1. Introduction
In the progress of personalised medicine, artificial intelligence has achieved significant breakthroughs through advanced imaging technology. Radiomics, a high-throughput mining technique for extracting quantitative image features from standard medical imaging, has shown its increasing importance in cancer research. Imagomics provides a powerful tool for modern medicine by rapidly developing and validating image-based signatures from medical imaging data through complex image analysis tools and applying them to clinical decision support systems to improve the accuracy of diagnosis, prognosis and prediction.
The imagomics process involves multiple steps, from image preprocessing and feature extraction to data analysis, each requiring precise and efficient processing. However, imaging omics also need help with some things, such as data standardisation problems, feature selection complexity, and clinical relevance validation. However, these challenges also present opportunities for further research, particularly in developing new analytical methods and validation techniques.
Although imaging has demonstrated great potential to improve clinical decision-making, particularly in diagnosing and treating cancer patients, the field currently lacks standardised assessments of the scientific integrity and clinical relevance of rapidly evolving imaging research. For imaging omics to become a mature discipline, strict evaluation criteria and reporting guidelines must be established. This paper aims to guide imaging omics research to meet this urgent need and promote its application and development in personalised medicine.
2. AI-Driven Data Visualization Technologies
2.1. Definition of Medical Imaging
Imaging omics originated in the field of oncology and was first formally proposed by Dutch scholar Lambin P et al. in 2012; which is the high-throughput extraction of many image features describing tumour characteristics. In the same year, Kumar V et al. further improved the concept, that is, imaging omics is a high-throughput extraction of many high-dimensional quantitative image features from MRI, PET and CT images and analysis. Imaging omics transforms traditional medical images into minable high-throughput image features, which can be used to describe spatial and temporal heterogeneity in images quantitatively, reveal image features that the naked eye cannot recognise, effectively transform medical images into high-dimensional identifiable feature Spaces, and conduct statistical analysis on the generated feature Spaces, to establish models with diagnostic, prognostic or predictive value. Provide valuable information for personalised treatment.
The emergence of imaging is derived from the development of radiology. With the rapid growth of precision and quantitative medical imaging technology and the continuous update of image recognition technology and data algorithms, the mining and analysis of medical image big data have been realised, greatly expanding the information volume of medical images.
Based on the characteristics that high-throughput features can be obtained after texture analysis of image information and inspired by genomics and tumour heterogeneity, Dutch scholar Lambin proposed the concept of Radiomics in 2012 based on previous scholars’ work. Lambin believes that “high-throughput extraction of a large number of features from medical images, through automatic or semi-automatic analysis methods to transform imaging data into high-resolution mining data space” Medical images can be a comprehensive, non-invasive, quantitative observation of tumour spatial and temporal heterogeneity. Kumar et al. extended the definition of imaging omics, which refers to the high-throughput extraction and analysis of many advanced and quantitative imaging features from medical images such as CT, PET or MRI. In the following seven years, this idea was rapidly improved and perfected by more and more scholars.
In essence, imagomics is an analytical approach that starts from the clinical problem and finally returns to the clinical problem being solved.
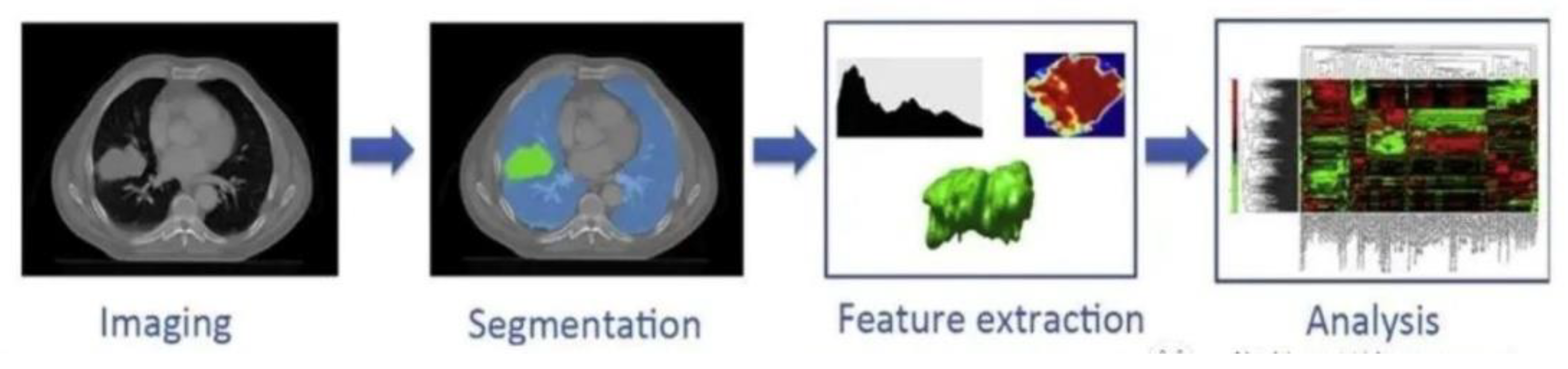
Figure 1.
