1. Introduction
Parkinson’s disease (PD) is the fastest growing neurological disorder with 6.2 million individuals currently affected worldwide and 14.2 million individuals projected to be affected by 2040 [
1]. PD prevalence studies are heterogeneous and often underestimate current and projected cases of PD [
2,
3]. Limited capabilities surrounding screening resources may contribute to underestimated global PD burden [
4].
PD is a multidomain disease traditionally characterized by motor symptoms via in-clinic evaluations [
5], which are subjective, rater-dependent, and infrequent [
6,
7]. Diagnostic accuracy using these methods, particularly in early-stage PD, ranges between 58% and 85% [
8,
9]. Population-based studies have found that 12% to 83% of individuals presenting with PD symptoms are undiagnosed [
10]. Detecting patients transitioning from prodromal to early-stage PD remains challenging [
11]. Thus, developing capabilities that provide easier access to more efficient and accurate screening resources promises to facilitate more frequent symptom monitoring, thereby enabling earlier diagnostic and treatment interventions and overall better long-term quality of life.
Wearable devices and sensors generate information-rich continuous data streams that can measure disease- and behavior-relevant signals [
12]. Digital measures engineered from wearable devices and sensor data offer opportunities to more precisely, objectively, and frequently monitor patient disease burden relative to traditional clinical endpoints [
13,
14,
15]. Numerous studies have evaluated wearable devices and sensors in PD symptom monitoring [
16,
17,
18,
19,
20,
21,
22,
23,
24]. Existing studies, however, have been insufficient in capturing the multi-domain sequelae of PD, focusing instead on capturing data from a singular device or sensor modality. Furthermore, despite emerging interests in using machine learning capabilities to develop multivariate predictive models of PD [
25], large-scale studies of wearable devices and sensors in clinically confirmed PD patients have yet to develop a classification model of PD status that can be implemented globally as a widely accessible screening tool.
In the current work, our goal was to develop a PD screening tool using consumer-grade wearable devices and sensors. To this end, we implemented feature engineering and multivariate machine learning modeling in data generated from the large-scale multicenter WATCH-PD (Wearable Assessments in The Clinic and at Home in PD) study (NCT03681015). Briefly, WATCH-PD was a one-year longitudinal study of clinically confirmed early-stage PD and age-matched comparison (HC) participants who completed a multidomain (i.e., cognitive, psychomotor, speech, and mobility) battery of assessments that acquired data from multiple sensors equipped on consumer-grade smartwatch and smartphone devices. Using this high-dimensional multi-sensor, multidomain dataset, we sought to: (1) engineer a robust library of features using a combination of time- and frequency-domain signal processing; (2) evaluate reliability, validity, and selectivity patterns of features for PD status; and (3) develop a machine learning model that maps engineered features onto PD status and evaluate model performance metrics across environmental and temporal contexts.
2. Materials and Methods
Study Design and Sample
Participants living with Parkinson’s disease (PD) and healthy controls (HC) were recruited to complete the multi-center (n=17) WATCH-PD (Wearable Assessments in The Clinic and at Home in PD) (NCT03681015) observational study at a designated Parkinson Study Group research site. The WCGTM Institutional Review Board approved the procedures used in the study, and there was full compliance with human experimentation guidelines.
Criteria for enrollment into the PD group included: (1) diagnosis clinically documented by a movement disorder specialist; (2) older than 30 years at diagnosis; (3) disease duration less than two years; (4) Hoehn & Yahr stage < 3; (5) no baseline use of dopaminergic or other PD medications; and (6) no alternative Parkinsonian diagnosis. Approximately half of the PD group underwent DaTscan screening to confirm their diagnosis. Criteria for enrollment into the HC group included: (1) age-matched to the PD group; and (2) no previous PD diagnosis; and (3) no other significant neurologic disease. All participants provided written informed consent before study participation. HC participants underwent clinical assessment, including MDS-UPDRS screening, to confirm lack of Parkinsonism.
Participants completed a one-year longitudinal study comprised of both traditional clinic visits (n=6) and remotely monitored home visits (n=24). Clinic visits were completed at baseline and months 1, 3, 6, 9, and 12. Home visits were completed every other week. Clinical visits served to establish traditionally optimal environmental conditions for collecting performance data from the mobile assessment battery; by contrast, home visits served to evaluate whether similar levels of performance could be observed when completed remotely without direct clinical supervision. Participants completed the same mobile assessment battery in both environments.
