1. Background
A rapidly evolving outbreak of MPOX disease caused by a mutant clade I virus (now dubbed 1b) that started around Sept 2023 has persisted in the South and North Kivu Provinces of Eastern DRC, with confirmed isolated cases now reported in the neighboring countries of Burundi, Rwanda and Uganda [
1,
2]. Epidemiological profiling in DRC shows a predilection for (a) children <15 yrs, (b) PLWHA and (c) pregnant women [
1,
3,
4]. Although the observed general case fatality rate is an acceptable 4.6%; this is higher among children <5yrs, PLWHA with uncontrolled viremia, and pregnant women. The East And Central African Great lakes region represents one of the global regions with poorly controlled HIV viremia; lagging behind the UN-SDG 3 targets of 95:95:95 by 2030. Only upto 75% PLWHA here have optimally controlled viral loads. This increases the risk of having severe cases of MPOX in this region, necessitating the use of medical countermeasures (MCMs) to combat the epidemic [
1,
2,
3].
Whereas there are several MCMs with demonstrated prior efficacy against MPOX, not only is the global capability to rapidly manufacture large doses of vaccines and therapeutics limited, but the impact of the mutations that have been observed in the circulating clade 1b remains unknown. That said, history has proven that—in the face of a global pandemic, the needs of the resource-limited countries of the South will not be prioritized, over those of the wealthier North. The lack of transparency and equitable access to COVID19 vaccines, revealed Africa’s vulnerabilities and limited capacity for research and development (R & D) of its own homegrown MCMs against priority pathogens. This forced the African Union (AU) to establish the Partnerships for Vaccine Manufacturing (PAVM) under the Africa Centers for Disease Control [
5,
6] . Although positive, the impact of the PAVM is yet to be realized, necessitating more agility in tackling the MPOX needs of MCM on the continent [
7].
Our group has over the past 2 decades acquired and built expertise to identify, map and validate epitopes against high consequence, priority pathogens for diagnostic, monoclonal antibody (mAbs) therapeutic and vaccine research and development ((R & D) [
8,
9,
10,
11,
12,
13,
14]. As an emergency local response to the on-going MPOX challenge, we set out to identify ideal epitopes of clade 1b MPXV as targets for the rapid research and development (R & D) of Mpox medical countermeasures.
Two MPXV proteins:— H3L (OPG108–heparin binding protein) and A17L (OPG143–myristylated protein), were chosen as targets by virtue of their surface localization, stability and known role in MPXV host cell fusion entry [
16,
17].
2. Methodology
Design: Immuno-informatics
Site: Division of Pathogen OMICS, Unit of Genetics & Genomics, Depts of Immunology, Molecular Biology & Pathology, School of Biomedical Sciences, College of Health Sciences, Makerere University, Kampala, Uganda
Materials and accession: Two MPXV proteins:— H3L (OPG108–heparin binding protein) and A17L (OPG143–myristylated protein) obtained from Uniprot, with the following swissport accession #s:
- -
>sp|A0A7H0DN80|PG108_MONPV Envelope protein OPG108 OS=Monkeypox virus OX=10244 GN=OPG108 PE=2 SV=1
- -
>sp|A0A7H0DNB5|PG143_MONPV Virion membrane protein OPG143 OS=Monkeypox virus OX=10244 GN=OPG143 PE=3 SV=1
Interventions: Within the immune epitope database analysis resource (IEDB-AR), the two epitopes were examined for surface accessibility, antigenicity and 3-D localisation (DiscoTope) [
15].