Medical realisation idea of imaging omics.
Figure 1.
Medical realisation idea of imaging omics.
2.2. Application of Artificial Intelligence (AI) Technology in Imaging Omics Research
According to the National Institutes of Health (NIH), medical imaging technology plays an important role in the U.S. health care system. According to reports, in recent years, medical institutions at all levels in the United States have widely adopted advanced medical imaging technologies such as CT scanning, MRI, and X-ray to help doctors carry out accurate disease diagnosis and treatment planning. These technologies greatly improve the efficiency and quality of medical care and provide critical data support for personalized treatment and surgical planning. In addition, the United States is committed to promoting innovation in medical imaging technology through investment and research and development support to promote the integration of imaging and biomedical engineering to improve the scientific and technological level of research and clinical applications.
1. Automatic image analysis and interpretation
AI technology can automatically identify and label focal areas in medical images, reducing the burden on doctors and improving diagnostic efficiency. By training deep learning algorithms and neural networks, AI can achieve accurate image segmentation, classification, and feature extraction, helping doctors make accurate diagnoses quickly.
2. Disease prediction and personalized treatment
With big medical data and AI technology, imaging omics research can uncover disease patterns and risk factors hidden in large-scale imaging data. Through the comprehensive analysis of patient images and relevant clinical data, AI can predict the progression and outcome of the disease and guide individualized treatment.
3. Assist decision-making and surgical planning
AI technology plays an essential role in surgical planning and assisting decision-making. Through three-dimensional reconstruction and simulation of patient images, combined with comparison and reference of big data, AI can help doctors develop the best surgical plan, reduce risks, and improve surgical results. In medical diagnosis, doctors usually need to combine clinical manifestations, medical images, and other aspects of information to identify and diagnose. Medical imaging AI technology can effectively help doctors diagnose diseases, and its application range is extensive.
For example, AI technology can diagnose tumors, fractures, and lung diseases in radiology and medical imaging by analyzing data from X-rays, CT, and MRI images. In ultrasound imaging, medical imaging AI technology can distinguish between tumors and benign lesions by extracting the characteristics of normal and abnormal structures.
3. Medical Imaging Group Personalized Medical Diagnosis
3.1. Analyze the Specific Impact of AI-Driven Data Visualization on User Experience
1. Standard medical image data acquisition and screening:
Before data collection, data should be screened according to a clear research direction, such as differential diagnosis of a tumor or pneumonia classification and whether the selected image data has the gold standard of pathological or etiological detection for comparison. When evaluating the efficacy of imaging, whether or not there is a matching of image data corresponding to multi-stage treatment is also essential.
Image data include CT, MRI, PET, ultrasonic images, etc. In an imaging omics study, the objective acquisition method of image data is preferably constant: the same equipment, the same sequence, the same parameters, and maintaining the same state during scanning, but this is almost impossible. Therefore, we must screen the data according to the research direction to perform corresponding differential diagnosis, laboratory control and image data matching.
2. Image segmentation:
The technique and process of dividing an image into specific areas with unique properties and extracting objects of interest. Depending on the purpose of the study, the target of image segmentation can be a lesion, normal reference tissue or tissue anatomical structure, and it can be a three-dimensional or two-dimensional region. The subsequent analysis and research of imagomics are carried out around these segmented regions from the image.
3. Feature extraction:
Traditional imaging doctors generally read images by the naked eye and rely on their own intuitive and long-term experience to diagnose diseases, so there will be different degrees of differentiation due to different experiences. However, these image data contain a lot of objective potential information; at this time, it is necessary to extract high-throughput features, the core step of imagomics, to analyse the substantial attribute of the region of interest so that the diagnosis based on this substantial attribute can achieve a high degree of identity.
