as the field develops, eventually leading to better patient outcomes and survival rates [
23,
24].
Chen et al. [
25] have recently tackled the difficulties presented by the intricate ultrasound patterns and the varying shapes and sizes of tumors in the segmentation of breast tumors. An advanced selective kernel convolution was developed for the segmentation of breast tumors. This technique incorporates several representations of feature map sections and dynamically adjusts the weights of these areas based on both spatial and channel dimensions. The implementation of this region recalibration approach permits the network to concentrate more efficiently on region features that have a significant impact while minimizing the influence of less valuable regions. To increase the accuracy of segmentation, they integrate the enhanced selective kernel convolution into a U-net architecture with deep supervision restrictions. This allows for the adaptive capture of strong representations of breast cancers. By tackling the complexities of breast tumor segmentation from ultrasound images, this method shows a considerable improvement in terms of robustness and accuracy.
Tagnamas et al. [
4] introduced an innovative approach for segmenting and classifying breast tumors. They utilized a two-encoder architecture that integrates EfficientNetV2 with a modified Vision Transformer (ViT) encoder. The EfficientNetV2 backbone is employed to maintain local information in breast ultrasound (BUS) images, while the Transformer encoder utilizes a self-attention (SA) method to capture a broad spectrum of high-level and intricate features. To efficiently combine the features obtained from both encoders, the authors devised a Channel Attention Fusion (CAF) module. This module selectively enhances significant features, hence enhancing the integration of local and high-level information. These feature maps that have been combined are subsequently reconstructed employing a decoder to create segmentation maps. In addition, a straightforward and efficient classifier based on the MLP-Mixer is used to categorize the segmented tumor regions into benign and malignant categories. This classifier, utilized for the initial time in lesion classification in BUS images, showcases a groundbreaking and efficient method for breast tumor segmentation and classification.
Zhu et al. [
12] propose a new method for segmenting breast tumors by utilizing a Swin-Net architecture that combines CNNs and Transformer. This integration improves the accuracy of breast ultrasound image segmentation. The technique employs the swing-transformer model due to its robust global modeling capability and accurate feature extraction capabilities. The Swin-Net framework has two novel modules: the feature refinement and improvement module (RLM) and the hierarchical multi-scale feature fusion module (HFM). The RLM module improves and strengthens the characteristics acquired by the transformer encoder, while the HFM module handles multi-scale low-level details and high-level semantic characteristics to provide efficient integration of features across layers, reduction of noise, and enhanced segmentation performance.
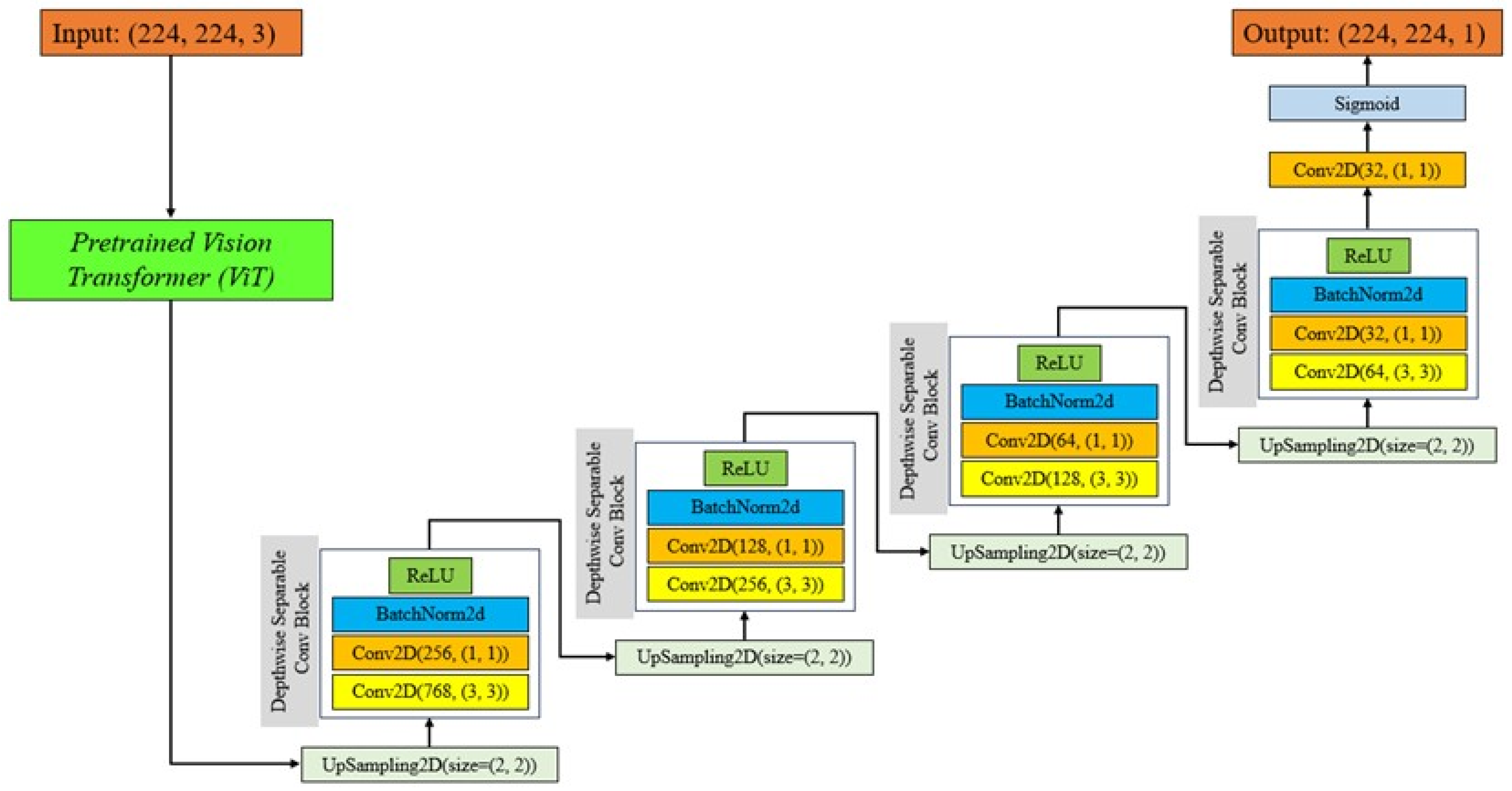
To optimize the effectiveness and precision of tumor segmentation and overcome the problems of varying shapes and sizes of tumors, this work presents the EfficientUNetViT model, which is a hybrid architecture that combines the advantages of U-Net, Depthwise Separable Convolutions, and Transformer (ViT).
The paper’s structure is arranged as follows: We go over each of our approach’s components in