Measurable variables: positive > 9-nomer epitopes
3. Results & Discussion
We identified three (3) and six(6) candidate >9-nomer epitopes for H3L and A17L respectively, that met the criteria for surface accessibility, antigenicity and 3-D colocalization. For details, see
Table 1 and
Table 2,
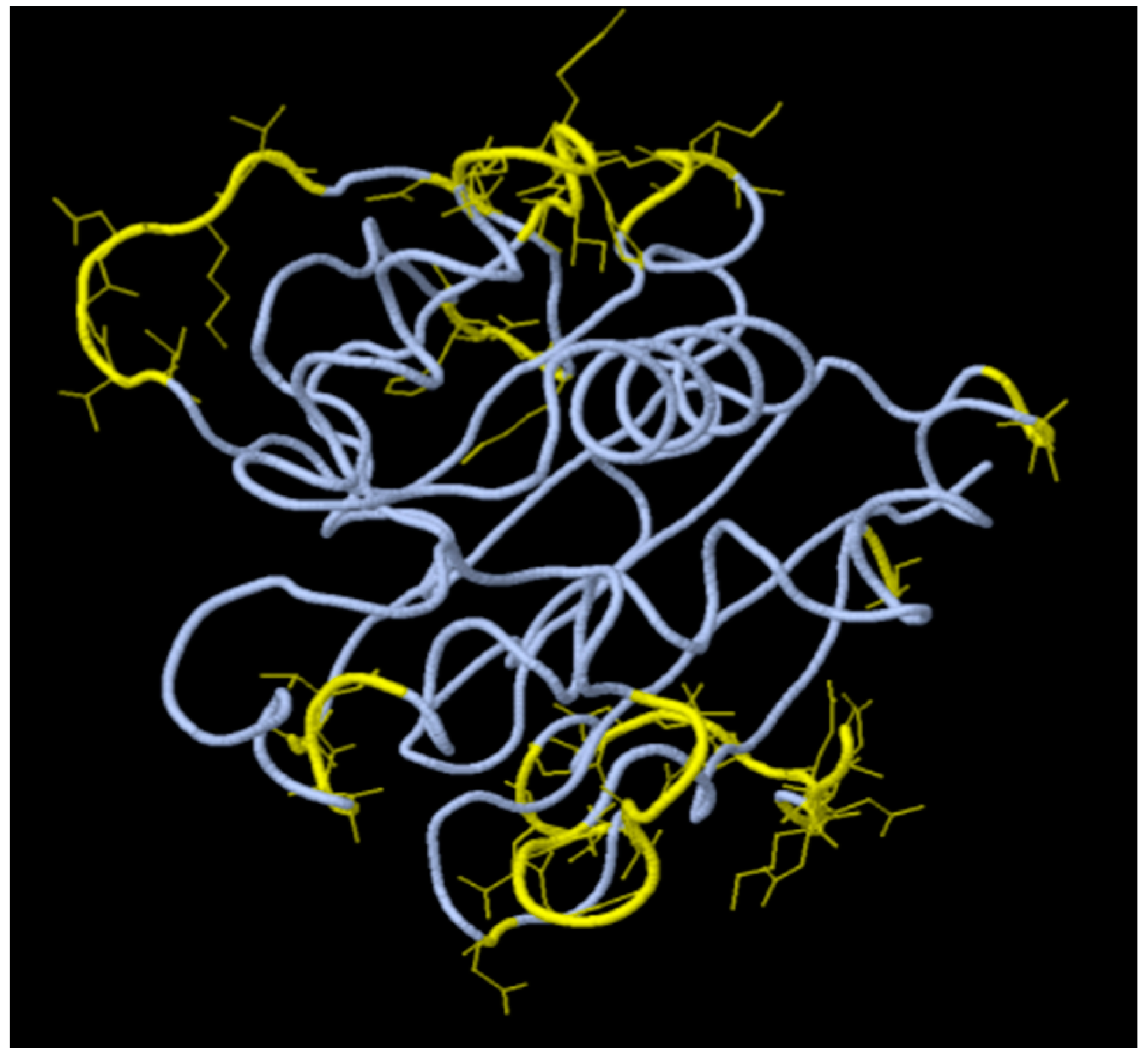
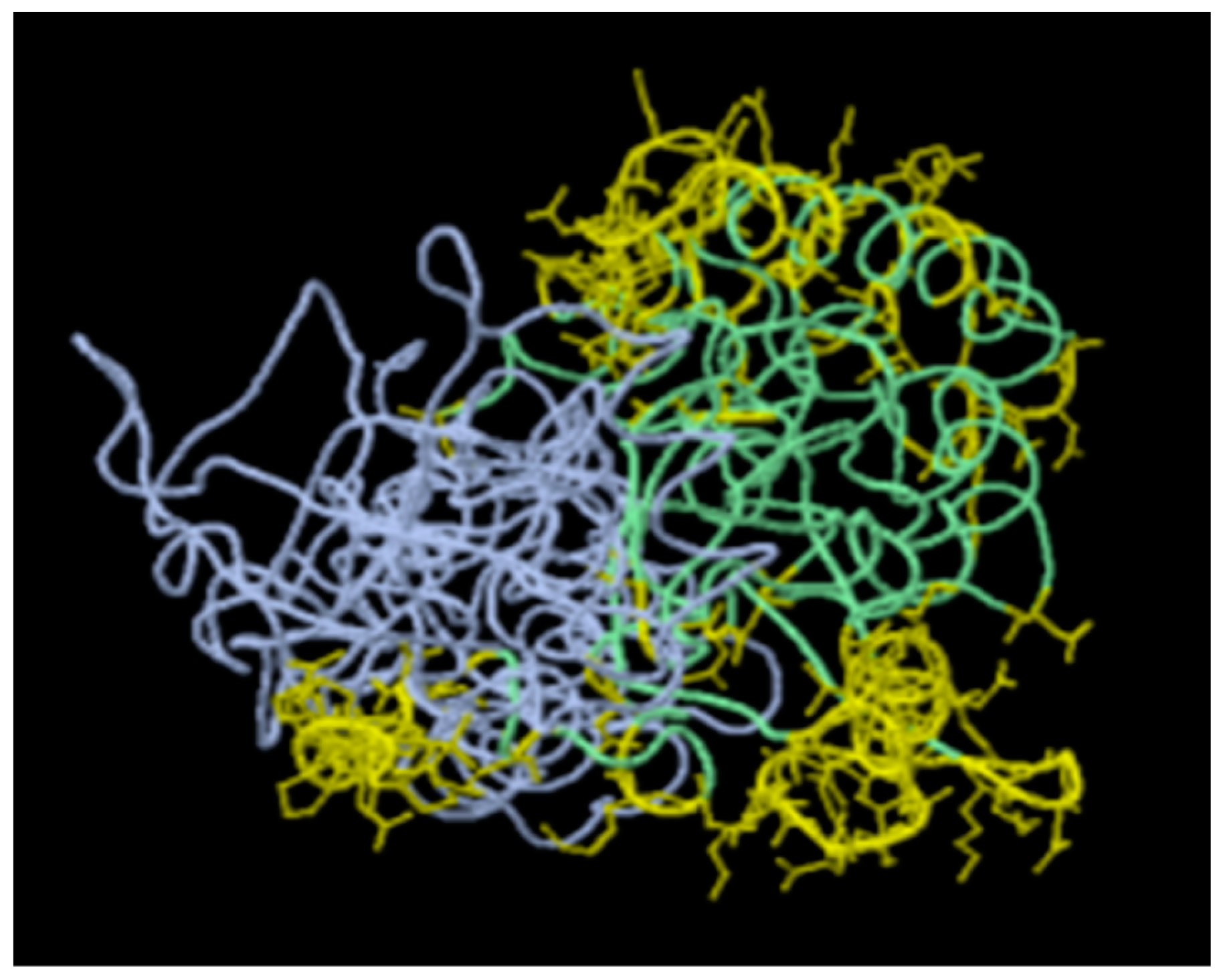
Figure 1 and
Figure 2, and Supplementary File S1, respectively. Overall, across all 3 analyzed parameters, the two best identified epitopes for each target were as follows:—H3L (8-
HINDQKFDDVKDNEVMQEKR-472 and 236-
YVEHDPRLVAEH-247) and A17L (104-
QNEVTPEYIKDLKYATDYIA-123;) 270-
KKDYLLGDSDSVAKCCSKTNTKHCPKIFNNNYKTEHC-290).
Interestingly, prior studies have highlighted H3L and A17L as ideal vaccine candidates, and separate epitopes of both surface proteins have been derived [
16,
17]. de Araújo LP, et
al. (2024) listed the A17L as one of the 6 host cell entry proteins for MPXV (alongside A26L/A30L, A33R, H2R, L1R) that can be targeted for vaccine R & D. This team identified 138 epitopes for A17L, all of which overlap with our 6 target epitopes. Some of these epitopes are shared with Alaskapox viruses [
18]. Disturbingly, Masirika LM, et
al. (2024) [
1] found A17L to be one of the 7 proteins emerging as hot spots mutations based on the consensuses inframe deletions, frameshift variants, synonymous variants, and amino acids substitutions among isolates of clade 1b taken from Kamituga area, South Kiv, DRC. The location of most of the referenced mutations, however, lay outside the listed epitopes in
Table 2, Supplementary File S1. That said, it remains to be examined, what the impact of these mutations will be on the real world effectiveness of derivative MCMs.