The core step of imaging omics is to extract high-throughput features to quantitatively analyse the fundamental properties of ROI. Based on the Image Biomarker Standardization Initiative (IBSI) [
4], image omics features are usually divided into shape features, texture-based features, high-order features, and features based on model transformation.
4. Feature selection:
Through the above feature extraction, the number of features extracted may range from several hundred to tens of thousands, and only some features are associated with the clinical problem to be solved. On the other hand, in practice, due to the relatively large number of features and the small number of samples, it is easy to cause the phenomenon of overfitting in the subsequent model, thus affecting the accuracy of the model. Feature selection directly selects appropriate subsets from feature sets according to some evaluation criteria or generates new feature sets through linear/nonlinear combinations of original features and selecting appropriate subsets from new feature sets.
5. Modeling and application:
(1) For doctors’ specific clinical problems, establish a prediction model based on the critical features screened by the above features or further combine features other than imaging omics (such as clinical signs, pathology, and genetic test data).
(2) Prediction model: For the analysis, screening, research and establishment of more than hundreds to 1,000 features extracted, the established model can predict data by comparing features.
(3) most imaging omics are still in the research stage and have yet to be used in clinical practice.
3.2. Imaging Omics Infrastructure
Imaging omics is a discipline that uses high-throughput feature extraction technology to extract a large amount of quantitative data from medical images, which shows great potential in deepening medical knowledge and expanding the application of imaging technology. Through imaging omics, researchers and clinicians can extract more biological information from traditional images, which has important implications for achieving higher precision diagnosis and treatment. A shared vision of interdisciplinary precision medicine must be constructed to exploit the potential of imaging omics fully.
In this shared vision, extracted imaging features must be stored in a searchable database. These databases will not only enable the unprecedented potential of radiological imaging data (RLHC), represented by routine standard treatment images, but also advance the entire field of medicine by dynamically capturing multimodal data and sharing knowledge across departmental and institutional boundaries. With such an RLHC network, researchers can accumulate enough data sets to provide a solid statistical foundation for developing and validating imaging models.
3.3. Realization of Personalised Medicine
The key to achieving personalized medicine is to utilize the characteristics of “big data” to optimize the application of imaging omics and RLHC (Radiology-Led Health Care). This includes the amount of data, the kind of data, the speed of data acquisition, and the accuracy of data. The following are the importance of these characteristics and their specific applications in the realization of personalized medicine:
1. Data Volume
Data volume is crucial in advancing personalized medicine through imaging omics and Radiology-Led Health Care (RLHC). The quality of knowledge derived from medical studies is directly proportional to the number of patients included in the dataset. Larger patient cohorts enable a more comprehensive exploration of medical phenomena and patterns, enhancing research accuracy and reliability. Moreover, increased data volume allows for acquiring a broader range of variables during model development. This richness of information not only facilitates robust model training and validation but improves the model’s ability to generalize findings across diverse patient populations. Additionally, large datasets are invaluable for studying rare disease variants, as they provide sufficient data points to understand and treat these conditions effectively.
2. Variety of Data
The diversity of data types is critical in tailoring treatment decisions in personalized medicine. By incorporating various treatment modalities and patient characteristics—such as genetics, lifestyle factors, and medical histories—into analyses, healthcare providers gain a comprehensive perspective for personalized treatment planning. Interdisciplinary collaboration is essential in managing diverse datasets, as it involves expertise from different fields to analyze and interpret multifaceted data effectively. This collaboration fosters a holistic approach to patient care, ensuring that treatments are tailored to individual needs based on a nuanced understanding of diverse data inputs.
3. Velocity of Data Acquisition and Data Veracity
The velocity of data acquisition is vital for timely knowledge generation and real-time decision support in clinical settings. Rapid data acquisition enables healthcare professionals to collect and analyze information promptly, facilitating quick insights that can guide immediate clinical interventions. Real-time data processing capabilities enhance clinical efficiency by providing timely support for decision-making processes, thereby improving patient care outcomes. Furthermore, data veracity, or the accuracy and reliability of data, is crucial for developing precise treatment strategies. High-quality data ensures the validity of analytical results, reducing the likelihood of misdiagnoses or treatment errors. This precision is essential in personalized medicine, where accurate data-driven insights lead to tailored treatment plans that optimize patient outcomes.
