1. Statement of Need
In recent years, sequencing technology has advanced such that DNA sequencing for ‘omics is faster, easier and cheaper than ever before. Subsequently these ‘omics experiments increasingly produce large (up to terabyte size) datasets and their analysis requires researchers to have access to both specialist tools and a robust high performance computing (HPC) infrastructure; metagenomics analyses are particularly resource intensive. This presents a steep learning curve for biologists who may not have any previous experience using HPC or command line tools.
There is therefore a clear demand for training in this area [
1,
2]. However, training provision is complicated by the heterogeneity of individuals’ computer setups and the many dependencies demanded by software packages. Furthermore, access to HPC clusters varies depending on institution and field of study.
Cloud computing services such as Amazon Web Services (AWS) offer a novel way to provide training in genomics and metagenomics analysis workflows without the need for participants to manage complex software installation or store large datasets in their computers. Each participant can be provided with an identical AWS instance (virtual machine) pre-configured with the software and data needed for a course. The Cloud-SPAN project has been using this approach, based on the Data Carpentry (DC) [
3] model [
4], to deliver highly successful genomics and metagenomics courses for almost three years. This model is known as
Infrastructure as a Service (IaaS) [
5], and is rather flexible in that
training providers can configure virtual machines as to compute, storage and networking capacities, and also as to the data and software analysis tools required by a course, while
cloud providers are responsible for managing actual compute, storage and networking hardware resources and virtualisation. IaaS has also been deployed for bioinformatics training on national HPC clusters [
6,
7] using OpenStack (Open Source Cloud Computing Infrastructure) [
8] for managing hardware resources and virtualisation.
Platform as a Service (PaaS) is another cloud computing service model that has been successfully used in bioinformatics training [
9,
10]. A PaaS comprises a software environment for data management and analysis tasks using a programming language such as Python and R. Examples include Google Colab [
11] and Posit [
12], a cloud-based RStudio. These environments are readily accessible through a web browser and simplify sharing code and data.
The main advantages of Cloud computing for training based on the Data Carpentry model are low-cost and flexibility. There is no need to manage nor invest in hardware resources nor physical space. Instead, an instance in the Cloud is first configured with all the data and software tools required by a course. This instance is then configured as a template, or Amazon Machine Image (AMI) in AWS terminology. Then, as many instances are created from the AMI as the number of participants in a course. Once the course is over, the instances are deleted in order to stop incurring costs. The AMI is typically preserved to serve as the starting point either (1) to create new instances for a new run of the course, or (2) to create a new AMI with updated data or software, or both, through creating an instance, updating the data or software, and creating a new AMI out of the newly configured instance. In addition, it is rather easy to change instances capacity in terms of number of processors, main memory size, and communication bandwidth to match the processing requirements of the analysis tasks to be taught.
However, managing multiple instances through a graphical user interface such as the AWS Console is cumbersome and error-prone. As the number of participants increases the problem is magnified. The nature of running workshops means participants may drop out, join the course late or not turn up, resulting in further manual management being required.
We therefore developed a set of Bash scripts to automate the management of AWS instances for use in training workshops. The scripts automate the creation and deletion of AWS instances and related resources, namely: encrypted login keys, Internet Protocol (IP) addresses, and domain names. We have also developed an accompanying online tutorial [
13] detailing how to open and configure an AWS account, and how to install, configure and run the scripts in a
terminal on Linux, Windows, and MacOS, or in the AWS CloudShell (browser-based)
terminal. The tutorial assumes that learners have no prior experience with the AWS concepts and tools covered in the tutorial. However, learners are expected to have some experience with both the Linux/Unix
terminal and Bash shell programming. Windows users need to install and configure the Git Bash
terminal and Mac users need to install or update the Bash shell as instructed in the
Precourse Instructions section of the tutorial [
14].
We use the scripts to manage Ubuntu Linux AWS instances configured for training in genomics and metagenomics. However, the scripts are broadly applicable to manage instances configured for any training purpose. The tutorial demonstrates how to customise AMI templates with other software tools and data [
15].
2. The Scripts and How to Use Them
The scripts are listed below. There are three types of scripts. The primary scripts, "csinstances_*.sh", are the topline scripts run by the person in charge of managing instances for workshops. The secondary scripts, "aws_*.sh", are invoked by the scripts "csinstances_create.sh" or "csinstances_delete.sh" to either create or delete instances and related resources: login keys, IP addresses, and domain names (if managed). The third script type corresponds to scripts that provide utility functions to the primary and secondary scripts. The only script in this category is "colours_utils_functions.sh", which provides text colouring functions and utility functions that validate the invocation and results of the primary and secondary scripts.