Section 2. The U-Net architecture with Depthwise Separable Convolutions and Vision Transformer (ViT) is described in
Section 2.1. The datasets utilized for training and evaluation, together with preprocessing techniques, are described in
Section 2.2. The augmentation approaches used to improve dataset variety are covered in
Section 2.3. The training setup, settings, and optimization algorithms are described in depth in Section 2.4. In
Section 3, the metrics used to evaluate the model’s performance are defined in
Section 3.1. Experimental results, comparisons with other approaches, and a discussion of the results are presented in
Section 3.2. The main contributions, clinical practice implications, and future research prospects are outlined in
Section 4.
2. Model Structure
In this work, we used a pretrained Vision Transformer (ViT) model with a U-Net architecture to perform breast tumor segmentation. The U-Net architecture is a widely recognized neural network specifically developed for the purpose of segmenting biological images. The system comprises a contracting pathway for context acquisition and a symmetric expanding pathway for reliable localization. The contracting route follows the conventional structure of a convolutional network, wherein each stage comprises two 3x3 convolutions, succeeded by a rectified linear unit (ReLU) and a 2x2 max pooling operation. Conversely, the expanding approach involves increasing the resolution of the feature map by upsampling and then using a 2x2 convolution (up-convolution) to reduce the number of feature channels by half. This is followed by concatenating the resulting feature map with the equivalent cropped feature map from the contracting path.
Our U-Net utilizes Depthwise Separable Convolutional Blocks, which are a type of convolution developed to decrease the computational cost and parameters of the model while maintaining performance. Depthwise Separable Convolutions divide the convolution operation into two distinct layers: depthwise convolutions, which utilize one filter for each input channel, and pointwise convolutions, which use a 1x1 convolution to merge the outputs of the depthwise layer [
26,
27]. This method greatly decreases the quantity of parameters and computing burden, resulting in a more efficient and expedited training process for the network, while yet retaining a high level of accuracy [
28,
29].
The U-Net’s Depthwise Separable Convolutional Blocks offer numerous benefits. Firstly, they aid in mitigating overfitting by reducing the number of parameters, which is particularly important when working with tiny datasets that are frequently seen in medical imaging. Additionally, the decreased computational complexity enables the utilization of more complicated networks or larger batches, thereby enhancing the model’s performance even further. Finally, these blocks maintain the spatial hierarchies and characteristics acquired at various layers, guaranteeing that the crucial details for segmentation tasks are conserved across the network [
26,
30].
The main objective of adding a ViT to the U-Net architecture is to make use of the transformers’ strong feature extraction capabilities, which have lately demonstrated exceptional performance on a range of computer vision applications [
31,
32]. Vision Transformers provide a major improvement over conventional CNNs by using self-attention processes to capture global context and dependencies inside an image. ViTs are a strong substitute for CNNs in image analysis due to their capacity to represent long-range interactions and contextual linkages [
33,
34].
