1. Introduction
Human physiology has co-evolved with the microbial life that make up the colonic microbiome, and their presence is essential to human health[
1]. The microbiome includes the bacteria, archaea, fungi, virus, protists, as well as their genomic material and metabolites. They provide a vast repertoire of symbiotic function, with particular importance to metabolic and immunogenic health. Our pre-agrarian ancestors would have, phylogenetically, evolved in close contact with bacterial species typically found in soil, un-treated water, and domesticated animals – for example, members of the phyla Bacillota, Proteobacteria, Actinobacteria and Verrucomicrobia[
2,
3]. Indeed, spore-forming bacteria (i.e. Bacillota) make up 60% of the colonic microbiota[
4]. Moreover, recent studies have suggested that the environmentally acquired aerobic spore-formers (EAS-Fs) are highly viable and transmissible to the colonic microbiome, and play an important role in health and maintaining a diverse microbiome[
5,
6,
7,
8,
9]. These EAS-Fs, often cultured from healthy human faecal samples, have been negatively correlated to the genesis of several human pathologies – such as, inflammatory bowel disease (IBD), asthma, and diabetes[
8,
10,
11,
12,
13]. The Old Friend’s Hypothesis is a leading theory that explains the pathogenesis of many chronic diseases seen in the West, links the absence of these EAS-Fs with the “diseases of affluence. The theory posits that an absence of the bacteria we have co-evolved with, leads to an increased risk of metabolic and immunogenic pathology. The modern “Western” lifestyle and diet specifically restricts exposure to these species[
14,
15,
16].
Interestingly, the evidence to date suggests that the aerobic EAS-Fs are
transient colonisers, and without regular replenishment the number of viable cells quickly decline[
17]. Thus, regular exposure is required to maintain colonies, making the environment a vital reservoir for allochthonous transmission[
9,
18]. Despite the growing scientific interest in the importance of these environmentally acquired bacteria, very few animal studies have been conducted.
Due to the ubiquitous nature of spore forming bacteria, controlling for them on surfaces is difficult, and may have been an overlooked variable in animal models of disease and are often present in standard animal husbandry conditions[
19]. Moreover, due to their heat-stability they can persist despite pasteurisation[
20]. Germ-free (GF) animal models have typically been used to model for the effect of specific microbiomes. These conditions allow for the controlled introduction of specific species to observe the clinical impact, providing valuable insights into numerous pathogenic processes[
21]. However, GF modelling has the significant limitation of being critically removed from the normal (non-sterile) physiology of humans, and as such the applicability of the findings becomes limited, and hard to interpret.
Our hypothesis is simple: living in hygienic conditions that restrict the exposure of these transient spore-forming bacterial Old Friends will increase the risk of developing diseases typified in the West (e.g. obesity, IBD, diabetes, hypercholesterolaemia, and non-alcoholic fatty liver disease (NAFLD)). To address this, we have designed and validated a new animal model that restricts exposure to environmentally acquired bacteria without the need for sterile germ-free conditions. This has been termed
Super Clean, to distinguish it from the true sterile conditions of GF animal studies. The purpose is to exclude environmentally acquired bacteria and represent the hygienic conditions that is typical of the Western Lifestyle, allowing a new technique to model health conditions whilst not hindering the normal physiological development of the animal[
22]. We will compare this new model to standard laboratory conditions over an 18-month period and report on several clinical, histological, genomic expression, and biochemical parameters for the aforementioned pathologies. We will comment on how this model can give insights into the role of environmentally acquired bacteria on health and future research potential.
2. Materials and Methods
The
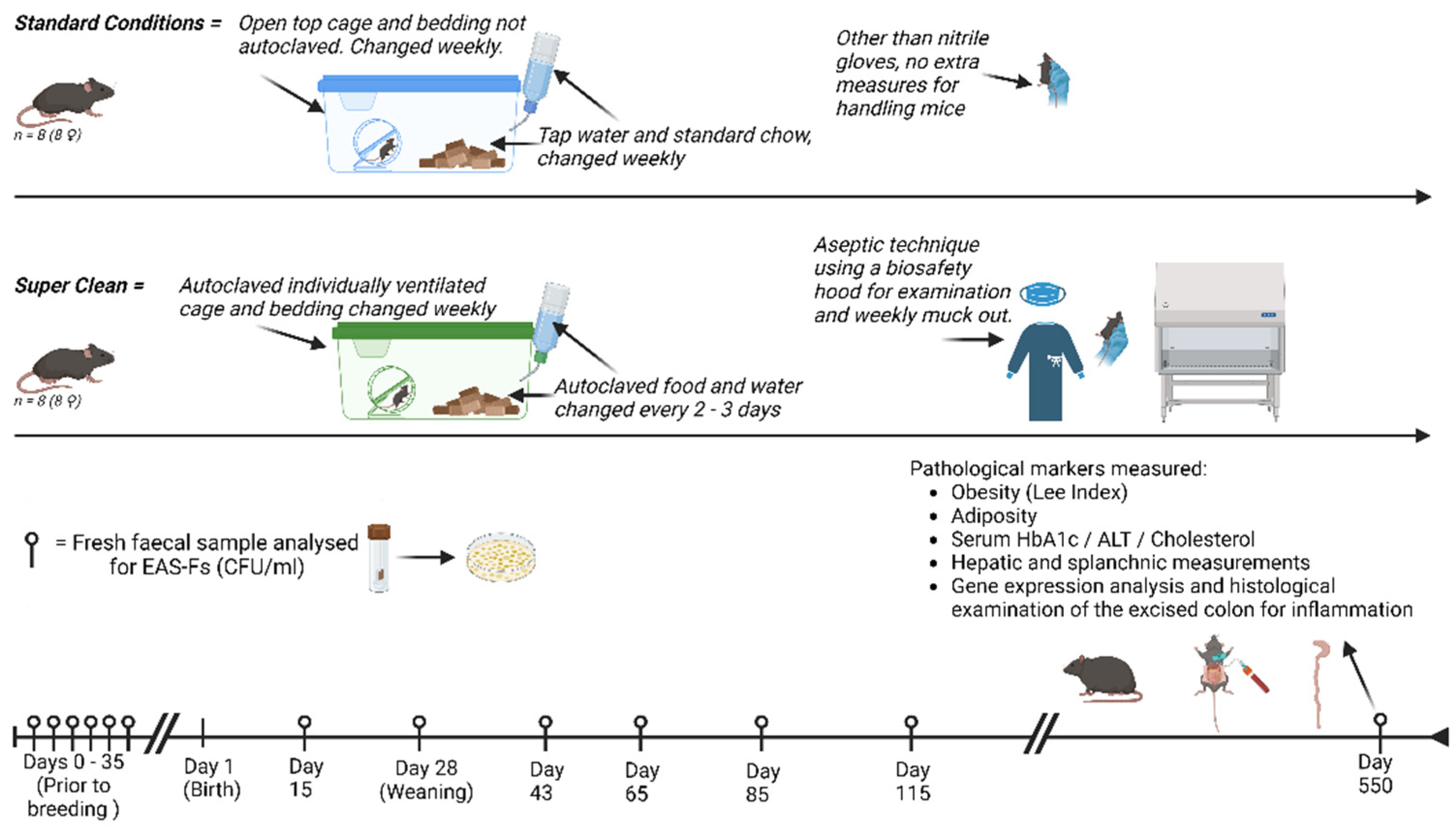
Animals. C57 BL/6 mice (Charles River, UK) were bred in-house, with the mothers being acclimatised to either super clean (SC) or standard laboratory (SL) conditions prior to breeding. Female pups were selected and randomly allocated to either SC or SL conditions from the day of birth, with eight pups per group. After reaching weaned maturity (28 days), the mothers were removed, and the litter remained as a group until the end of the study,
Figure 1. Due to the length of this study, females were exclusively selected to avoid in-group violence and stress that is typically seen with male-only litter mates[
23]. Ethical approval was from our institution’s Animal Welfare and Ethics Review Board (AWERB) and approved by the Home Office (UK).