The secondary scripts can each be run directly in the same way the primary scripts are run (as described shortly), but this is not recommended except for the purpose of improving a script or troubleshooting a failed step in creating instances and related resources. The section
Troubleshooting [
16] of the tutorial describes the conditions under which we have had to run some secondary scripts directly.
aws_domainNames_create.sh aws_instances_terminate.sh csinstances_create.sh
aws_domainNames_delete.sh aws_loginKeyPair_create.sh csinstances_delete.sh
aws_instances_configure.sh aws_loginKeyPair_delete.sh csinstances_start.sh
aws_instances_launch.sh colour_utils_functions.sh csinstances_stop.sh
2.1. Managing Instances for Workshops
Before running the scripts, an AWS account and a terminal environment must be configured (as described in the tutorial [
17]). Then, prior to creating instances for a workshop, three files must be created and organised as follows:
instancesNamesFile.txt — contains the names of the instances to be created and managed. Only the name of this file can be changed if preferred. This file must contain only one instance name per line, and each instance name must start with an alphabetic character followed by alpha-numeric characters, hyphens (-) or underscores (_) only.
resourcesIDs.txt — contains a set of space-separated "key value" pairs that specify the AWS resources to use in creating each instance and related resources. This is the contents of the
resourcesIDs.txt file we use for the Cloud-SPAN Genomics course [
18]:
| KEYWORD |
VALUE examples (Cloud-SPAN’s for Genomics course using instance domain names) |
|
|
## NB: "key value" pairs can be specified in any order |
| imageId |
ami-07172f26233528178 |
## NOT optional: instance template (AMI) id |
| instanceType |
t3.small |
## NOT optional: processor count, memory size, bandwidth |
| securityGroupId |
sg-0771b67fde13b3899 |
## NOT optional: should allow ssh (port 22) communication |
| subnetId |
subnet-00ff8cd3b7407dc83 |
## optional: search vpc in AWS console then click subnets |
| hostZone |
cloud-span.aws.york.ac.uk |
## optional: specify to use instance domain names |
| hostZoneId |
Z012538133YPRCJ0WP3UZ |
## optional: specify to use instance domain names |
As shown in this example, a resourcesIDs.txt file can have comments in addition to the "key value" pairs to specify. The "key value" pairs can be specified in any order, but each key word must be the first item in a line and its corresponding value the second item in the same line. The key words in the example must be used but they are NON-case sensitive. The three Not optional "key value" pairs must be specified.
The values all are validated. The value of imageId is validated to correspond to an AMI in your AWS account or to a public AMI available in the AWS region on which your are running the scripts. The value of instanceType is validated to be a valid AWS instance type. The values of securityGroupId, subnetId, hostZone and hostZoneId are validated to exist in your AWS account.
The key word subnetId and its value are optional. If not specified, the scripts will try to obtain a subnetID from your AWS account. We have successfully tested the scripts to obtain and use a subnetID running the scripts with a personal AWS account and with an institutional AWS account (see details in section Validating the Workshop Environment below).
The key words hostZone and hostZoneId and their values are optional. If specified and valid, each instance will be accessed using a domain name which will look like this: instance01.cloud-span.aws.york.ac.uk, where instance01 is just an example of a specified instance name and cloud-span.aws.york.ac.uk is the base domain name (hostZone) in the example. If hostZone and hostZoneId and their values are not specified, each instance will be accessed using the public IP address or the generic domain name allocated by AWS which will look like this: 34.245.22.106 or ec2-34-245-22-106.eu-west-1.compute.amazonaws.com.
The three files must be placed inside a directory called inputs, and the inputs directory must be placed within at least one other directory, whose name you can choose, and to which we refer to as Workshop Environment (WE). We use this directory structure to manage instances for our workshops:
courses ### you can omit this directory or use other name
genomics01 ### workshop/course WE name; you can use other name
inputs ### you CANNOT use other name
instancesNames.txt ### you can use other name
resourcesIDs.txt ### you CANNOT use other name
tags.txt ### OPTIONAL - you CANNOT use other name
outputs ### created automatically by the scripts - don’t modify
genomics02 ### another WE: inputs and outputs directories inside
metagenomics01 ### another WE: inputs and outputs directories inside
...
The outputs directory inside a WE is created automatically by the scripts to store the results of invoking AWS services as described below.
2.2. Running the Scripts
Running the scripts requires only the path of the file that contains the names of the instances to create, stop, start or delete. Login keys, IP addresses and domain names used by the instances are created or deleted automatically.