By integrating ViT into the U-Net architecture, the model may take advantage of the advantages of both worlds: the global context knowledge offered by ViT and the accurate localization and segmentation capabilities of U-Net. This combination improves the model’s ability to capture both local and global information, which improves the model’s ability to appropriately segment breast cancers [
32,
35].
The ViT functions by partitioning an image into a series of patches, seeing each patch as a token similar to words in natural language processing. The series of patches is subsequently fed into a transformer model, which utilizes self-attention to record the interconnections among the patches. By employing this method, ViT is able to effectively capture extensive connections and contextual details throughout the entire image, surpassing the capabilities of conventional CNNs that typically depend on limited receptive fields. ViT has demonstrated exceptional performance in image classification challenges by capturing global context. This success has inspired us to apply ViT to segmentation tasks in our study [
31,
35,
36].
Our U-Net architecture utilized a pre-trained ViT model as the encoder. The choice to using a pretrained model was motivated by numerous benefits. Pretrained models have acquired extensive feature representations from vast datasets, allowing them to exhibit superior generalization capabilities when faced with novel problems and limited data. This transfer learning strategy greatly diminishes the requirement for abundant labeled data, which is especially advantageous in the field of medical imaging where annotated datasets are frequently limited. In addition, pretrained models accelerate the training process by leveraging existing information instead of starting from the beginning.
Integrating a pretrained ViT into our U-Net model has several advantages. The ViT encoder captures salient information from the input images, yielding a resilient representation for the succeeding decoder stages. These characteristics collect both specific and overall information, improving the model’s capacity to accurately define tumor boundaries. The ViT encoder utilizes the self-attention mechanism to identify important areas in the image that are crucial for accurate segmentation. This enhances the overall performance of the model [
35,
36].
Figure 1 indicates Structure of the proposed EfficientUNetViT model.
2.1. Dataset
This study utilizes the Breast Ultrasound Image (BUSI) dataset to assess the effectiveness of our breast tumor segmentation model [
37]. The BUSI dataset, gathered in 2018, consists of 780 ultrasound images obtained from 600 female patients between the ages of 25 and 75. The photos are classified into three categories: malignant, benign, and normal. The images initially exist in PNG format and exhibit a diverse range of dimensions, contours, and tumor characteristics, which presents considerable difficulty for automatic segmentation approaches. An image typically has dimensions of 500 pixels by 500 pixels. To ensure consistency in the input for the model, all images in the dataset have been scaled to a uniform dimension of 224x224 pixels, with three color channels (224, 224, 3). The act of resizing guarantees uniformity and achieves a harmonious balance between computational effectiveness and the retention of adequate data for precise segmentation. In addition, the pixel values are rescaled to a range of 0 to 1 to expedite the convergence process during model training.
The BUSI dataset contains meticulous annotations for each image, accurately outlining the precise boundaries of the tumors. The annotations are presented as binary masks, with tumor locations indicated by a pixel value of 1 and background pixels represented by a value of 0. These masks function as the ultimate point of reference for both training and assessing the segmentation model. To guarantee an equitable evaluation of the model’s effectiveness, the dataset is partitioned into three distinct subsets: training, validation , and test sets. The training set consists of 80% of the images, the validation set contains 10% of the images, and the test set includes the remaining 10% of the images. This division guarantees that the model is trained on a varied assortment of images and evaluated on unfamiliar data to assess its ability to generalize.
2.2. Data Augmentation
This study utilizes a comprehensive data augmentation technique to enhance the resilience and generalization capabilities of our EfficientUNetViT model. Data augmentation is an essential method in deep learning, especially when working with restricted datasets [
38,
39]. It involves artificially increasing the training data by applying several modifications. We employed several augmentation approaches, such as random rotations, zooms, vertical and horizontal flips, shifts, and elastic deformations, for the purpose of segmenting breast tumors. These adjustments replicate the diversity observed in actual clinical situations, aiding the model in becoming immune to alterations in tumor location, dimensions, and alignment [
40,
41].
2.3. Training Procedure
The training approach of our EfficientUNetViT model is specifically intended to achieve optimal performance and enhance generalization capabilities. At first, the dataset is partitioned into training, validation, and test sets. Data augmentation is applied to the training and validation sets to artificially increase the dimension of the dataset and create variability. This technique is used to prevent overfitting.
There is an early termination criterion if the validation loss does not show progress after 10 iterations in the 50 iterations of the training method. When the model has adequately learned the underlying patterns in the data, the training process is stopped, preventing overfitting to the irrelevant details. The model keeps its capacity to generalize on unknown data by doing this. To balance the requirement for adequate learning progress without significantly affecting the model’s performance, the learning rate is set at 1e-4.