Mobile Assessment Battery
Participants completed the BrainBaseline Movement Disorders mobile assessment battery (Clinical ink; Horsham, PA) as part of their study participation. The BrainBaseline Movement disorders mobile assessment battery was completed on provisioned Apple smartphone (iPhone XS Max or 12 Pro; OS versions 12.1-15.1) and smartwatch (Apple Watch 4 or 5; OS versions 5.1.2-8.0) devices. Smartwatches were worn on either the most affected wrist (PD group) or the non-dominant wrist (HC group). Smartphones were primarily held with both hands during the battery apart from the Gait & Balance task, in which the phone was worn at the trunk. Brief assessment descriptions are as follows:
Visual Short-Term Memory (VSTM): Participants were instructed to remember four colored squares and to respond, after a brief blank display screen, as to whether a single probe color matched one of the previously remembered squares. Response accuracy was the primary outcome measure.
Symbol Digit Modalities Test (SDMT): Participants completed a modified SDMT in which they were presented with a symbol, matched the symbol to an appropriate number within a symbol-number key, and then verbalized the appropriate number before proceeding onto the next symbol. Total completed was the primary outcome measure.
Trails: Participants completed a digital version of the Trail Making Test, in which they were instructed to draw lines between target objects in either numerical order (i.e., 1-2-3-4) or in alternating number-letter order (i.e., 1-A-2-B-3-C). Total time to completion was the primary outcome measure.
Fine Motor: Participants were presented with a randomly positioned and oriented major circular sector and instructed to drag and rotate the object to match position and orientation of a target sector. Total completed was the primary outcome measure.
Finger Tapping: Participants were presented with two circular target locations and instructed to rapidly tap the center of each location with their pointer and middle fingers in alternating order. Total number of taps was the primary outcome measure.
Gait & Balance: During the Gait task, participants were instructed to walk as they normally would for 60 seconds. During the Balance task, participants were instructed to stand with feet should-width apart and remain motionless for 30 seconds. Motion data were captured from smartphone and smartwatch sensors (see
Table 1) during Gait and Balance tasks.
Tremor: Tremor was comprised of Postural and Resting Tremor sub-tasks. During the Postural Tremor sub-task, participants were seated and instructed to maintain their arms in a straight line at a 90-degree angle with respect to their body for 10 seconds. During the Resting Tremor sub-task, participants were seated and instructed to maintain their arms at rest by their sides for 10 seconds. Motion data were captured from smartwatch sensors (see
Table 1) during Tremor tasks.
Phonation: Participants were instructed to speak and sustain the phoneme ‘ahh’ for as long as possible within a 15 second measurement window. Speech data were captured from the smartphone microphone and encoded in .wav format.
Articulation: Participants were instructed to speak and repeat the phoneme sequence ‘pa-ta-ka’ as many times as possible within a 15 second measurement window. Speech data were captured from the smartphone microphone and encoded in .wav format.
Reading: Participants were instructed to read three sentences sampled from the Harvard sentences bank at a rate reflective of their typical reading speed. Speech data were captured from the smartphone microphone and encoded in .wav format. In the present work, Reading task data were excluded from analysis.
Clinic visits were initiated by study site personnel and self-administered by participants. For home visits, participants received reminder notifications at 9:00 AM local time and, once initiated, were given one hour to complete their self-administered scheduled activities.
Feature Engineering
Features were engineered from all continuous data sources available from each assessment (see
Table 1). Our library of features was modeled after previous work extracting features from cognitive [
26,
27], speech [
28,
29], and accelerometer-based mobility assessments [
30,
31,
32]. Details regarding feature engineering are as follows.
VSTM. Two features were extracted from VSTM: mean response accuracy and working memory capacity (K).
SDMT. Two features were extracted from SDMT: total completed symbols-digit pairs and mean response accuracy.
Trails. Twelve features were extracted from Trails. Total completion time was extracted from A and B sub-tasks. Additional features were engineered from the reconstructed path drawn during each subtask: (1) total reconstructed path length; (2) root mean squared error between reconstructed and idealized paths between target locations; and total spectral energy in the 4-10 Hz band observed within: (3) reconstructed path tracings in the vertical screen dimension; (4) reconstructed path tracings in the horizontal screen dimension; and (5) residual errors between reconstructed and idealized paths.