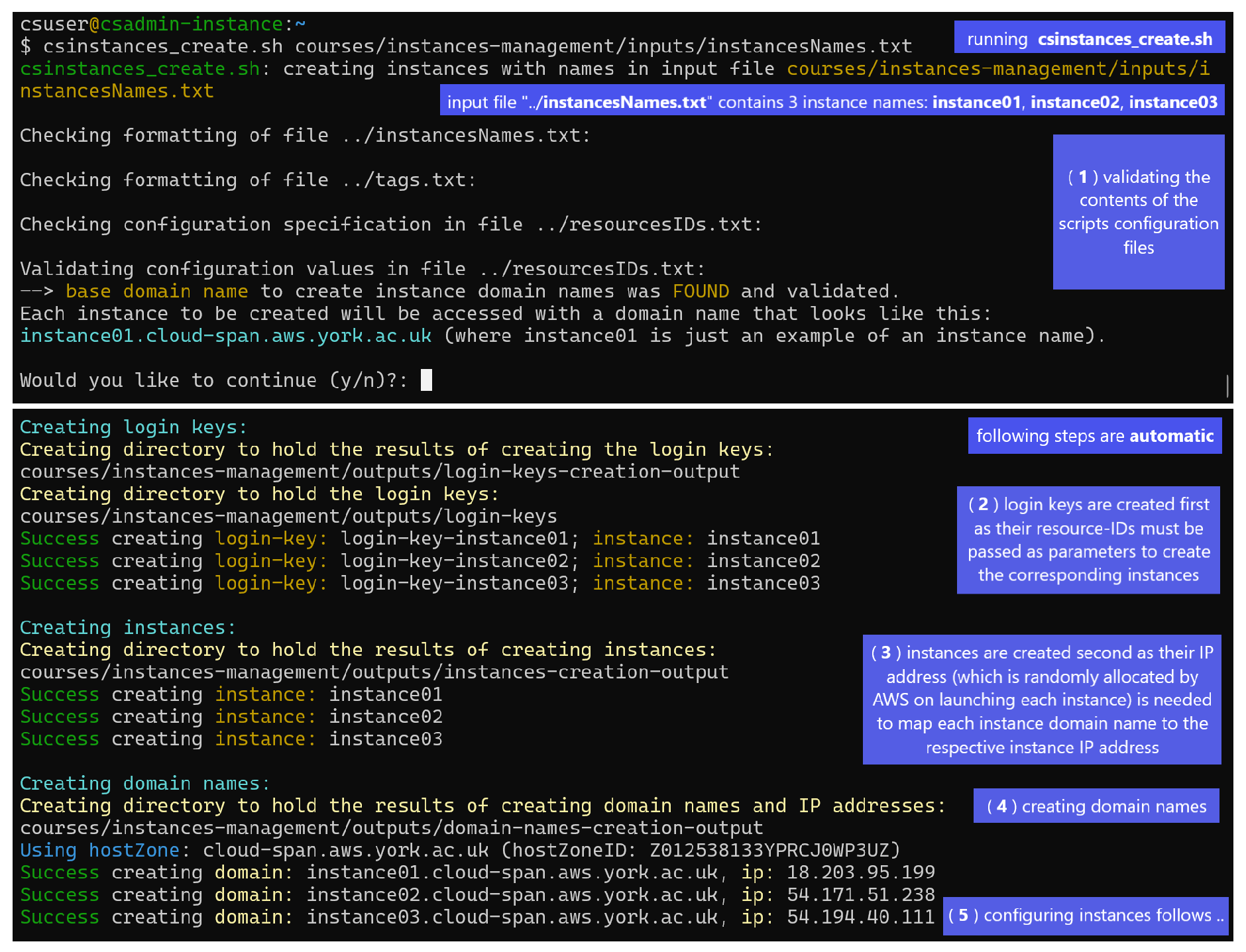
Figure 1 shows a Linux terminal where the script
csinstance_create.sh has been run thus (second line in the figure):
$ csinstances_create.sh courses/instances-management/inputs/instancesNames.txt
Creating instances involves 5 or 4 steps depending on whether domain names to access instances are to be managed or not:
-
(
1) validating the contents of the scripts configuration files (
instancesNames.txt passed as parameter,
tags.txt if found, and
resourcesIDs.txt) as described above and shown in
Figure 1.
If no problem is found in these files, the
option to continue with the
configuration detected, regarding managing or not managing domain names to access instances,
is displayed for the user to confirm or cancel the run.
If there is a problem with the files, messages (not shown) are displayed that point out the specific problem/s in each file and the run is aborted.
In
Figure 1, the configuration detected corresponds to managing domain names to access instances, that is,
hostZone and
hostZoneId and valid values were specified and found in the
resourcesIDs.txt file and were validated.
If hostZone and hostZoneId are not specified, the option to continue looks like this:
--> NO base domain name was FOUND.