The training procedure uses the Adam optimizer, which is renowned for its effectiveness and flexible learning rate capabilities. Adam excels at handling noisy situations and sparse gradients because it incorporates the best features of AdaGrad and RMSProp, two additional extensions of stochastic gradient descent. Its adjustable learning rate facilitates more rapid and stable convergence by dynamically modifying the learning rate during training. In order to minimize the risk of overfitting and maintain the model’s performance and reliability, the combination of an early stopping, robust optimizer, and a controlled learning rate helps the model learn from the data effectively [
42,
43].
Throughout each epoch, the model undergoes training using batches of augmented training data. The model’s parameters are changed for each batch via backpropagation to minimize the binary cross-entropy loss between the true and predicted segmentation masks. Following every epoch, the model’s performance is evaluated on the validation set. The validation loss and metrics are utilized to ascertain whether early stopping should be initiated and to ascertain the optimal model based on validation performance. The optimal model parameters are stored to guarantee the utilization of the most efficient model for testing.
Ultimately, the optimal model is loaded and its performance is assessed on the test set to determine its ability to generalize. This systematic training method guarantees that the model not only achieves high performance on the training data but also exhibits effective generalization to novel, unseen data.
3. Experiments
3.1. Evaluation Metrics
The study assessed our breast tumor segmentation models’ performance employing a range of established measures to ensure a thorough evaluation of their efficacy and accuracy. Intersection over union (IoU), recall, precision, and dice similarity coefficient (Dice) were the main metrics that were employed. Precision gauges how well positive predictions come true, including important information about how well the model reduces false positives. To prevent the model from mistakenly classifying non-tumorous areas as tumors, this metric is crucial. Recall, on the other hand, indicates how well the model detects accurate tumor locations by measuring its accuracy in identifying all pertinent occurrences [