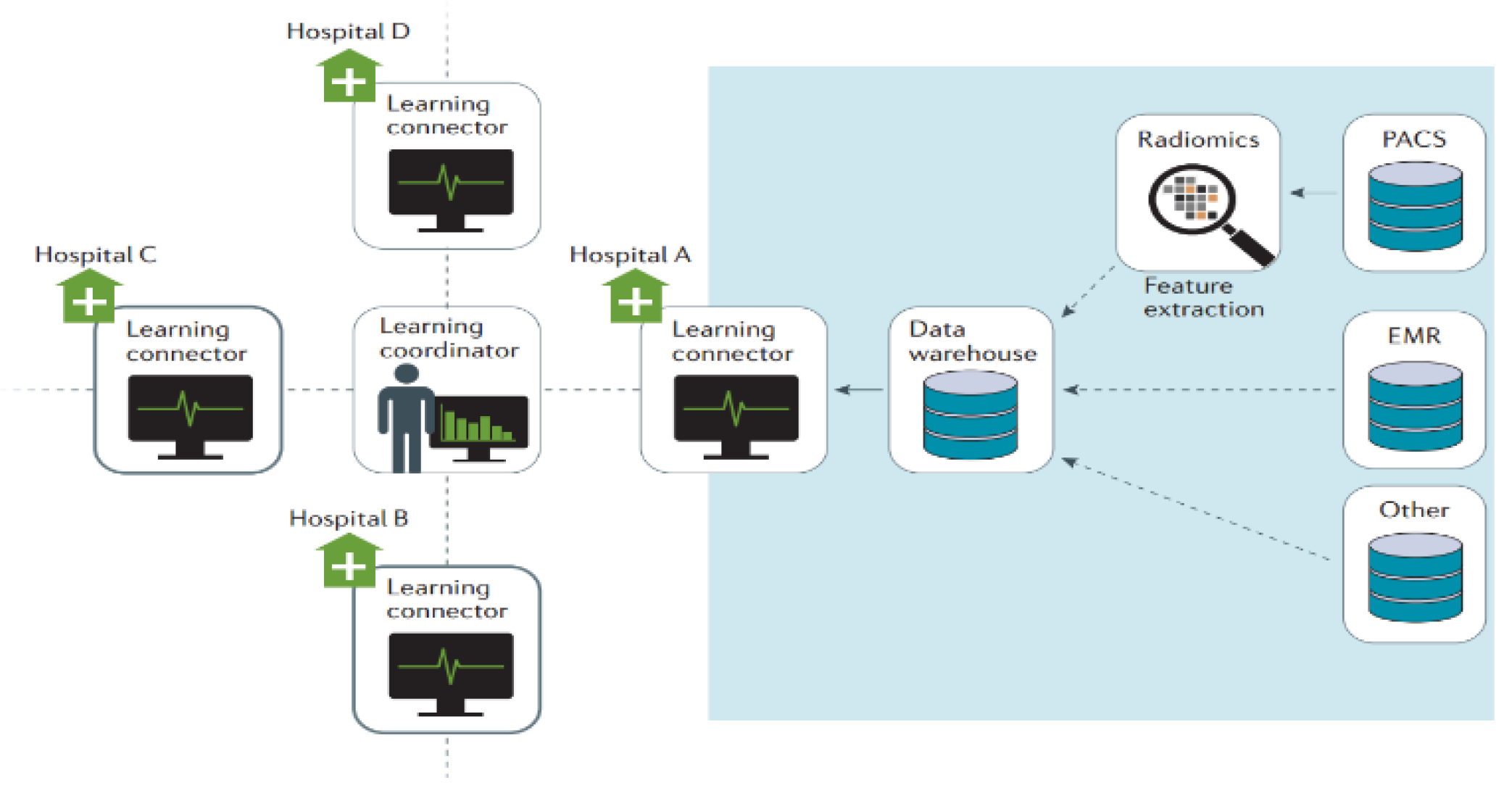
Figure 1.
Architecture diagram of Velocity of data acquisition.
Figure 1.
Architecture diagram of Velocity of data acquisition.
3.4. Challenges and Solutions to Data Sharing
1. Barriers to Data Sharing
Data sharing in healthcare faces numerous challenges, including insufficient human resources dedicated to data management, cultural and linguistic differences affecting data interpretation and exchange, inconsistent methods for recording and storing data, and concerns over the political and academic value of data. Additionally, issues such as reputational risks associated with data breaches and legal complexities surrounding privacy regulations further complicate efforts to share healthcare data effectively.