Elsewhere, Wang Y, et al. (2023) [
17] highlighted several potential vaccine targets for MPVX, emphasizing the relevance of 6 potent immunogens inclusive of H3L (alongside
M1R, E8L, A29L, A35R, and B6R). Interestingly, residues of the epitopes identified by this group ( 13-VIDRLPSETFPNVHE
HINDQKF-34; 231-DNAAK
YVEH-239) overlap and traverse the target regions of the priority epitopes we identify for the H3L ( 8-
HINDQKFDDVKDNEVMQEKR-472 and 236-
YVEHDPRLVAEH-247) (
Table 1). They conclude that these targets can be integrated into diverse vaccine platforms to elicit powerful B-cell and T-cell responses, thereby providing protective immunity against MPXV infection [
17].
Lastly, we have mapped residues of these epitopes on the available 3-D crystal structures of the corresponding proteins in Uniprot. Notable is that, only the structure of the vaccinia virus H3L (PDB entry # 5EJ0) exists. No 3-D structure corresponding to the VACV A17L protein was found, except the structure of the vaccinia virus A16/G9 sub-complex from the orthopoxvirus entry-fusion complex (PDB entry # 8GP6) [
19]. The predictions by DiscoTope are shown in
Figure 1 and
Figure 2 in color yellow, revealing surface accessibility for presentation as vaccine immunogens. This is consistent with findings by Yefet R, et al. (2023) who found that Monkeypox infection elicits strong antibody and B cell response against A35R and H3L antigens [
20]. Indeed, Zuiani A, et
al. (2024) recently found that multivalent mRNA monkeypox virus vaccine (BNT166) that expresses H3L as one of the antigens protects mice and macaques from orthopoxvirus disease [
21]. Moreover, there is evidence for cross reacting protective immunity against other members of the genera
orthopoxviridae [
20,
21].
In conclusion, we identify epitopes of two MPXV surface proteins involved in fusion entry which, whether expressed alone, multiplexed as nano-spiked virus-like particles (VLPs), or fused as chimeric recombinants form immunogens for the production of protein sub-unit vaccines. Inversely, derivative monoclonal antibodies (mAbs) from the same are precursors for therapeutic cocktails and or antigen, rapid diagnostic tests (RDT).
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, S1: Details of methodology for epitope mapping presented.
Funding
No specific funding was received towards this work. MW is currently funded by a GH-EDCTP3-JU ‘ Mobilisation of Emergency funding for Mpox outbreak research response’, DECIPHER-MPOX.
Conflicts of Interest
The author declares no sources of competing interests to declare.
References
- Masirika LM, et al. Complete Genome Sequencing, Annotation, and Mutational Profiling of the Novel Clade I Human Mpox Virus, Kamituga Strain. J Infect Dev Ctries 2024, 18:600–608. [CrossRef]
- Katoto PDMC, et al. Shifting transmission patterns of human mpox in South Kivu, DR Congo. Lancet Infect Dis. (2024). [CrossRef]
- Ritter JM, et al. Mpox Pathology Working Group. Pathology and Monkeypox virusLocalization in Tissues From Immunocompromised Patients With Severe or Fatal Mpox, The J of Infect Dis, 229 (2), (2024).
- Duarte-Neto AN, et al. Main autopsy findings of visceral involvement by fatal mpox in patients with AIDS: necrotising nodular pneumonia, nodular ulcerative colitis, and diffuse vasculopathy. Lancet Infect Dis. 23(11), (2023). [CrossRef]
- PAVM Framework-for-Action Available at the following url: https://africacdc.org/download/partnershipsfor-african-vaccine-manufacturing-pavm-framework-for-action.
- Vaccine R&D and Vaccine Manufacturing Competency Frameworks – Africa CDC. Available at the following url: https://africacdc.org/download/vaccine-rd-nd-vaccine-manufacturing-competencyframeworks/.
- Business and Operational Models for African Biomanufacturing Training Initiatives – Africa CDC. Available at the following url: https://africacdc.org/download/business-and-operational-models-forafrican-biomanufacturing-training-centres/.