Normal Laboratory Conditions. As per our standard animal house protocol – mice (n = 8), were housed in upto five mice per cage (MB1, NKPisotec), with ad libitum water (tap) and chow (5LF5 LabDiet). They were exposed to twelve hourly light / dark cycles. Standard bedding, environmental stimulants, and bottles were used (not autoclaved). They were mucked on a weekly basis. (n = 8).
Super Clean Design. The mice (n = 8) were housed in group of up to five mice per unit. They were housed in individually ventilated, HEPA filtered, cages (IVC) (Tecniplast Group, Italy), with a reported 99.9999937% efficiency at restricting contamination from the environment. To maintain relative sterility, the cage, bedding materials, environmental stimulants, drinking bottles were all autoclaved prior to use each week. Tap-water was autoclaved, and irradiated sterile chow (5L0D, LabDiet) was changed every 2 – 3 days, both ad libitum. An aseptic protocol was designed for handling the mice during mucking out – employing sterile gloves with work carried out under a biosafety extraction cabinet. The mothers were housed under SC conditions for 35 days prior to breeding, in the attempt to reduce aberrant exposure to environmental bacteria in their offspring. For quality control of the SC model, intermittent samples of the cage surface, food and water were performed to ensure the absence of environmental contamination, growth was assessed by plating the heat-treated samples on DSM agar, as described below. This demonstrated no contamination.
Detection of aerobic spore forming bacteria. Faecal samples were taken, using the clean catch method, at several points throughout the first 100 days of the experiment, and at the end of the experiment. These were heat treated (65oC, 45 min.), serially diluted, and plated on complex media agar (Difco Sporulation Medium (DSM), 37oC, 24h). Colony forming units (CFU) were calculated and inspected under the microscope to inspect the morphological appearance.
Clinical Measurements. All measurements reported are taken at the end of the experiment when the mice reached 18 months of age, the point at which they are regarded as an
old adult[
24]. Each mouse was weighed (g), and the naso-anal length (mm) was measured. To classify obesity, the cube root of body weight was divided by the naso-anal length to give the Lee Index (LI)[
25], analogous to the body mass index (BMI) used to measure human obesity. Subcutaneous fat was measured (mm) from the thickest part of the posterior inguinal white adipose tissue (WAT), measurements were an average of the left and right adipose deposits per mouse. To measure visceral fat, the gonadal fat pad was completely excised and weighed (g). A pan-proctocolectomy was then performed and the caecal-anal length was measured (cm). The caecum was then transected from the colon, being careful not to spill its faecal content, and weighed (g). The liver and spleen where harvested and individually weighed (g).
Biochemical and Histological Measurements. A serum sample, from a cardiac puncture, was taken using paediatric K3EDTA tubes and sent for total cholesterol (mmol/l) and Alanine Transaminase (ALT)(U/L) analysis (Royal Veterinary College diagnostic laboratory, UK). A second serum sample was taken for HbA1C (%) measurement (BHR Pharmaceuticals, UK). Samples of the liver were taken and fixed in 10% (v/v) neutral buffered formalin. They were histologically processed and stained with H&E and inspected for evidence of non-alcoholic fatty liver disease (NAFLD).
Gene expression analysis. The rectosigmoid was sectioned to provide a 100 mg sample, and placed in 1 ml TRIzol (Invitrogen), and stored at -80oC for later work. RNA was extracted using chloroform / isopropanol precipitation, as per the manufacturers protocol. Residual gDNA was removed and reverse transcription performed (QuantiTect Reverse Transcription, Quigen). Quality was checked using spectrophotometry (Nanodrop, Thermo Scientific). RT-qPCR followed as per the manufacturers protocol (QUantiNova Sybr Green PCR, Qiagen & RotorGene 6000 Thermocycler, Corbett Research). Primer sequences, Supp. Figure S1, were chosen from our previous work on colonic inflammation. To allow comparative expression fold-change between the SC and SL groups, a third set of samples were taken from 5 female control mice, 14 weeks old, that were housed under SL conditions.
Statistical Analysis. All animals were included in the analysis. GraphPad Prism (Version 9.4.1 for Windows, Dotmatics) was used. Statistical significance was accepted as p < 0.05. Independent-samples t-test was employed to determine statistical difference between the two groups. There were no outliers in the data, as assess by inspection of boxplots. All results were normally distributed, as assessed by Shapiro-Wilk’s test. Data is presented as mean ± standard deviation, against the “standard laboratory” group. For RT-qPCR results the data was transformed logarithmically for statistical analysis, and are presented as an expression fold-change, to the log2.