Each instance to be created will be accessed with the IP address or the generic domain name provided
by AWS, which look like this: 34.245.22.106 or ec2-34-245-22-106.eu-west-1.compute.amazonaws.com.
Would you like to continue (y/n)?:
(2) creating login keys.
(3) creating instances, each configured to use one of the login keys created in the previous step.
(4) creating instances domain names as mapped to the respective instance IP addresses (AWS randomly allocates IP addresses to instances when instances are launched for the first time or started after having been stopped) — THIS STEP is only run if hostZone and hostZoneId and valid values were specified in the resourcesIDs.txt file.
(5) configuring each instance both to enable the csuser account (used by workshop participants) to be logged in and to change the instance host name to the instance name (the default host name is the instance IP address) regardless of whether domain names are to be managed or not. This step is not shown in the figure.
To stop or start the instances created with the command above, or to delete them along with all the related resources, you would run the scripts csinstances_stop.sh, csinstances_start.sh or csinstances_delete.sh using the same input file:
$ csinstance_stop.sh courses/instances-management/inputs/instancesNames.txt
$ csinstance_start.sh courses/instances-management/inputs/instancesNames.txt
$ csinstance_delete.sh courses/instances-management/inputs/instancesNames.txt
2.3. Using Instances and Customising AMIs
Each instance has two user accounts:
csuser and
ubuntu. You login to these accounts using the
ssh program as in the examples below, which correspond to how you would login to those accounts on the instances created in the example in
Figure 1, wherein the instance names specified were
instance01,
instance02, and
instance03.
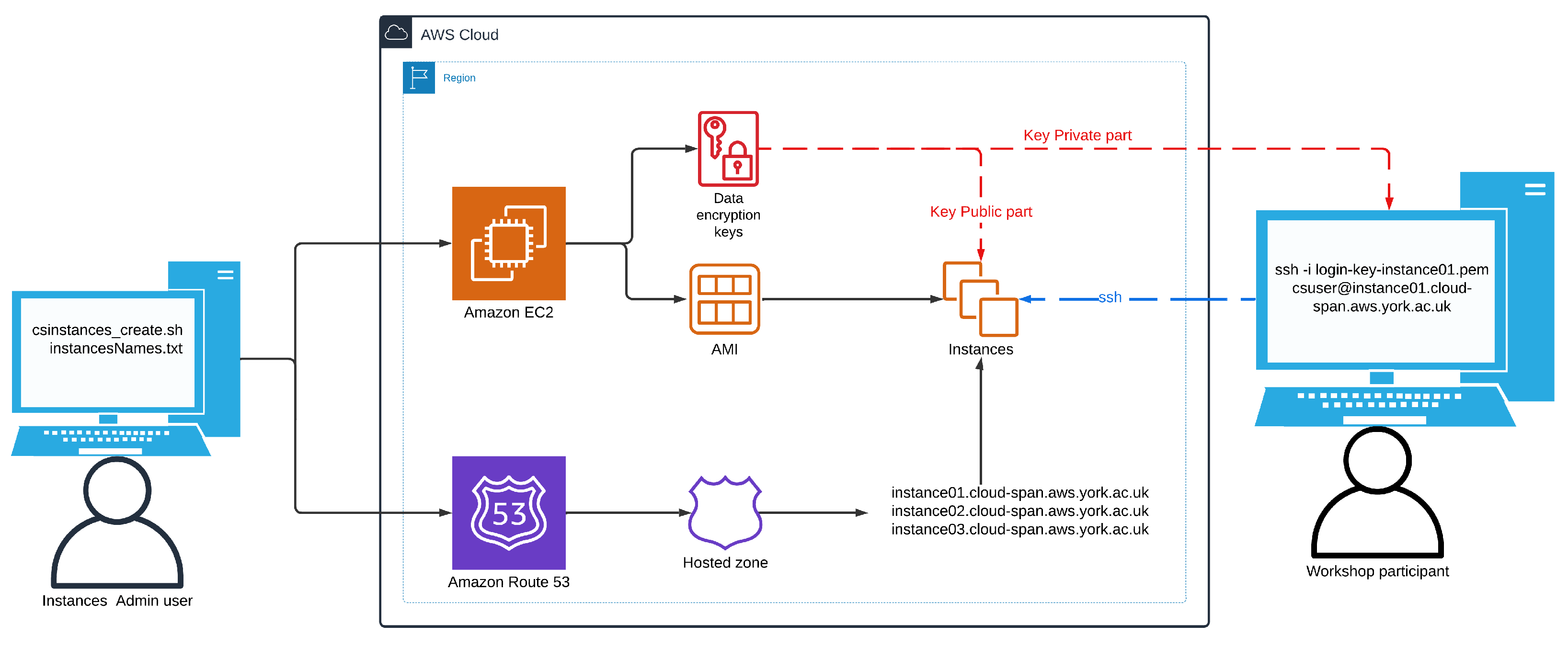
ssh -i login-key-instance01.pem csuser@instance01.cloud-span.aws.york.ac.uk
ssh -i login-key-instance01.pem ubuntu@instance01.cloud-span.aws.york.ac.uk
...