2. Strategies and Success Stories
Despite these challenges, several successful initiatives demonstrate effective strategies for overcoming barriers to data sharing. For instance, CancerLinQ, initiated by the American Society of Clinical Oncology (ASCO), has streamlined data centralization efforts in oncology by facilitating the sharing of comprehensive cancer treatment data among healthcare providers and researchers. This centralized approach has significantly advanced both research and clinical practice in cancer treatment, enabling insights from large datasets to inform personalized patient care strategies.
Another successful example is the WorldCat project, which has connected radiotherapy facilities across multiple countries through innovative data integration techniques. By harmonizing data from diverse sources, WorldCat promotes global collaboration and knowledge sharing in radiotherapy practices, demonstrating the potential of novel data combination approaches to overcome cultural and logistical barriers to data sharing.
3. Technological Solutions and Collaborative Initiatives
Advanced Information and Communication Technology (ICT) solutions offer standardized, simplified approaches to synchronizing Radiology-Led Health Care Systems (RLHCS) across different healthcare centers without compromising sensitive patient data. These ICT solutions facilitate secure and efficient data sharing practices, ensuring that healthcare providers can collaborate seamlessly while adhering to strict privacy and security standards.
Moreover, collaborative projects such as the Cancer Imaging Archive (TCIA), the Quantitative Imaging Network (QIN), the Quantitative Imaging Biomarker Consortium (QIBA), and Quantitative Cancer Imaging (QuIC-ConCePT) exemplify global efforts to enhance data sharing in healthcare. These initiatives focus on developing standardized protocols for imaging data collection, analysis, and sharing, fostering interdisciplinary collaboration, and accelerating advancements in diagnostic imaging and personalized medicine.
In conclusion, while challenges persist, innovative strategies and technological advancements are paving the way for improved data-sharing practices in healthcare. By leveraging successful case studies and collaborative frameworks, healthcare systems can overcome barriers to data sharing, promote transparency, and harness the collective power of data to advance patient care and research outcomes globally.
Principle of personalised medicine
1. Data Integration and Sharing: Establish a global data-sharing network to integrate diverse medical data across institutional and national borders.
2. Interdisciplinary cooperation: Multiple data types for comprehensive analysis are used to develop accurate diagnosis and treatment plans.
3. Advanced Technical support: Use advanced information and communication technologies and data analysis tools to ensure efficient data acquisition, processing and application.
4. Regulations and Privacy Protection: Develop and comply with strict regulations to ensure data privacy and security and build patient and institutional trust in data sharing.
Through these measures, imaging and RLHC can better realise their potential in personalised medicine to provide patients with more accurate and efficient medical care.
4. Conclusion
The future Image Archiving and Imaging Omics Knowledge System (PARKS) demonstrates the great potential of imaging technologies for personalised and precision medicine. The PARKS system can automatically identify, segment, and extract features from areas of interest, especially when previous images related to the same patient are accessible. This automated process improves the efficiency of image analysis and significantly enhances the role of clinical decision support systems (CDSS) in diagnosis, prognosis, and treatment. By uploading quantitative image features to a shared database and comparing them with historical images, the PARKS system can automatically extract and analyse these features to provide more precise diagnosis and treatment recommendations. This capability has been proven at the technical, scientific and clinical levels, driving the development of personalised medicine.
Today, most image archiving and communication systems can register current images with previous images and perform user interactive segmentation. On this basis, the PARKS system achieves higher automation and intelligence, improving data processing and analysis efficiency and accuracy. In the future, imaging omics will focus on creating the infrastructure for a robust radio-image-guided health care (RLHC) network. This infrastructure will support the development and validation of models, ensuring that imaging omics technology plays a more significant role in clinical practice. Through advanced ICT and data analysis tools, the PARKS system will overcome traditional barriers to data sharing, enabling synchronisation and data sharing of RLHC across centres.
In summary, the PARKS system provides powerful technical support for personalised and precision medicine by automating the identification and processing of historical patient image data. This improves the diagnostic accuracy and consistency of medical images and promotes the progress of personalised medicine. In the future, the profound combination of imaging omics and artificial intelligence will continue to promote the development of personalised medicine and provide patients with more accurate and efficient medical services.