3. Results
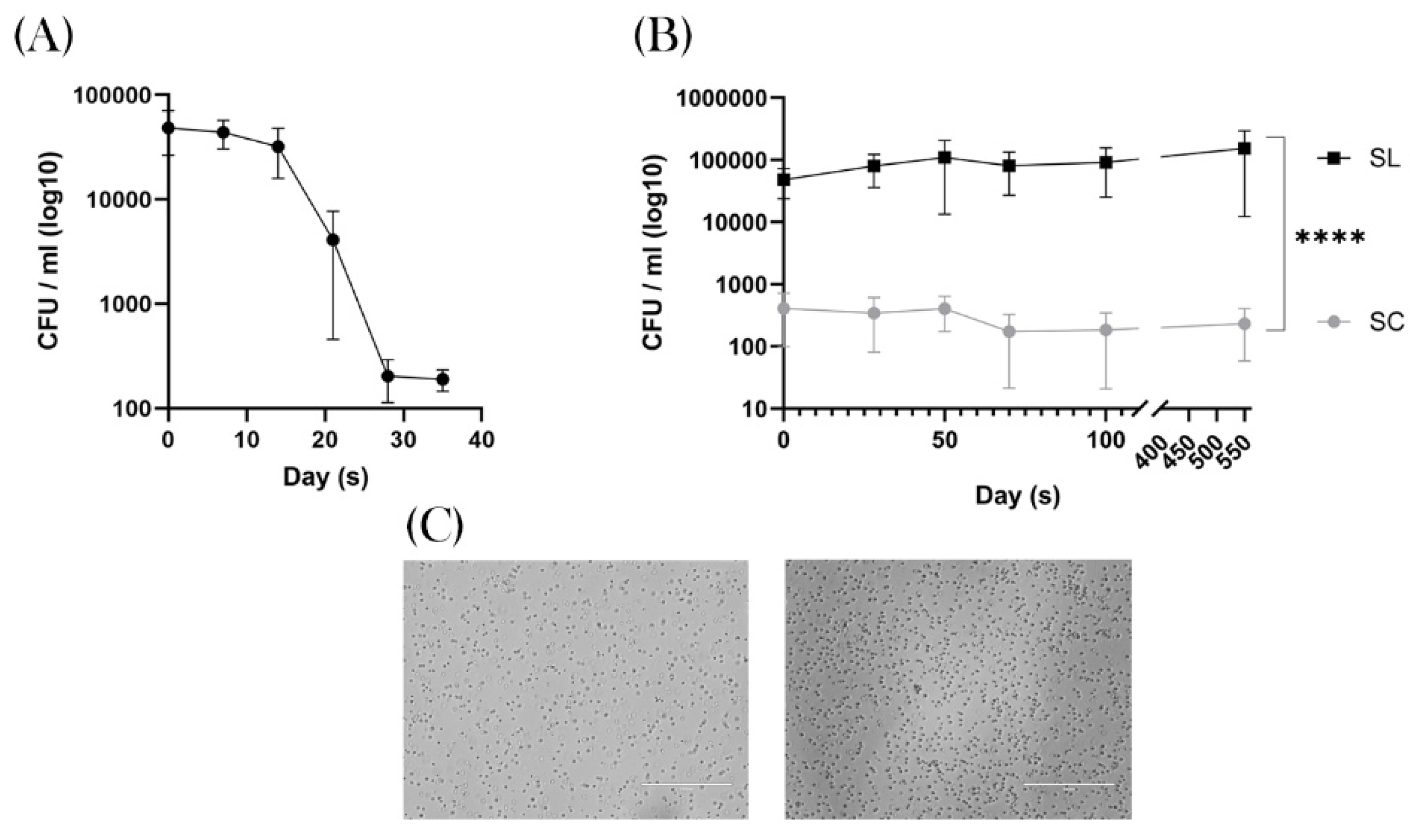
Super Clean conditions effectively removed colonic spore-forming bacteria
The adult breeding mice (prior to breeding) demonstrated a mean of 5.5x10
4 CFU/ml (95% CI = 8.4x10
4 – 2.7x10
4). For the group who were then placed under SC conditions this started to decrease after 21 days, reaching a nadir of 2.06x10
2 CFU/ml (95% CI = 293 – 119) after 35 days of SC exposure,
Figure 2A.
After breeding, once the pups were mature for faecal sampling (~15 days), the SL mice demonstrated a spore count ranging from 4.42 x 10
4 – 1.06 x 10
5 CFU/ml. In contrast the SC group had a significantly lower level of 2.36 - 4.22 x 10
2 CFU/ml (
p = <0.0001 ; 95% CI 55320 – 102814), with two mice demonstrating 0 spores during the experiment,
Figure 2B. This quantitative divergence of spore-forming bacteria between the two groups remained significant throughout the experiment, demonstrating that SC conditions successfully removed this group of bacteria from the experiment. When examining the bacteria under the microscope these were predominantly rod-like bacillus, in keeping with the phylum
Bacillota, Figure 2C. 16SrRNA analysis was not performed, so we are unable to describe the genus or species composition of this microbiomic shift.