Fine Motor. Nineteen features were extracted from Fine Motor. Total completed with dominant hand, total completed with non-dominant hand, and the ratio of total completed with dominant to non-dominant hand were extracted from summary data. For each hand, additional features were engineered from raw time series data reflecting the reconstructed position and orientation of target objects: (1) total path length; (2) ratio of total path length to idealized path length; (3) movement speed (pixels/millisecond); (4) total rotation; (5) ratio of total rotation to idealized rotation; (6) rotation speed (degrees/millisecond); and total spectral energy observed within the 4-10 Hz band: (7) reconstructed position tracings in the vertical screen dimension; (8) reconstructed position tracings in the horizontal screen dimension
Finger Tapping. Twenty-eight features were extracted from Finger Tapping. Number of alternating taps, number of total taps, and ratio of alternating to total taps for both dominant and non-dominant hands were extracted from summary data. For each hand, additional features were engineered from raw time series data reflecting tap position and duration: (1) median and inter-quartile range (IQR) of tap duration; (2) median and IQR of tap distance from target location; (3) median and IQR of tap onset asynchrony time; (4) median and IQR of inter-tap interval; (5) IQR of spatial variance in vertical and horizontal screen dimensions; and (6) peak spectral frequency of tap time series.
Tremor. Triaxial accelerometry data were generated with respect to the smartwatch reference frame (Figure). Prior to implementing feature engineering libraries on Tremor data, preprocessing routines linearly rotated triaxial accelerometry data to triaxial gravitational acceleration data to produce a watch-independent geospatial reference frame in which the z-axis forms a 90-degree angle with respect to the surface plane. Velocity and position were derived by calculating the first and second cumulative trapezoidal integral, respectively, of the raw accelerometer signal.
Two-hundred thirty-one unique features were engineered from Tremor tasks, yielding 462 total features. 20 univariate time-domain features were engineered separately from each accelerometer axis: (1) zero-cross rate; (2) summary statistics (median, mean, 25th percentile (Q1), 75th percentile (Q3), IQR, standard deviation (SD), kurtosis, skewness) of inter-cross interval time distributions; (3) SD and IQR of raw acceleration; (4) summary statistics (mean, median SD, Q1, Q3, IQR) of integrated velocity; (5) total path length of acceleration, velocity, and position. Univariate time-domain analyses yielded 60 unique features.
Two multivariate time-domain features (total path length, convex hull area) were engineered separately from each unique combination of accelerometer axes (x-y, x-z, y-z, x-y-z) for acceleration, velocity, and position. Multivariate time-domain analyses yielded 24 unique features.
Forty-nine univariate frequency-domain features were engineered separately from each accelerometer axis: (1) spectral energy within the following energy bands: 3.5-4.5 Hz, 4.5-5.5 Hz, 5.5-6.5 Hz, 6.5-7.5 Hz, 7.5-8.5 Hz, 8.5-9.5 Hz, 9.5-10.5 Hz, 4-10 Hz; (2) spectral roll-off at 1%, 25%, 50%, 75%, and 99% percentiles; (3) Mel-frequency cepstral coefficients (MFCC) 1-16, calculated within the 0-49 Hz range; and (4) summary statistics (entropy, SD, kurtosis, skewness, flatness) of total spectral energy (0-49 Hz), spectral energy within 4-10 Hz band, spectral energy outside 4-10 Hz band, and the ratio of energy within to outside the 4-10 Hz band. Univariate frequency-domain analyses yielded 147 unique features.
Gait & Balance. Triaxial accelerometry data were generated with respect to the smartwatch and smartphone reference frame. Prior to implementing feature engineering libraries on Gait & Balance tasks, several preprocessing routines were performed. Triaxial accelerometry data from both devices were linearly rotated into a common geospatial reference frame as performed on Tremor data. Smartphone and smartwatch data streams were temporally synchronized to ensure each sample was comparable in time. Step detection algorithms were implemented to identify periods in which foot strikes were detected in acceleration signals. First, a 10-Hz high-pass finite impulse response (FIR) filter was applied to the raw acceleration signal followed by the application of 5-Hz low-pass FIR filter was applied to the modulus of the high-pass filtered signal. Next, the second derivative of the signal was calculated and values exceeding a 15% increase in signal relative to the mean signal were counted as steps. Steps with an inter-step interval greater than 1000 milliseconds were excluded from analyses. Velocity and position were derived by calculating the first and second cumulative trapezoidal integral, respectively, of the raw accelerometer signal.
1053 unique features were engineered from Gait & Balance tasks, yielding 2106 total features. Time- and frequency-dependent feature engineering routines described in Tremor were implemented on both smartphone and smartwatch data from Gait & Balance, yielding 462 unique features.