1,
25,
44,
45].
The overlap between the anticipated segmentation and the ground truth is measured by the Intersection over Union (IoU) metric. IoU is a trustworthy indicator of spatial prediction accuracy since it shows how well the model’s segmentations match the real tumor regions. Similarly, when assessing the overlap between anticipated and true segmentations, the Dice coefficient highlights the harmonic average of accuracy and recall, providing a fair assessment of both measures. Because it takes into account both the accuracy and completeness of the segmentations, the Dice coefficient is very useful for evaluating the quality of segmentation outputs. By using these criteria, we make sure that the performance of our model in breast tumor segmentation is thoroughly and nuancedly evaluated, offering important insights into its therapeutic value [
8,
23,
46,
47].
3.2. Results and Discussion
This section focuses on the presentation and analysis of the performance of the EfficientUNetViT model in segmenting breast tumors. The effectiveness of the model is assessed by measuring many important metrics, such as Intersection over Union (IoU), precision, F1 score, recall, and dice coefficient, on both the validation and test datasets. In addition, we evaluate the outcomes by comparing them to baseline models to emphasize any enhancements. In addition, we analyze the influence of data augmentation and the incorporation of the ViT into the U-Net architecture on the accuracy of segmentation. The results indicate that our method greatly improves the accuracy of segmenting breast tumors, providing a reliable and efficient solution for automated segmentation.
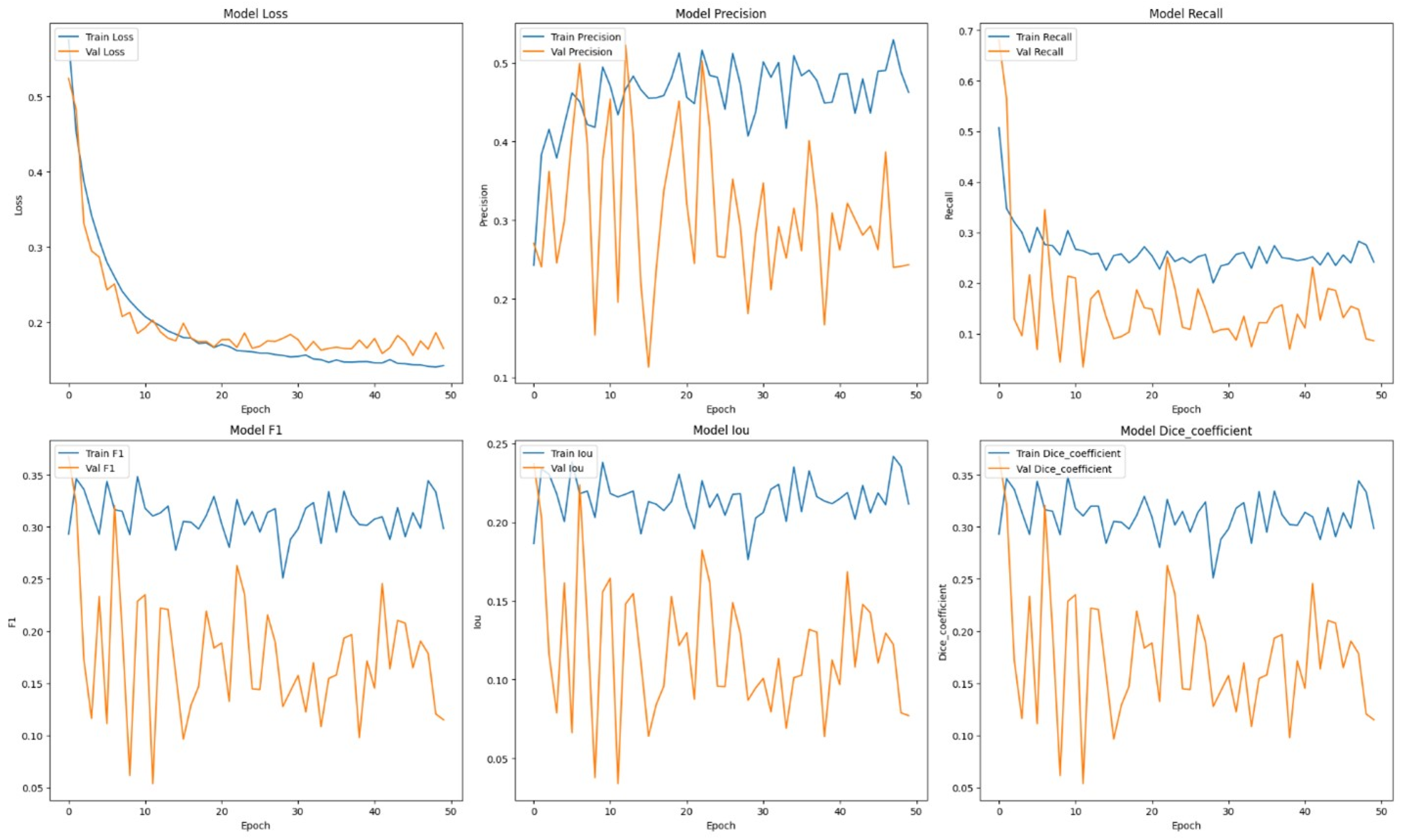
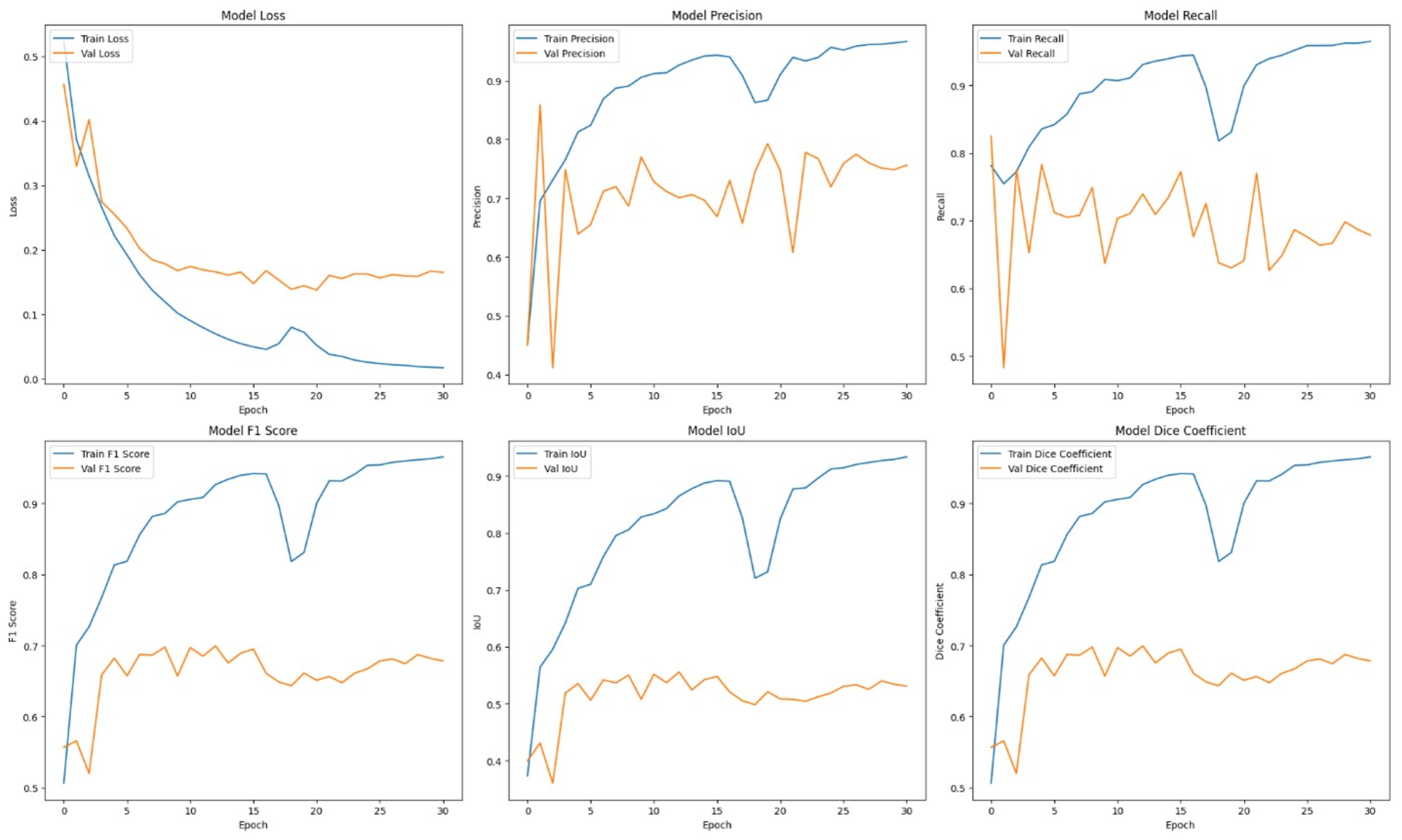
Table 1 presents comprehensive comparisons of the training time effectiveness between ViT + UNet model and ViT + Unet + Augmentation model. The Training dynamics over epochs of models are indicated in
Figure 2 and
Figure 3.