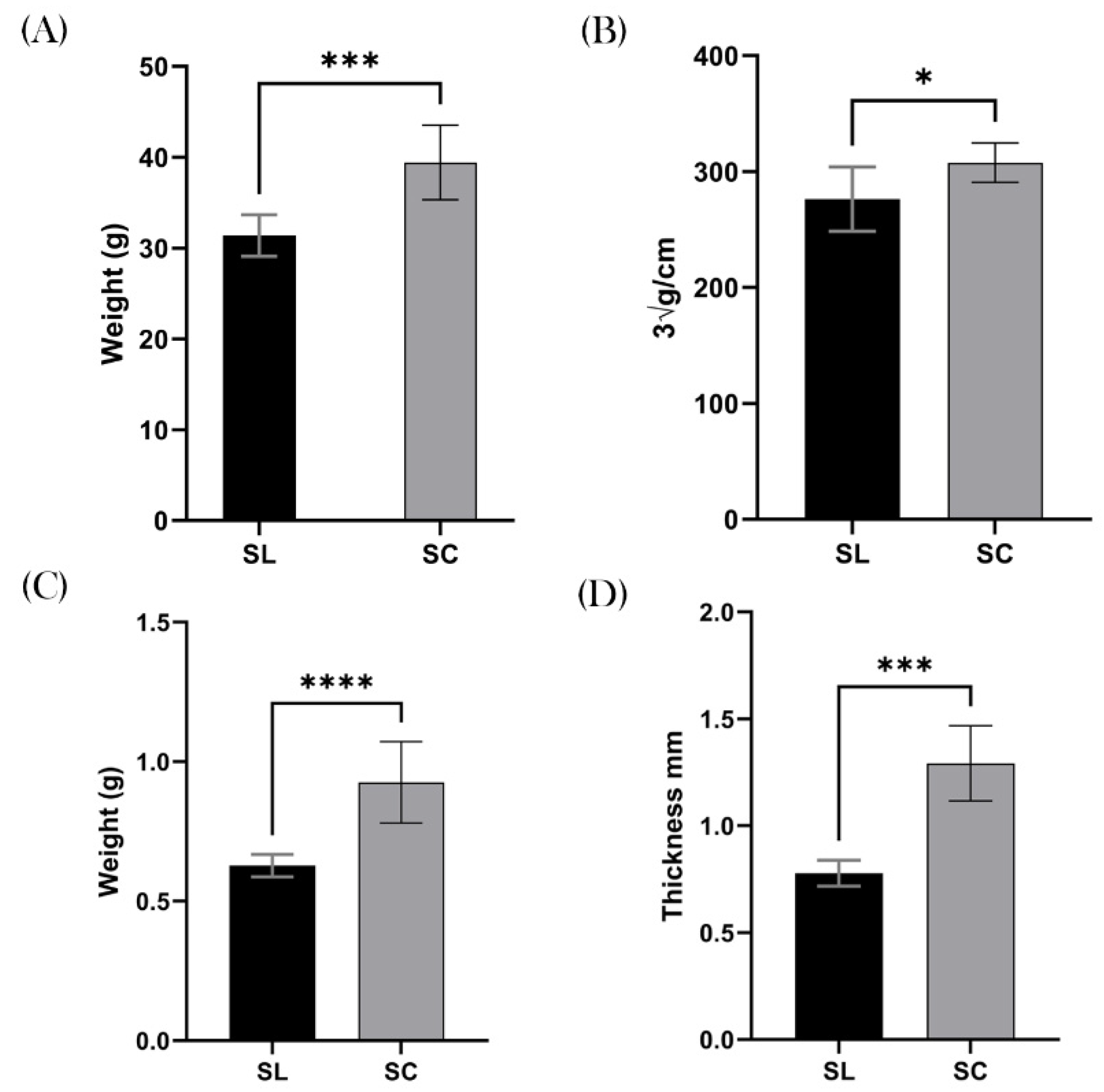
Weight & Obesity
The SC mice demonstrated a significant increase in total body weight compared to their SL counterparts, with a mean weight of 39.4 g and 31.4 g respectively (
p = 0.0003 ; 95% CI 4.466 – 11.61),
Figure 3A. When calculating the LI, the results remained significant with the SC group demonstrating a greater body mas index compare to SL. For example the mean SL was 276.3 ∛g/cm and SC was 307.7 ∛g/cm (
p = 0.0163 ; 95% CI 6.732 – 56.07),
Figure 3B.
In keeping with this finding, measurements of subcutaneous fat was markedly increased in the SC group, suggesting the increased weight was due to adipose tissue. The mean gonadal fat pad weight in the SC group was 0.93g, and mean for the SL group was 0.63g (
p = <0.0001 ; 95% CI 0.1843 – 0.4132),
Figure 3C. Similarly the posterior inguinal adipose thickness was greater in the SC group, with a mean of 1.3 mm, compared to 0.8 mm in the SL group (
p = 0.0003 ; 95%CI 0.3223 – 0.7057),
Figure 3D.
Colonic Measurements
The colonic length of the SL & SC groups were similar, with mean values being 12.3 cm and 12.6 cm respectively (
p = 0.2217),
Figure 4A. However, the weight of their respective “wet” caeca was divergent, with a mean value for SL of 2.7 g and SC 3.0 g (
p = 0.0252 ; 95% CI 0.03953 – 0.5105),
Figure 4B.
When analysing the transcriptome of the two groups, there was no difference in gene expression. The two group’s samples were compared against healthy 14-week-old BALB/c mice, allowing quantitative analysis. TNF-α was expressed at higher levels in both SL and SC compared to the comparator, with a 3.6 and 3.5 log2 fold increase (
p = 0.3975),
Figure 4C. IL-6 was also found to be upregulated by 3.1, and 3.6 respectively (
p = 0.6609),
Figure 4D. In contrast IL-1β was downregulated in the SL group by 3.4, and 3.8 in the SC group (
p = 0.6063),
Figure 4E. IL-10 was upregulated in the SL group by 4.3, and 3.1 in the SC group (
p = 0.2443),
Figure 4F. MUC-2 results SL & SC were 5.1 and 6.2 respectively (
p = 0.1297),
Figure 4G. Claudin-1 results for SL & SC were 2.0 and 3.1 respectively (
p = 0.902),
Figure 4H.