Nineteen spectral coherence features were engineered from all 15 combinations of accelerometer dimensions and device pairings: (1) peak spectral coherence frequency; (2) total spectral coherence across frequencies; (3) spectral coherence observed at 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, 30, 35, 40, and 45 Hz. Coherence analyses yielded 285 unique features.
Fifty-one gait-related features were engineered from each triaxial dimension from each device: (1) step count; (2) step frequency; (3) step distance; and summary statistics (mean, median, Q1, Q3, IQR, SD, kurtosis, skewness) features derived from data interleaving each step: (4) inter-step interval; (5) step velocity; (6) step distance; (7) path length of acceleration; (8) path length of velocity; and (9) path length of position. Gait analyses yielded 306 unique features.
Phonation & Articulation. 495 unique features were engineered Phonation & Articulation tasks, yielding 990 total features. Total speech duration was extracted from summary data. Voice waveforms were divided into 15 segments of equal length, and summary statistics (mean, median Q1, Q3, IQR, 1st percentile, 99th percentile, IQR99, SD, kurtosis, skewness, flatness, root mean square) were derived for 38 features generated within each segment: (1) fundamental frequency; (2) harmonic-to-noise (HNR); (3) jitter in both milliseconds and percentage; (4) shimmer in both decibels and percentage; (5) total energy within the 50-80, 80-120, 120-200, 200-360, 360-680, 680-1500, and 1500-4000 Hz spectral energy bands; (6) flatness, entropy, variance, skewness, and kurtosis of all spectral energies; (7) spectral roll-off at 25%, 50%, 75%, and 90% percentiles; and (8) MFCC1-16.
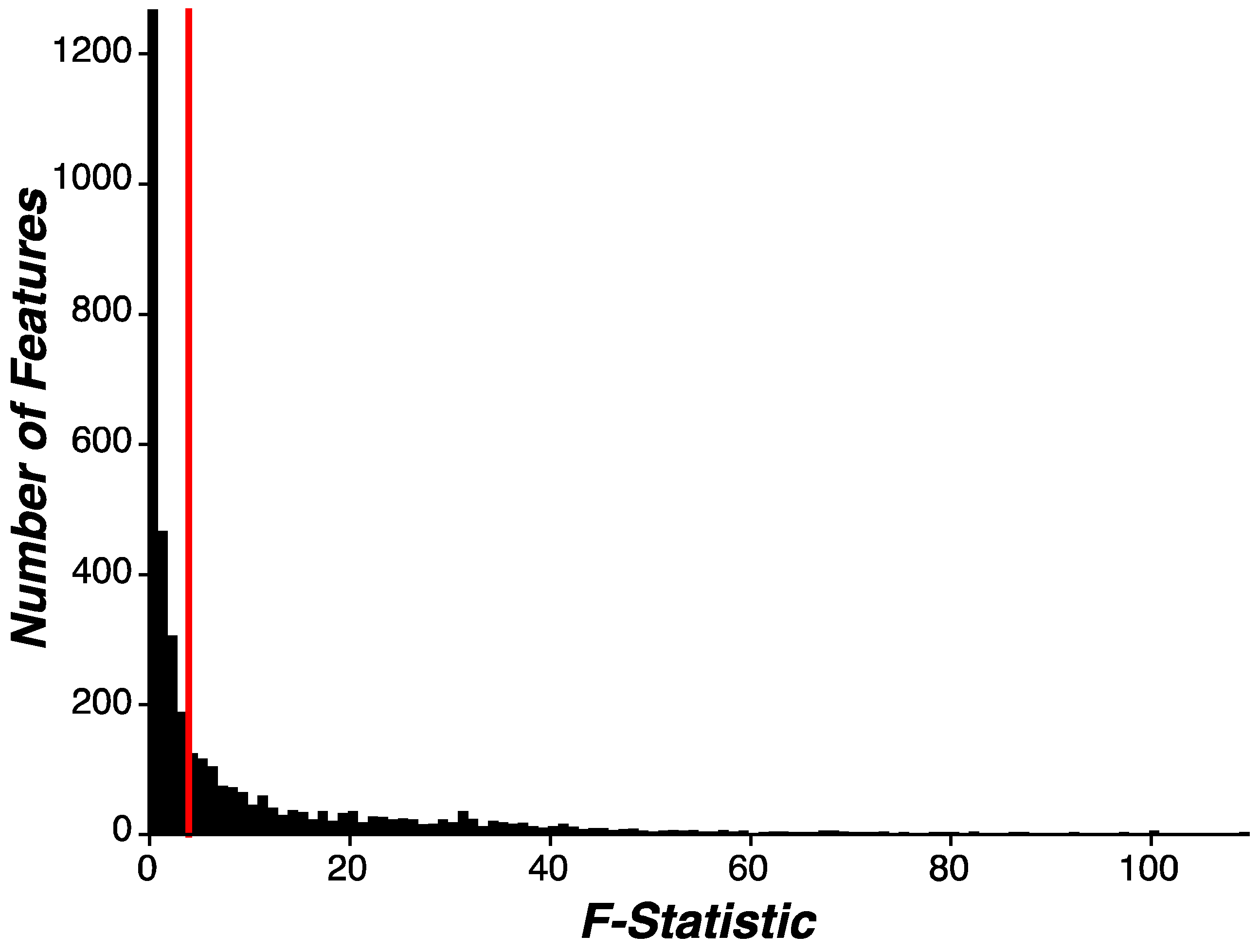
Evaluating Disease and Behavioral Relevance of Features. In an exploratory analysis, we evaluated whether our engineered features demonstrated disease or behavioral relevance by inspecting the proportion of all features that associated with early-stage PD status. Importantly, separate feature selection routines were implemented during the machine learning modeling phase of this analysis. To enumerate how many features showed significant distributional differences between PD and HC participants, univariate linear regression was performed on each feature independently, where a given feature was the dependent variable and participant group (PD, HC) was a categorical independent variable (Feature ~ Group [PD, HC]). F-statistics from each feature-wise univariate test were aggregated into a distribution. We then evaluated the overall proportion of features showing significance as well as the proportion of features within each assessment showing significance. Given the exploratory nature of this analysis, thresholds for feature-wise significance were set to p=0.05 and corrections for multiple comparisons were not implemented.
Machine Learning Modeling
To evaluate the robustness of machine learning models, all machine learning analyses were implemented using a Monte Carlo simulation (n=100) on data randomly sorted into training (90% of all participants) and test (10% of all participants) sets.
Model Comparison. To identify the model most predictive of PD status, model comparison analyses were conducted using the following steps in turn: feature wrangling, feature selection, feature reduction, model development, model evaluation, and model selection.