Initially, the ViT + U-Net model was trained and evaluated without the use of any data augmentation. In spite of this, the model’s performance measures were encouraging: 0.7682 for recall, 0.6196 for IoU, 0.7505 for F1, 0.8030 for precision, and 0.7505 for dice. These findings suggest that the model exhibits good recall and precision rates and can successfully differentiate between various forms of breast cancer. To ensure that as many cases as possible are recognized in medical diagnosis, a high recall model must be able to identify a significant fraction of true positive cases. Conversely, high precision indicates a low false positive rate, which means the model is quite precise in its predictions.
In the context of medical diagnosis, where it’s critical to strike a balance between identifying true positives and avoiding false positives and false negatives, the high recall and precision rates are very significant. Based on its performance measures, the model appears to be a viable clinical tool that could help detect breast cancer early and accurately. The viability of the ViT + U-Net model is further demonstrated by this early achievement without data augmentation, which raises the possibility of much bigger gains with the right augmentation techniques.
In order to enhance the model’s resilience, data augmentation techniques were implemented, and the ViT + UNet model underwent retraining. Despite what was expected, the performance measurements exhibited a substantial decrease: precision decreased to 0.5702, recall to 0.2471, F1 score to 0.3158, IoU to 0.2263, and Dice coefficient to 0.3158. This implies that the techniques used to enhance the data may have caused changes that the model was unable to successfully learn from, potentially because of the addition of random or unrealistic alterations that differed significantly from the true distribution of the data.
An in-depth examination of the training measures during the epochs offers valuable understanding into the model’s acquisition process. During the first epoch, the model’s training loss was 0.5231, while the validation loss was 0.4568. The precision and recall values during training were 0.4521 and 0.7811, respectively. during validation, the equivalent values were 0.4497 for precision and 0.8248 for recall. The measures demonstrated constant improvement throughout subsequent epochs. At the end of the training process, the model had a train loss of 0.1421 and a validation loss of 0.1650. Additionally, the training precision was 0.4626 and the recall was 0.2416. The consistent enhancement illustrates the model’s aptitude to acquire knowledge and apply it broadly from the provided training data.
The learning curves demonstrated that the model without augmentation consistently and significantly improved in performance, whereas the model with augmentation exhibited notable instability. The instability observed suggests that the augmentation strategies used may not have been well-suited for the dataset, perhaps resulting in a decrease in the model’s capacity to generalize from the enhanced data.
It was evident from the learning curves that the performance trajectories of the augmented and unaugmented models differed significantly. The augmented model showed notable instability, whereas the unaugmented model showed a consistent and significant gain in performance with time. This instability raises the possibility that the augmentation methods used weren’t ideal for the particular features of the dataset. As a result, the enhanced model encountered difficulties while attempting to make generalizations from the enhanced data, which could have limited its capacity to acquire strong characteristics that are essential for accurate segmentation. The steady growth of the unaugmented model emphasizes how important it is to use augmentation procedures that are appropriate and customized to the particular properties of the dataset.
Subsequent investigation suggests that the noise and distortions caused by the improper augmentation techniques may have confused the model and resulted in irregular learning patterns. This draws attention to a crucial factor in the training of models: augmentation techniques must improve the dataset without sacrificing its underlying organization and informational value. The instability of the enhanced model may be related to over-augmentation or the introduction of fictitious variations that don’t accurately represent the variety of breast tumor images.