ssh -i login-key-instance03.pem csuser@instance03.cloud-span.aws.york.ac.uk
ssh -i login-key-instance03.pem ubuntu@instance03.cloud-span.aws.york.ac.uk
Note that instance names are used by the scripts to "label" the names of the corresponding login key files, instances domain names, and other files. Each instance domain name is the previously configured base domain name (in our case cloud-span.aws.york.ac.uk) prefixed with the corresponding instance name.
Workshop participants use the csuser account — all ’omics data and most software analysis tools are installed in this account.
The ubuntu account has superuser privileges. We use it to update system software, software analysis tools that need to be installed at system level, etc. We use the csuser account to update ’omics data and software analysis tools that can be installed at user account level. We update an instance as just outlined in order to create a new AMI (virtual machine template) from which to create new updated instances.
Figure 1.
Running the script |csinstancescreate.sh| (second line) to create 3 instances with the names specified in the file +courses/.../instancesNames.txt+: instance01, instance02, and instance03 (one name per line). Creating instances involves 5 or 4 steps depending on whether domain names to access instances are to be managed or not. The run in the figure corresponds to domain names being managed but only the first 4 steps are shown. The last step (5) involves configuring each instance both to enable the csuser account (used by workshop participants) to be logged in and to change the instance host name to the instance name (the default host name is the instance IP address). If domain names are not to be managed, the step creating domain names (4 in the figure) is not run.
Figure 1.
Running the script |csinstancescreate.sh| (second line) to create 3 instances with the names specified in the file +courses/.../instancesNames.txt+: instance01, instance02, and instance03 (one name per line). Creating instances involves 5 or 4 steps depending on whether domain names to access instances are to be managed or not. The run in the figure corresponds to domain names being managed but only the first 4 steps are shown. The last step (5) involves configuring each instance both to enable the csuser account (used by workshop participants) to be logged in and to change the instance host name to the instance name (the default host name is the instance IP address). If domain names are not to be managed, the step creating domain names (4 in the figure) is not run.
2.3.1. Login to Instances When Domain Names Are NOT Managed
Assuming that
domain names were
not managed in the example in
Figure 1 (but the instance names specified were those in the example:
instance01, etc.), you would login to the
csuser and
ubuntu accounts thus:
| ssh -i login-key-instance01.pem csuser@34.245.22.106 |
#### where 34.245.22.106 is just an example IP address |
| ssh -i login-key-instance01.pem ubuntu@34.245.22.106 |
#### the IP address of each instance will vary |
| ... |
|
2.4. Customising the Login Account of Workshop Participants
The
csuser account is only available on instances created from a Cloud-SPAN AMI — "cs" in
csuser stands for Cloud-SPAN project [
19,
20]; the Data Carpentry AMI uses the
dcuser account for workshop participants.
The
ubuntu account is available on instances created from any AWS Linux Ubuntu AMI, and is the
only account that is
enabled to be logged in when an instance runs for the first time. This enabling is performed by the
AWS service that launches instances. When an instance boots for the first time, that service
adds the
public key part of the login key created to access the instance to
the file /home/ubuntu/.ssh/authorized_keys — thereafter the
private key part of the login key can be used with
ssh as shown above [
21] to access the
ubuntu account. The last step in creating instances, that is,
configuring instances, enables the
csuser account in each instance to be logged in by copying that file to the
csuser account in
/home/csuser/.ssh/authorized_keys. The copy is made through running a Bash script in the
ubuntu account that is remotely invoked by the script
aws_instances_configure.sh.
If instead of the
csuser account you would like to use an account with a name related to your project, institution, etc., you can follow the steps below to create a new AMI from which you will create instances with your new user account (see details in the section
Configure an Instance to Become AMI [
15] in the tutorial):
create an instance using the Cloud-SPAN (CS) Genomics AMI and login as the ubuntu user.
create and initialise your new user account (as described in the tutorial)
edit the script /home/ubuntu/bin/usersAuthorizedKeys-activate.sh to replace the string csuser with the name of your new account — this script copies the file /home/ubuntu/.ssh/authorized_keys to the csuser account.
delete or edit the file /etc/motd (message of the day) — it contains a screen welcome message that includes the Cloud-SPAN name.
create a new AMI from the updated instance.