Data wrangling was performed to combine multiple data sources into a single dataframe prior to implementing feature processing and modeling routines. Any given assessment produced a single array of unique features. Performing feature engineering routines across all sessions and participants produced an m x n matrix for each assessment, where m is the total number of sessions completed and n is the total number of features engineered for the assessment. Each feature matrix was merged into one dataframe, where each row was a unique participant and session. For example, if a participant partially completed some assessments in a single session, the row for that session would be missing some features. Features generated across all sessions – both home and clinic – were averaged together for each participant, producing a single row of features for each participant.
Feature selection routines were performed to reduce the dimensionality of features used during modeling by identifying features that best discriminate between PD and HC participants. To this end, we used univariate linear regression (as described in Evaluating Disease and Behavioral Relevance of Features) and sorted features by F-statistic value. Features (n=100) demonstrating the highest F-statistic were selected for modeling and the remaining features (n=3,521) were excluded from further analysis. To note, not all features showing a significant association with early-stage PD status were selected for further analysis. As will be described below, feature selection was parametrically included or omitted from the analysis routine to evaluate the impact of feature selection on model performance.
Feature reduction routines were performed to reduce the dimensionality of features used during modeling by eliminating multicollinearity between features. To this end, we used principal component analysis using the Python scikit-learn function decomposition.PCA. Principal components (n=10) explaining the greatest amount of variance in the input features were selected for further analysis and the remaining principal components were excluded from further analysis. A fixed number of principal components were selected to ensure data formats were consistent across models. As will be described below, feature reduction was parametrically included or omitted from the analysis routine to evaluate the impact of feature reduction on model performance.
To evaluate the impact of feature processing on model performance, feature selection and feature reduction routines were parametrically manipulated. For feature selection, either a subset of features (n=100) was selected for modeling (selection +) or all features were selected for modeling (selection -). For feature reduction, either principal components (n=10) were extracted from raw features and selected for modeling (reduction +) or raw features were selected for modeling (reduction -). Consequently, each model included four possible feature processing steps: (1) selection (+), reduction (+); (2) selection (+), reduction (-); (3) selection (-), reduction (+); and (4) selection (-), reduction (-). Prior to model development, features in both the training (90% of participants) and test (10% of participants) sets underwent one of these four processing routines. The performance of each model was compared against the context of the inclusion and exclusion of each of these feature processing steps.