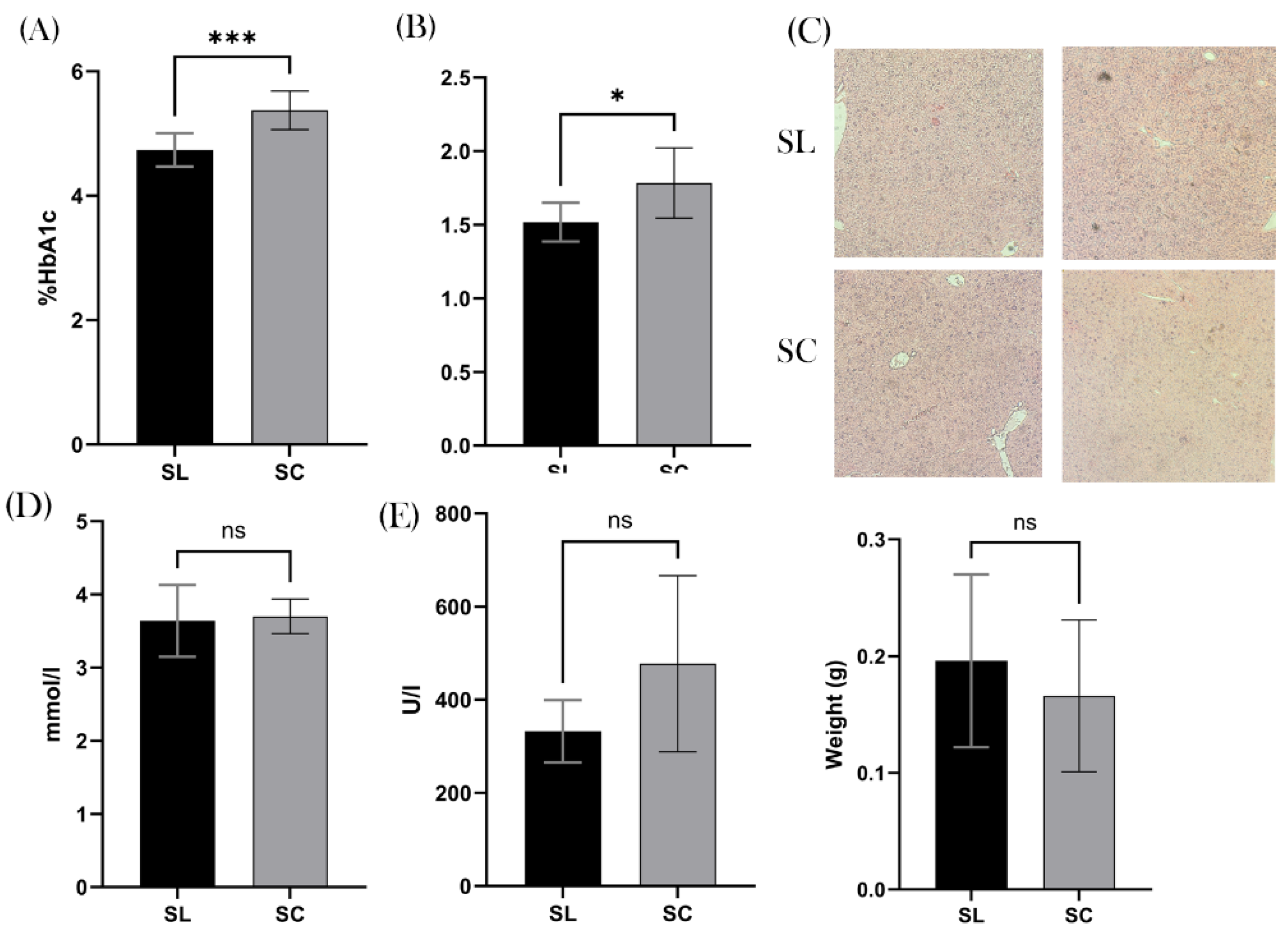
Metabolic Measurements
We found a significant difference in the % of HbA1C between the two groups, with the mean SL value of 4.74% and SC of 5.38% (
p = 0.0006 ; 95% CI 0.3270 – 0.9480),
Figure 5A. Furthermore, there was a significant difference in the average hepatic weight, with SL being 1.5 g and SC 1.8 g (
p = 0.0151 ; CI 95% 0.05998 – 0.4725),
Figure 5B. Interestingly, when inspecting the hepatic histology, none of the samples demonstrated non-alcoholic fatty liver disease (NAFLD), or indeed any other pathology,
Figure 5C. Thus, questioning the clinical relevance of this difference. We found no significant difference in serum total cholesterol or ALT levels (p = 0.8580 and
p = 0.2799),
Figure 5D-E. We also found no difference in splenic weight (
p = 0.5152),
Figure 5F.
4. Discussion
This pilot study has two pertinent findings. Firstly, our new animal model, designed to replicate hygienic conditions of the West by restricting exposure to EAS-Fs was successful. We found the Super Clean methodology significantly reduced the colony forming units of these bacterium from the faecal microbiome after 21 days. This partially corroborates former work that has suggested that these species may be transient colonisers[
17]. Previous studies have suggested that aerobic spore-formers germinate in the small intestine, and proliferate for a period of 5 - 27 days before culturable numbers start to significantly drop unless replenished[
26,
27,
28]. Instead, our data suggests that these species reduce to a “trough level” in the gastrointestinal tract, rather than an absolute eradication. This is demonstrated by the persistence seen in the SC group throughout the experiment, with a mean of 3.02x10
2 CFU/ml, compared to a mean of 8.178x10
4 CFU/ml in the mice housed under standard conditions. These species were morphologically in keeping with the spore formers of the Bacilli class (e.g.
Bacillus spp). This was expected, it has been estimated that over half of the colonic microbiome is comprised of spore forming bacteria, and environmental exposure
We believe there are two reasons why the spore counts in the SC group remained at a constant trophic level throughout the experiment, i) despite removal of external sources of these bacterial species, mice are a coprophagic animal and may be replenishing the Bacillota via faecal microbial self-reinoculation. We decided against using a typical grid-like device to prevent coprophagia as these can only limit coprophagia rather than eliminate it, furthermore it would add complexity to the SC mucking out protocol and finally there is evidence to suggest that mice have several detrimental health outcomes if coprophagia is prohibited[
29]. ii)
Bacillus, and similar genera, may not be true transient colonisers as previously thought. They may instead occupy a niche in the GIT allowing a constant attenuated presence, that is transiently increased after exposure to external sources. Indeed, several characteristics have been previously identified that would make them good candidates to fill this role, notably the formation of biofilms on the colonic epithelium[
30]. Furthermore, another finding from our study supports the observation of a reduced bacterial abundance in SC mice – the significantly increased size of the caecum compared to the SL group. It is well established that GF and antibiotic treated mice have enlarged caecum that inversely correlates with the level of bacterial abundance, due to an accumulation of mucopolysaccharides and water in the caecal lumen[
31].