The CS Genomics AMI is of size 30-Gigabyte (GB). It has the scripts installed along with a few other scripts that will help you customise your AMI. Two scripts automatically update system software and the Genomics software applications installed. The tutorial describes how to modify these scripts to install other applications and to remove the ones installed, and how to use another script to automatically increase the size of secondary storage and the file system up to 2 Terabytes.
Figure 2 shows an schematic of the scripts environment. We have described the left and right elements in that figure: the use of the scripts and the use of instances created with the scripts. How the scripts manage the relevant AWS services is described below.
3. The Scripts Design and Implementation
AWS services can be managed using (1) the AWS Console, (2) the AWS CLI (command line interface) program, (3) Software Development Kits (SDKs) or libraries with a programming language such as Python, Java, etc., or (4) infrastructure as code (IaC) blueprints. The level of automation increases from the AWS Console to IaC blueprints [
22].
The AWS Console is mostly used to open an AWS account, to do one-off configurations, and to browse the overall state of resources used by an account. The AWS CLI enables you to manage any AWS service but is complex to use on its own, yet combined with shell scripts is probably the fastest way to manage AWS services as shell scripts have been used for decades for such purpose. SDKs are mostly used to develop end user applications comprising a
front-end (mobile or browser-based) user interface and
back-end cloud-based server/s and databases. IaC blueprints are used to manage infrastructures (service architectures) that are complex [
23], that change often because of continuous improvements, or that require "zero-downtime deployments" where "changes must be made with live traffic" [
23]. Basically, a blueprint of the required infrastructure is written as code in a declarative language specifying “what we want”, as opposed to “how to do what we want” (which is typical of procedural languages). On “running” the blueprint using a software tool such as AWS CloudFormation or Terraform, the services making up the infrastructure will be created, configured to some extent, and launched. If later the blueprint is updated (say, to delete or add more services), the infrastructure will be updated accordingly when the blueprint is run again. As blueprints are simple text files, it is possible to use version control to roll-back to a previous version of the infrastructure. It may be convenient and practical to develop a system using the AWS CLI, SDKs and IaC.
The scripts use the AWS CLI to manage instances and related resources. Using Bash scripts and the AWS CLI was a design decision based on convenience and feasibility given the time constraints we had when confronted with the task of managing multiple AWS instances. We had used the Python AWS SDK little (compared to Bash and the AWS CLI) and had not used IAC before. With hindsight, we believe our decision is practical and adequate too. The AWS infrastructure we manage (login keys, instances, IP addresses and domain names) is relatively simple and has not changed since we developed the scripts almost three years ago. Also, one of the main goals of the Cloud-SPAN project was that our solution to manage AWS instances be shared with others and taught to others, and having used Bash scripting with the AWS CLI has made the online tutorial simpler and more readily accessible to more people out of Bash being more widely known and used than SDKs, Terraform or similar tools.
The scripts organisation is straightforward, with most communication between the scripts taking place through shared files. Some scripts perform a fair amount of pattern matching in preparing AWS service requests, processing results requests and other tasks.
3.1. The Scripts Execution Flow — Overview
We saw above how to run the scripts csinstances_create.sh and csinstances_delete.sh: passing the path of the file that contains the names of the instances to be created or deleted (along with related resources). Those two scripts do not invoke the AWS CLI directly; they only invoke the scripts "aws_*.sh", passing each such script the file path received, as shown in the main code of csinstances_create.sh below. In the code, the file path is in the script variable $1, as csinstances_create.sh receives the file path as the first (only) parameter:
| check_theScripts_csconfiguration |
"$1" || { message "$error_msg"; exit 1; } |
| aws_loginKeyPair_create.sh |
"$1" || { message "$error_msg"; exit 1; } |
| aws_instances_launch.sh |
"$1" || { message "$error_msg"; exit 1; } |
|
|
| if [ -f "${1%/*}/.csconfig_DOMAIN_NAMES.txt" ]; then ### %/* gets the inputs directory path |
| aws_domainNames_create.sh |
"$1" || { message "$error_msg"; exit 1; } |
| fi |
|
| aws_instances_configure.sh |
"$1" || { message "$error_msg"; exit 1; } |
| exit 0 |
|
In the code, the function check_theScripts_csconfiguration (which is in the script file colour_utils_functions.sh) is first invoked to validate the contents of the configuration files instancesNamesFile.txt, resourcesIDs.txt, and tags.txt if specified. The scripts "aws_*.sh" are only invoked if that function or the previous script runs successfuly; otherwise csinstances_create.sh prints an error message and aborts execution (exit 1;) — unsuccessful runs are discussed below in the section Validating the Workshop Environment. The script aws_domainNames_create.sh is only invoked if the file ../inputs/.csconfig_DOMAIN_NAMES.txt exists, which is created if, in validating the resourcesIDs.txt file, a base domain name and its host zone ID are found and are valid AWS resources within the AWS account being used.