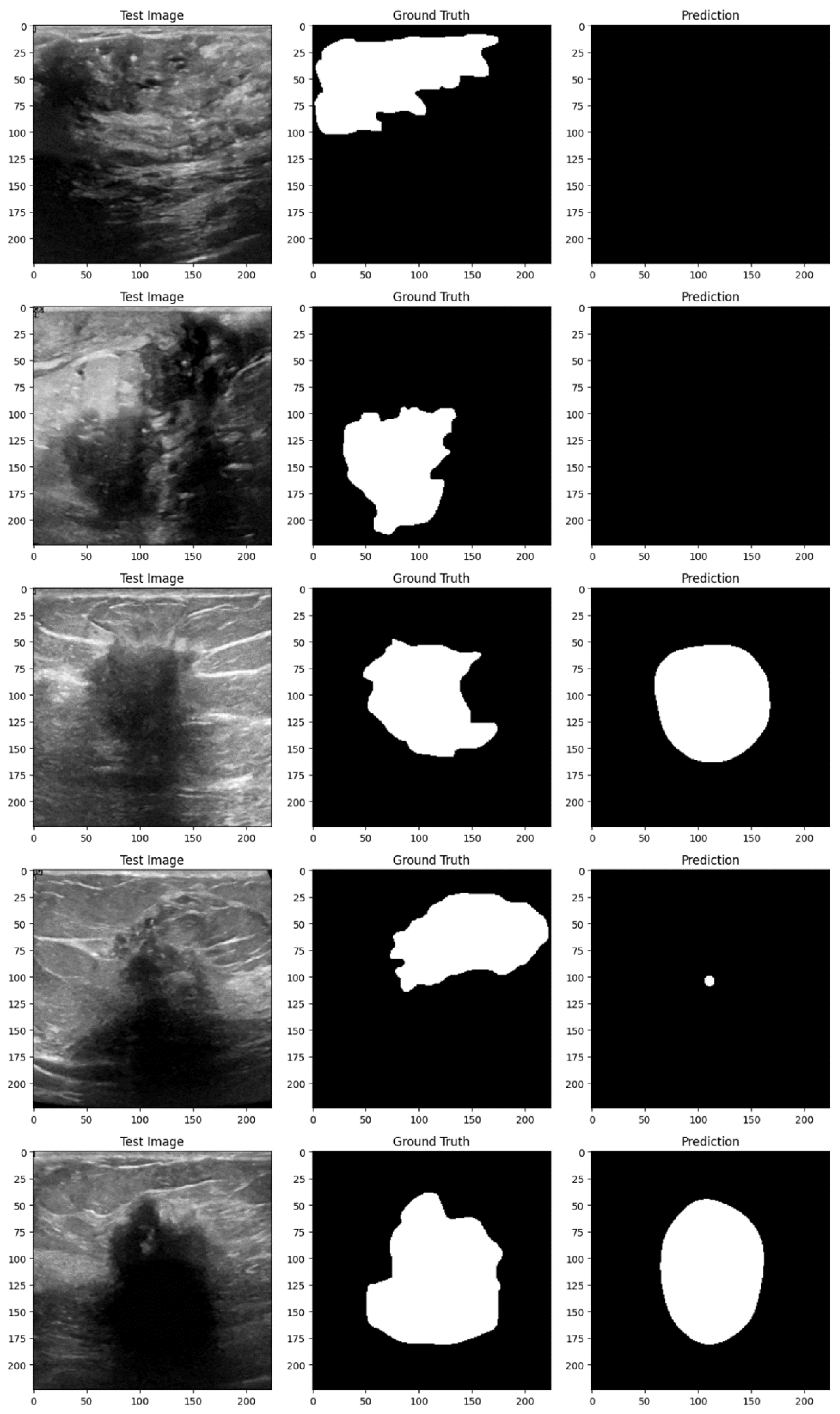
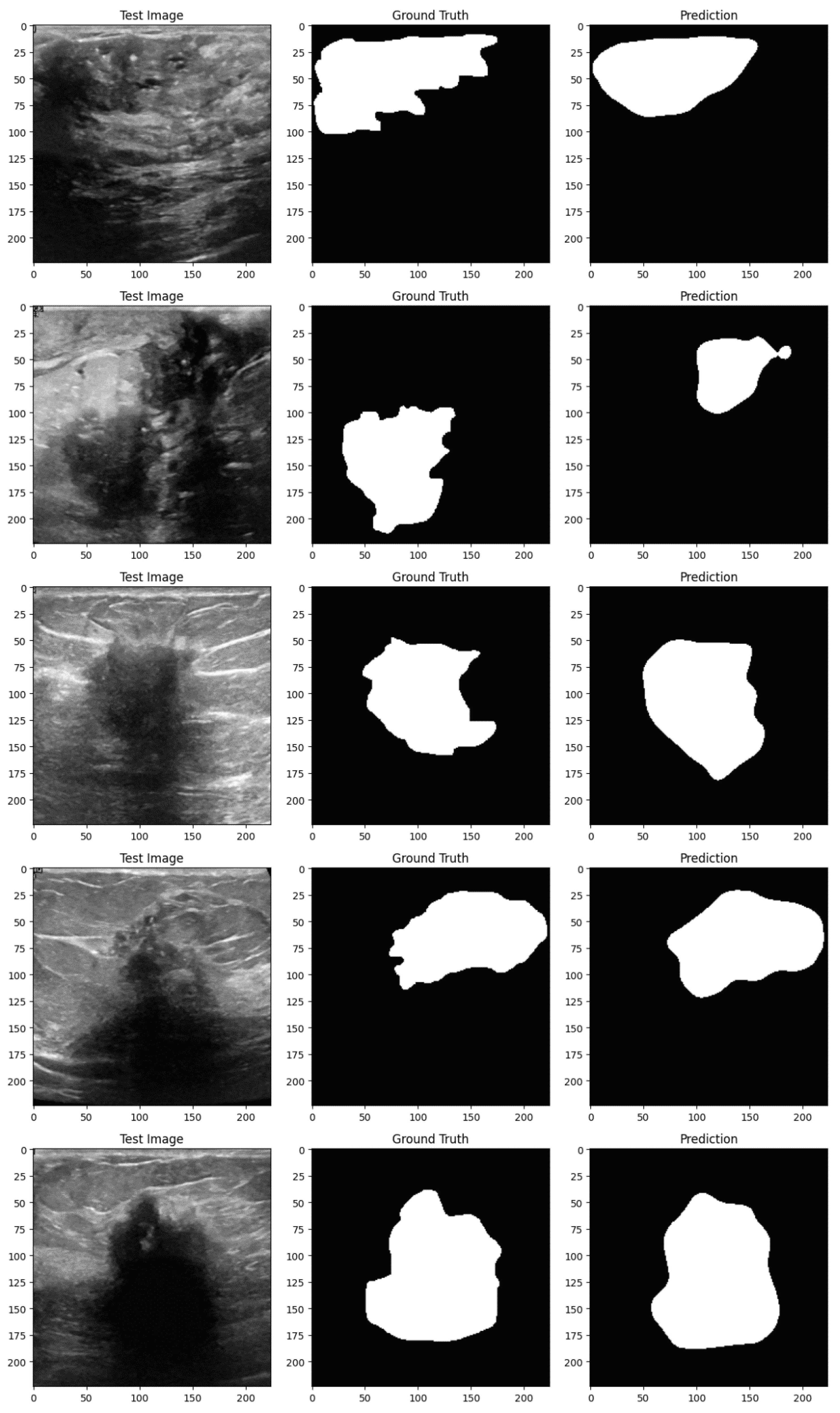
Figure 4 and
Figure 5 demonstrate the implementation of the proposed EfficientUNetViT model on test samples from the breast tumor segmentation dataset.
Conclusively, the EfficientUNetViT model exhibited robust performance in segmenting breast tumors, achieving high precision, recall, and overall accuracy without the need for augmentation. Nevertheless, the use of data augmentation in this scenario resulted in reduced performance, emphasizing the importance of meticulously devising augmentation strategies that are in line with the attributes of the medical images and the particular demands of the segmentation task. This study highlights the possibility of integrating Vision Transformers with UNet topologies for medical image segmentation. It also emphasizes the significance of employing suitable augmentation strategies to further improve the model’s resilience and performance.
4. Conclusion
This paper presents the development and evaluation of a sophisticated breast tumor segmentation model called EfficientUNetViT, which combines Vision Transformers (ViT) with a UNet architecture. The findings of our study clearly indicate that the suggested model is highly effective, as it achieves excellent levels of F1 score, precision, IoU, recall, and Dice coefficient when tested on a comprehensive dataset. The ViT + UNet model, when used without augmentation, demonstrated exceptional performance metrics, highlighting its potential for precise and dependable segmentation in medical imaging. The results confirm the efficacy of combining the characteristics of ViT for collecting overall context and UNet for precise localization, resulting in a strong solution for segmenting breast tumors.
Nevertheless, the utilization of data augmentation strategies in this investigation unveiled significant observations. Although augmentation is typically anticipated to improve the generalization of the model, our particular augmentation techniques resulted in a significant decrease in performance. This implies that the model had difficulty learning from the alterations generated by the augmentation, maybe because of the presence of noise or transformations that did not match well with the inherent characteristics of the breast tumor images. These findings emphasize the significance of meticulous design and selection of augmentation techniques in medical imaging tasks. Future research should prioritize the improvement of these techniques and the exploration of more advanced augmentation procedures to further enhance the model’s robustness and ability to generalize. In summary, this study highlights the potential of ViT-UNet hybrids in the field of medical image segmentation, while also identifying areas that require further investigation and enhancement.