The second striking finding is that the reduction in these environmentally acquired bacteria resulted in significant metabolic differences. We found that the SC mice had a mean LI, the murine equivalent to BMI, that was significantly greater than the SL mice. Using the definition for obesity as a LI ≥300 ∛g/cm, 75% of SC mice were obese, compared to 25% SL mice. Similarly subcutaneous and visceral fat were found to be significantly higher (both in thickness and weight) in the SC group compared to the SL mice. As with obesity and adipose deposits, we found glycosylated haemoglobin (HbA1C) to be significantly elevated in the SC group, suggesting impairment in glucose (5.4 vs 4.7%). HbA1C is a marker of serum glucose (mmol/l) over the prior 60 days in mice (120 in humans), and as such gives a general appreciation for periods of time with relative hyperglycaemia. Furthermore, the SC group mean was also greater than the murine reference range (<4.9%), implying that there could be clinically significant impaired glucose metabolism [
32,
33]. Interestingly, the obesity and increased visceral fat did not correlate with an increase in total cholesterol or the presence of NAFLD, with both groups having normal results. We believe these findings suggest that an absence of exposure to the environmentally acquired transient bacteria results in significant alterations to the microbiome and metabolome of the mice, leading to obesity and hyperglycaemia. This perhaps is not surprising given the increasing evidence base, and interest in, the use of spore-forming bacteria for managing obesity[
34,
35,
36]. We noted that there was an overall relative upregulation in the Th1 inflammatory cytokines, as well as genes related to colonic homeostasis, in both the SC and SL groups. We have attributed this to the aged nature of our experimental mice, a finding that has been published elsewhere[
37].
Spore-forming bacteria have the special characteristic of being highly resistant to environmental stress. This is due to their protective soluble proteins, core, cortex, coat, membranes, and soluble proteins[
38]. As a result, they can survive prolonged periods of: UV radiation, extremes of temperature, the absence of water and nutrition, predation, and the host’s immune system[
39]. They are consequently ubiquitous in the natural environment and are highly transmissible. They are less well adapted, however, to the relative anoxic conditions found in the gastro-intestinal tract. As such regular exposure is required to maintain numbers[
26,
28]. Others have hypothesised that a Western lifestyle restricts the exposure to EAS-Fs, and increases the likelihood of developing chronic metabolic and inflammatory diseases (the Old Friend’s Hypothesis)[
40]. The main genera of EAS-Fs –
Bacillus and
Clostridia are highly metabolically active, producing a plethora of secondary metabolites that have antibiotic, antifungal, antiviral, and immunomodulatory functions and appear to have efficacy in treating several pathologies[
7,
10,
41]. This might explain the epidemiological differences seen in human populations that have regular exposure to EAS-F bacteria, and those who do not. To date, we are unaware of any animal model that test this hypothesis, and believe this new model will provide invaluable insights, allowing researchers to test and find mechanistic pathways behind the Old Friends hypothesis.
There are several limitations of this pilot study that deserve mention. To culture the aerobic spore-forming bacteria from faeces, heat treatment is required[
28]. As such, we are killing all other bacterial and fungal species, and might be ignorant to important shifts in the microbiome. As we did not perform metagenomic analysis on the faecal pellets, we are unable to comment for certain that the clinical effects observed are purely down to these aerobic spore-formers rather than an un-observed species. Due to well documented inefficiency in extracting DNA from bacterial spores, and the subsequent underrepresentation of them in metagenomic analysis, we deliberately chose not to measure 16SrRNA[
4]. Furthermore, in our approach to replicate the hygienic conditions of the West by using the SC protocol we have created an extreme example of relative sterility that may not fully translate to clinical practice. However, we believe that the model is substantially more generalisable than otherwise using germ-free mice. Moreover, there is significant improvements in economic and practical viability for the researcher.
5. Conclusions
After several weeks of strict adherence to the “Super Clean” experimental conditions, environmentally acquired spore-forming bacteria are significantly reduced from the murine colonic microbiome, becoming evanescent. These experimental conditions, designed to reflect the Western lifestyle, allow a more accurate examination on the long-term health outcome from this environmental variable than has been possible with previous animal models. We found that the lack of these bacteria directly correlates to poor long-term health, resulting in an analogous metabolic syndrome that is commonly seen in the West.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper post on:
Preprints.org, Figure S1: Primer sequences for RT-qPCR.
Author Contributions
EH: Conceptualization, methodology, investigation, formal analysis and writing of the original draft. WF: Conceptualization, methodology, investigation. HH: Methodology, review and editing of the paper. PB: Methodology, review and editing of the paper. SC: Methodology, supervision, review and editing of the paper.
Funding
This work was supported by funding from the UK Medical Research Council (MRC) under grant number MR/R026262/1 to SC. Doctoral fellowship of EH was paid for by Ashford and St Peter’s NHS Foundation Trust (UK).
Institutional Review Board Statement
All experimental work on animals conformed to the Guide for the Care and Use of Laboratory Animals and the ARRIVE guidelines. The project was granted approval by the UK Home Office (Project Licence Number = Project Licence Number = 70/8276) as well as our institution’s Animal Welfare and Ethical Review Body (AWERB) (Royal Holloway University of London).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data is in the manuscript and figures. The raw data generated from this study is available upon request to the corresponding author.
Conflicts of Interest
Professor Simon Cutting is SEO of SporeGen LTD.