The code of csinstances_delete.sh is similar to the code above but instead invokes the scripts that delete AWS resources.
3.2. Creating and Deleting Instances and Related Resources
Each "aws_*.sh" script (except aws_instances_configure.sh) can create or delete one or multiple AWS resources of a single type, for example: one or more login keys, or one or more instances, etc. Each script invokes the AWS CLI with a specific AWS service request for each instance name specified in the input file received as parameter. Each script makes a list with the instances names in the input file and then loops through the list, in each loop invoking the relevant AWS service "for the instance name" in the current loop as outlined next. These are some of the AWS services invoked by the "aws_*.sh" scripts:
| aws ec2 create-key-pair --key-name $loginkey --key-type rsa ... |
### invoked by aws_loginKeyPair_create.sh |
| aws ec2 run-instances --image-id $resource_image_id ... |
### invoked by aws_instances_launch.sh |
| aws ec2 delete-key-pair --key-name $loginkey ... |
### invoked by aws_loginKeyPair_delete.sh |
| aws ec2 terminate-instances --instance-ids $instanceID ... |
### invoked by aws_instances_terminate.sh |
The AWS CLI program is called aws once installed. The first parameter, ec2 (elastic compute cloud), is the AWS service being invoked; the second parameter, create-key-pair, or run-instances, etc., is the operation being requested; the following parameters are key-value pairs required by the operation. Some values of the key-value pairs are specified as script variables, for example: $login_key and $resource_image_id. Some of these variables are updated within each loop just before invoking the AWS CLI, so that each such invocation uses the values (in those variables) that correspond to the relevant instance name.
3.3. Scripts Communication
Each invocation of the AWS CLI returns a result that states whether the AWS service request was successful or not along with other information. In the case of a successful request that creates or allocates a resource, the result includes the resource-id assigned by AWS to uniquely identify the resource. As resource-id’s are needed to further manage the corresponding resources, for example, to delete them, the scripts store the results of each AWS invocation to a file inside the outputs directory in the workshop environment (WE) being used. The name of each file has, as a sub-string, the instance name used to invoke the AWS service to create the resource. This file naming convention enables the other scripts to later recover the resource-id of the resources to delete, stop, etc.
The names of login key files and domain names are managed similarly, by including the relevant instance name as a sub-string, as shown in the login examples with ssh shown earlier.
3.4. Configuring, Stopping and Starting Instances
The scripts aws_instances_configure.sh, csinstances_stop.sh, and csinstances_start.sh are organised and work similarly to the other "aws_*.sh" scripts that create or delete AWS resources. They build a list of instance names and loop through the list in order to invoke the AWS services required to configure, stop, or start each instance in the list.
However, these scripts are somewhat more complex than the others, as configuring, stopping and starting instances require different handling depending on whether domain names are managed or not, and these scripts handle both scenarios.
The relevant point to admin users of the scripts is that not managing domain names requires knowing where to find the IP addresses allocated to instances in order to provide them to workshop participants. Each of these IP addresses is saved to a file in the directory outputs/instances-creation-output in the WE being used. For example, the contents of this directory after creating 3 instances (instance01, instance02, instance03) not managing domain names would be as shown in this file listing:
| csuser@csadmin-instance:~ |
| $ ls courses/instances-management/outputs/instances-creation-output/ |
| instance01-ip-address.txt |
instance02-ip-address.txt |
instance03-ip-address.txt |
| instance01.txt |
instance02.txt |
instance03.txt |
The contents of the files "instance*-ip-address.txt" is just the instances IP addresses:
csuser@csadmin-instance:~
$ cat courses/instances-management/outputs/instances-creation-output/instance*-ip-address.txt
3.252.164.131
34.242.5.243
3.249.248.80
The files "instance??.txt" are created by the script aws_instances_launch.sh; the files "instance*-ip-address.txt" are created by the script aws_instances_configure.sh. This is because an instance public IP address is not available in the results of invoking aws_instances_launch.sh (that is, in the files "instance??.txt"), but is available until each instance is actually running, a condition that the script aws_instances_configure.sh waits for and detects in order to recover each instance IP address to start configuring the instance, and finally save the IP address to the file instanceName-ip-address.txt.