- Wayengera, M.et al. Proteomics of Marburg and Ebola Glycoproteins: Insights into Their Physicochemical Similarities and Irregularities. African Journal of Biotechnology 2009, 8.
- Babirye, P., et al. . Identity and Validity of Conserved B Cell Epitopes of Filovirus Glycoprotein: Towards Rapid Diagnostic Testing for Ebola and Possibly Marburg Virus Disease. BMC Infect Dis 2018, 18, 498. [CrossRef]
- Wayengera M, et al.. Cloning, Expression and Purification of recombinant forms of Full Length and Extracellular Domain EBOV Glycoprotein within Mammalian Cell-Lines. J Antivir Antiretrovir. 2019, 11:187. [CrossRef]
- Bongcam-Rudloff E, et al. Consensus DNA and amino acids sequences of the recombinant panfilovirus glycoprotein specific mAb 4B3B5 and its expression in CHO mammalian cell lines. Research Square. 2023. DOI: https://doi.org/10.21203/rs.3.rs-2532962/v1.
- Vlachakis D, et al. Coding DNA of the recombinant panfilovirus mAb 7C8F1: cloning and expression in CHO mammalian cell lines. Research Square. 2023. [CrossRef]
- Plewczynski D, et al. Sequencing, cloning and expression of the panfilovirus glycoprotein specific recombinant mAb 8C12F11 in a CHO mammalian cell line. Research Square. 2023. DOI: https://doi.org/10.21203/rs.3.rs-2532766/v1.
- Wiśniewski M,et al. Use of in silico approaches, synthesis and profiling of Pan-filovirus GP-1,2 preprotein specific antibodies. Brief Funct Genomics. 2024 Apr 11:elae012. doi: 10.1093/bfgp/elae012. Epub ahead of print. PMID: 38605526.
- Dhanda SK, et al. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019 Jul 2;47(W1):W502-W506. doi: 10.1093/nar/gkz452. PMID: 31114900; PMCID: PMC6602498.
- de Araújo LP, de Melo Santos NC, Corsetti PP, de Almeida LA. Immunoinformatic Approach for Rational Identification of Immunogenic Peptides Against Host Entry and/or Exit Mpox Proteins and Potential Multiepitope Vaccine Construction. J Infect Dis. 2024 Mar 26;229(Supplement_2):S285-S292. [CrossRef] [PubMed]
- Wang Y, Yang K, Zhou H. Immunogenic proteins and potential delivery platforms for mpox virus vaccine development: A rapid review. Int J Biol Macromol. 2023 Aug 1;245:125515. doi: 10.1016/j.ijbiomac.2023.125515. Epub 2023 Jun 21. PMID: 37353117; PMCID: PMC10284459.
- de Araújo LP, Silva EN, Corsetti PP, de Almeida LA. Shared immunogenic epitopes between host entry and exit proteins from monkeypox and Alaskapox viruses. Lancet Microbe. 2024, 5(7):624-625. doi: 10.1016/S2666-5247(24)00095-8. Epub 2024 Apr 20. PMID: 38653317.
- Lu GW, et al. Structure of the vaccinia virus A16/G9 sub-complex from the orthopoxvirus entry-fusion complex. (2023) Emerg Microbes Infect 2023, 12: 2179351-2179351.
- Yefet R, et al. Monkeypox infection elicits strong antibody and B cell response against A35R and H3L antigens. iScience. 2023, 26(2):105957. doi: 10.1016/j.isci.2023.105957. Epub 2023 Jan 13. PMID: 36687315; PMCID: PMC9838220.
- Zuiani A, et al. A multivalent mRNA monkeypox virus vaccine (BNT166) protects mice and macaques from orthopoxvirus disease. Cell. 2024, 187(6):1363-1373.e12. doi: 10.1016/j.cell.2024.01.017. Epub 2024 Feb 15. PMID: 38366591.
- Ahmed SF, Sohail MS, Quadeer AA, McKay MR. Vaccinia-Virus-Based Vaccines Are Expected to Elicit Highly Cross-Reactive Immunity to the 2022 Monkeypox Virus. Viruses. 2022, 14(9):1960. doi: 10.3390/v14091960. PMID: 36146766; PMCID: PMC9506226.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).