References
- Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med 2011;3:14. [CrossRef]
- Blum WEH, Zechmeister-Boltenstern S, Keiblinger KM. Does Soil Contribute to the Human Gut Microbiome? Microorganisms 2019;7:287. [CrossRef]
- Anthony MA, Bender SF, van der Heijden MGA. Enumerating soil biodiversity. Proceedings of the National Academy of Sciences 2023;120. [CrossRef]
- Egan M, Dempsey E, Ryan CA, Ross RP, Stanton C. The Sporobiota of the Human Gut. Gut Microbes 2021;13. [CrossRef]
- Saggese A, Baccigalupi L, Ricca E. Spore Formers as Beneficial Microbes for Humans and Animals. Appl Microbiol 2021;1:498–509. [CrossRef]
- Nicholson WL. Roles of Bacillus endospores in the environment. Cell Mol Life Sci 2002;59:410–6. [CrossRef]
- Horwell E, Vittoria M, Hong HA, Bearn P, Cutting SM. A Family of Cyclic Lipopeptides Found in Human Isolates of Bacillus Ameliorates Acute Colitis via Direct Agonism of Toll-Like Receptor 2 in a Murine Model of Inflammatory Bowel Disease. Dig Dis Sci 2024. [CrossRef]
- Ferreira WT, Hong HA, Adams JRG, Hess M, Kotowicz NK, Tan S, et al. Environmentally Acquired Bacillus and Their Role in C. difficile Colonization Resistance. Biomedicines 2022, Vol 10, Page 930 2022;10:930. [CrossRef]
- Yahya G, Ebada A, Khalaf EM, Mansour B, Nouh NA, Mosbah RA, et al. Soil-Associated Bacillus Species: A Reservoir of Bioactive Compounds with Potential Therapeutic Activity against Human Pathogens. Microorganisms 2021;9:1131. [CrossRef]
- Ilinskaya ON, Ulyanova V V., Yarullina DR, Gataullin IG. Secretome of Intestinal Bacilli: A Natural Guard against Pathologies. Front Microbiol 2017;8 SEP. [PMID: 28919884. [CrossRef]
- Benchimol EI, Kaplan GG, Otley AR, Nguyen GC, Underwood FE, Guttmann A, et al. Rural and Urban Residence During Early Life is Associated with Risk of Inflammatory Bowel Disease: A Population-Based Inception and Birth Cohort Study. American Journal of Gastroenterology 2017;112:1412–22. [CrossRef]
- Dagenais GR, Gerstein HC, Zhang X, McQueen M, Lear S, Lopez-Jaramillo P, et al. Variations in Diabetes Prevalence in Low-, Middle-, and High-Income Countries: Results From the Prospective Urban and Rural Epidemiological Study. Diabetes Care 2016;39:780–7. [CrossRef]
- Wong GWK, Chow CM. Childhood asthma epidemiology: Insights from comparative studies of rural and urban populations. Pediatr Pulmonol 2008;43:107–16. [CrossRef]
- Tasnim N, Abulizi N, Pither J, Hart MM, Gibson DL. Linking the Gut Microbial Ecosystem with the Environment: Does Gut Health Depend on Where We Live? Front Microbiol 2017;8. [CrossRef]
- Schnorr SL. The soil in our microbial DNA informs about environmental interfaces across host and subsistence modalities. Philosophical Transactions of the Royal Society B: Biological Sciences 2020;375:20190577. [CrossRef]
- Rook GAW. 99th Dahlem Conference on Infection, Inflammation and Chronic Inflammatory Disorders: Darwinian medicine and the ‘hygiene’ or ‘old friends’ hypothesis. Clin Exp Immunol 2010;160:70–9. [CrossRef]
- Hong HA, To E, Fakhry S, Baccigalupi L, Ricca E, Cutting SM. Defining the natural habitat of Bacillus spore-formers. Res Microbiol 2009;160:375–9. [PMID: 19589385. [CrossRef]
- Hong HA, To E, Fakhry S, Baccigalupi L, Ricca E, Cutting SM. Defining the natural habitat of Bacillus spore-formers. Res Microbiol 2009;160:375–9. [CrossRef]
- Gopal N, Hill C, Ross PR, Beresford TP, Fenelon MA, Cotter PD. The Prevalence and Control of Bacillus and Related Spore-Forming Bacteria in the Dairy Industry. Front Microbiol 2015;6. [CrossRef]
- Gómez-Jódar I, Ros-Chumillas M, Palop A. Effect of heating rate on highly heat-resistant spore-forming microorganisms. Food Science and Technology International 2016;22:164–72. [CrossRef]
- Park JC, Im S-H. Of men in mice: the development and application of a humanized gnotobiotic mouse model for microbiome therapeutics. Exp Mol Med 2020;52:1383–96. [CrossRef]
- Broussard JL, Devkota S. The changing microbial landscape of Western society: Diet, dwellings and discordance. Mol Metab 2016;5:737–42. [CrossRef]
- Weber EM, Zidar J, Ewaldsson B, Askevik K, Udén E, Svensk E, et al. Aggression in Group-Housed Male Mice: A Systematic Review. Animals 2022;13:143. [CrossRef]
- Yanai S, Endo S. Functional Aging in Male C57BL/6J Mice Across the Life-Span: A Systematic Behavioral Analysis of Motor, Emotional, and Memory Function to Define an Aging Phenotype. Front Aging Neurosci 2021;13. [CrossRef]
- Lee MO. DETERMINATION OF THE SURFACE AREA OF THE WHITE RAT WITH ITS APPLICATION TO THE EXPRESSION OF METABOLIC RESULTS. American Journal of Physiology-Legacy Content 1929;89:24–33. [CrossRef]
- Spinosa MR, Braccini T, Ricca E, De Felice M, Morelli L, Pozzi G, et al. On the fate of ingested spores. Res Microbiol 2000;151:361–8. [CrossRef]
- Leser TD, Knarreborg A, Worm J. Germination and outgrowth of Bacillus subtilis and Bacillus licheniformis spores in the gastrointestinal tract of pigs. J Appl Microbiol 2008;104:1025–33. [CrossRef]
- Tam NKM, Uyen NQ, Hong HA, Duc LH, Hoa TT, Serra CR, et al. The intestinal life cycle of Bacillus subtilis and close relatives. J Bacteriol 2006;188:2692–700. [PMID: 16547057. [CrossRef]
- Sha H, He X, Yan K, Li J, Li X, Xie Y, et al. Blocking coprophagy increases the levels of inflammation and depression in healthy mice as well as mice receiving fecal microbiota transplantation from disease model mice donors. APMIS 2023;131:351–68. [CrossRef]
- Auger S, Ramarao N, Faille C, Fouet A, Aymerich S, Gohar M. Biofilm Formation and Cell Surface Properties among Pathogenic and Nonpathogenic Strains of the Bacillus cereus Group. Appl Environ Microbiol 2009;75:6616–8. [CrossRef]
- Kennedy EA, King KY, Baldridge MT. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front Physiol 2018;9. [CrossRef]
- Glastras SJ, Chen H, Teh R, McGrath RT, Chen J, Pollock CA, et al. Mouse Models of Diabetes, Obesity and Related Kidney Disease. PLoS One 2016;11:e0162131. [CrossRef]
- Wang M-Y, Yan H, Shi Z, Evans MR, Yu X, Lee Y, et al. Glucagon receptor antibody completely suppresses type 1 diabetes phenotype without insulin by disrupting a novel diabetogenic pathway. Proceedings of the National Academy of Sciences 2015;112:2503–8. [CrossRef]
- Kim B, Kwon J, Kim M-S, Park H, Ji Y, Holzapfel W, et al. Protective effects of Bacillus probiotics against high-fat diet-induced metabolic disorders in mice. PLoS One 2018;13:e0210120. [CrossRef]
- Wang P, Gao X, Li Y, Wang S, Yu J, Wei Y. Bacillus natto regulates gut microbiota and adipose tissue accumulation in a high-fat diet mouse model of obesity. J Funct Foods 2020;68:103923. [CrossRef]
- Cao GT, Dai B, Wang KL, Yan Y, Xu YL, Wang YX, et al. Bacillus licheniformis , a potential probiotic, inhibits obesity by modulating colonic microflora in C57BL/6J mice model. J Appl Microbiol 2019;127:880–8. [CrossRef]
- Wu D, Ren Z, Pae M, Guo W, Cui X, Merrill AH, et al. Aging Up-Regulates Expression of Inflammatory Mediators in Mouse Adipose Tissue. The Journal of Immunology 2007;179:4829–39. [CrossRef]
- Christie G, Setlow P. Bacillus spore germination: Knowns, unknowns and what we need to learn. Cell Signal 2020;74:109729. [CrossRef]
- Tetz G, Tetz V. Introducing the sporobiota and sporobiome. Gut Pathog 2017;9:38. [CrossRef]
- Rook GAW, Raison CL, Lowry CA. Microbial ‘old friends’, immunoregulation and socioeconomic status. Clin Exp Immunol 2014;177:1–12. [CrossRef]
- Gao Z, Zhao X, Yang T, Shang J, Shang L, Mai H, et al. Immunomodulation therapy of diabetes by oral administration of a surfactin lipopeptide in NOD mice. Vaccine 2014;32:6812–9. [PMID: 25239487. [CrossRef]
Figure 1.