If instances are stopped and eventually started through running csinstances_stop.sh and csinstances_start.sh, the IP address of each instance will change because AWS randomly allocates IP addresses when instances are launched for the first time or when they are started after having been stopped. Therefore, csinstances_start.sh overwrites the contents of the files "instanceNames*-ip-address.txt" with the newly allocated IP addresses.
By the way, the admin user of the scripts does not need to use IP addresses to login to the respective instances (only workshop participants need to use IP addresses if domain names are not managed). The admin user can instead use the script lginstance.sh providing the path of the instance login key file and the user account to login (csuser or ubuntu):
| csuser@csadmin-instance:~ |
| $ lginstance.sh courses/instances-management/outputs/login-keys/login-key-instance01.pem csuser |
| logging you thus: |
### this and the next line are displayed by lginstance.sh |
| ssh -i courses/instances-management/outputs/login-keys/login-key-instance01.pem csuser@3.253.59.74 |
| ... |
### instance welcome message |
| csuser@instance01:~ |
### instance prompt |
| $ |
|
Note that that long command is rather easy to enter using the Tab key for the shell to complete the name of the script lginstance.sh, the names of the intermediate directories and the name of the login key .pem file. Note also that, as mentioned above, the host name of an instance is the instance name and not its IP address, regardless of whether domain names are managed or not. These two points will help troubleshooting if domain names are not managed.
3.5. Validating the Target Workshop Environment
We have already discussed above most of the validating of the contents of the scripts configuration files: instancesNames.txt, tags.txt (if specified), and resourcesIDs.txt. The following points will help understand better how the scripts work.
The scripts configuration files are all validated only the first time any of the scripts is run against a WE; thereafter only the file instancesNames.txt is validated (below is outlined why and how you may want to handle multiple instancesNames.txt files in the same WE). Specifically, once it is determined, validated, and confirmed (by the user of the scripts) whether domain names are to be managed or not, either the file inputs/.csconfig_DOMAIN_NAMES.txt or the file inputs/.csconfig_NO_DOMAIN_NAMES.txt is created in the WE being used. These files are empty. It is only their existence that is used to drive the execution of the scripts accordingly.
Recall that, in the file resourcesIDs.txt, the key word subnetId and its value are optional and that if they are not specified, the scripts will try to obtain a subnetID from the AWS account being used. As mentioned above, we have successfully tested the scripts to obtain and use a subnetID running the scripts with a personal AWS account and with an institutional AWS account. Our personal account uses the default subnets allocated by AWS. Our institutional account is configured not to use the default subnets but other subnets configured by the Information Technology (IT) Department at our institution. If the scripts cannot obtain a subnetID, they will display a message telling you to search for vpc (virtual private cloud) in the AWS Console (in the search box at the top), then click on subnets on the left menu. Copy one of the subnetIDs displayed and paste in the resourcesIDs.txt file. If no subnetID is displayed in the AWS Console, you need to contact your IT department.
In addition to the validating already discussed, the scripts follow an ALL or NOTHING policy in managing resources, as follows. When creating, deleting, stopping, starting, or configuring instances or related resources, any such operation must be possible for all the instances (or resources implicitly) specified in the file "instanceNamesFile.txt" passed to the scripts. If there is a problem the scripts cancel their operation. The policy is implemented by checking whether any of the specified instances/resources exists already, that is, whether any of the files containing the AWS resource-id of the specified instances/resources exists in the outputs directory of the target workshop environment. No such file should exist when creating instances/resources. Conversely, all relevant files should exist when deleting instances/resources, or stopping, starting or configuring instances.
The policy was implemented to avoid overwriting the files with AWS resource-id’s accidentally, as when this happened the corresponding AWS resources had to be deleted manually using the AWS Console.
Note that you can manage multiple "
instanceNamesFiles.txt" in the
inputs directory of any workshop environment. We do so to handle late registrations and cancellations to our workshops. The section
Unforeseen Instance Management [
24] in the tutorial describes our approach to naming multiple "
instanceNamesFiles.txt" so we can easily track all the scripts runs performed on a workshop environment.
3.6. Overview of the Online Tutorial
How to configure and use the scripts is described in detail in the tutorial
Automated Management of AWS Instances [
13] which covers these main topics:
how to open an AWS account and how to configure it both with programmatic access with the AWS CLI and with a base domain name from which to create instances (sub) domain names.
how to configure a terminal environment with the scripts and the AWS CLI on Linux, MacOS, and Windows (Git Bash), or in the AWS CloudShell, a browser-based Linux Bash terminal.
how to configure and run the scripts to manage instances for a workshop, manage late registrations and cancellations, and some troubleshooting.
how to create, manage and configure Amazon Machine Images (AMIs) which serve as templates to create AWS instances.
the organisation and workings of the scripts.