Schematic of experimental design and timeline.
Figure 1.
Schematic of experimental design and timeline.
Figure 2.
Colony Forming Units. A, Faecal samples taken during a five-week acclimatisation period. The graph shows SC conditions produces a decline in heat-resistant aerobic spore formers after three weeks, reaching a plateau of 2 x 102 CFU/ml. B, Samples taken at regular intervals over a 100-day period in the weaned mice that were born into SC (grey) and SL (black) conditions. The results show a significant difference in CFU / ml that is constant throughout the experiment. ****p ≤.0001. C, Shows representative images of the typical morphology of the observed spore forming bacteria, in keeping with Bacillota.
Figure 2.
Colony Forming Units. A, Faecal samples taken during a five-week acclimatisation period. The graph shows SC conditions produces a decline in heat-resistant aerobic spore formers after three weeks, reaching a plateau of 2 x 102 CFU/ml. B, Samples taken at regular intervals over a 100-day period in the weaned mice that were born into SC (grey) and SL (black) conditions. The results show a significant difference in CFU / ml that is constant throughout the experiment. ****p ≤.0001. C, Shows representative images of the typical morphology of the observed spore forming bacteria, in keeping with Bacillota.
Figure 3.
Obesity. All clinical measurements for obesity and adipose tissue demonstrated increased quantities in the SC group compared to SL. A, Mean total body weight (g) of SL was significantly lower than SC. B, This graph shows that SC had significantly higher levels of obesity, as measured by the Lee Index (∛g/cm). C, Demonstrates the mean weight of the gonadal fat pad (g) was higher in the SC group. D, The mean thickness of the inguinal adipose tissue was also markedly increased in the SC group compared to SL. ns = non-significant, *p ≤ .05, **p ≤.01, ***p ≤.001, ****p ≤.0001.
Figure 3.
Obesity. All clinical measurements for obesity and adipose tissue demonstrated increased quantities in the SC group compared to SL. A, Mean total body weight (g) of SL was significantly lower than SC. B, This graph shows that SC had significantly higher levels of obesity, as measured by the Lee Index (∛g/cm). C, Demonstrates the mean weight of the gonadal fat pad (g) was higher in the SC group. D, The mean thickness of the inguinal adipose tissue was also markedly increased in the SC group compared to SL. ns = non-significant, *p ≤ .05, **p ≤.01, ***p ≤.001, ****p ≤.0001.
Figure 4.
Colonic Inflammation. A, colonic length (cm), a surrogate marker for inflammation in mice, showed no significant difference between the groups. B, Wet caecal weight (g) was however increased in the SC group, suggesting a decreased microbial diversity. RTq-PCR results demonstrated no difference between the groups, with C, representing TNF-α ; D, IL-6 ; E, IL-1β; F, IL-10 ; G, MUC-2 and H, Claudin-1. ns = non-significant, *p ≤ .05, **p ≤.01, ***p ≤.001, ****p ≤.0001.
Figure 4.
Colonic Inflammation. A, colonic length (cm), a surrogate marker for inflammation in mice, showed no significant difference between the groups. B, Wet caecal weight (g) was however increased in the SC group, suggesting a decreased microbial diversity. RTq-PCR results demonstrated no difference between the groups, with C, representing TNF-α ; D, IL-6 ; E, IL-1β; F, IL-10 ; G, MUC-2 and H, Claudin-1. ns = non-significant, *p ≤ .05, **p ≤.01, ***p ≤.001, ****p ≤.0001.
Figure 5.
Metabolic Health. A, Demonstrates a significantly increased HbA1C in the SC group (5.38%) compared to SL (4.74). B, The mean weight of the liver was also significantly increased in the SC group. C, histological slides of the liver with H&E staining demonstrates however that this increased weight was not due steatosis. D, shows that there was no difference in serum total cholesterol (mmol/l). E, similarly shows no significant difference in serum alanine transaminase (ALT) (U/l). F, Mean weight of the spleen (g) was also the same between the two groups. ns = non-significant, *p ≤ .05, **p ≤.01, ***p ≤.001, ****p ≤.0001.
Figure 5.
Metabolic Health. A, Demonstrates a significantly increased HbA1C in the SC group (5.38%) compared to SL (4.74). B, The mean weight of the liver was also significantly increased in the SC group. C, histological slides of the liver with H&E staining demonstrates however that this increased weight was not due steatosis. D, shows that there was no difference in serum total cholesterol (mmol/l). E, similarly shows no significant difference in serum alanine transaminase (ALT) (U/l). F, Mean weight of the spleen (g) was also the same between the two groups. ns = non-significant, *p ≤ .05, **p ≤.01, ***p ≤.001, ****p ≤.0001.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).