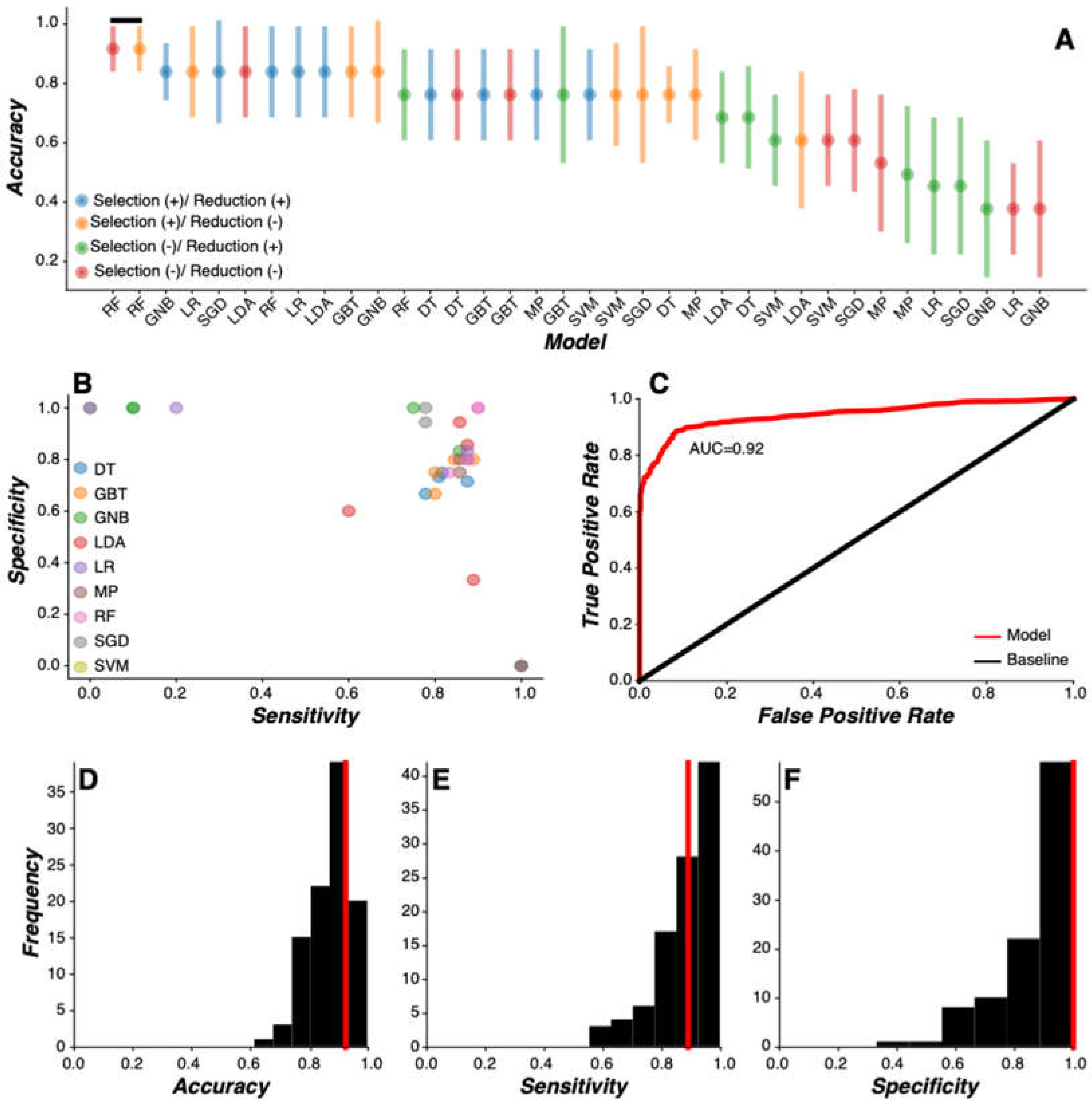
Nine unique models were developed and evaluated: logistic regression (LR), linear discriminant analysis (LDA), support vector machine (SVM), decision tree (DT), gradient boosted tree (GBT), random forest (RF), stochastic gradient descent (SGD), gaussian native bayes (GNB), and multilayer perceptron (MP). Each model underwent four feature processing routines through separate iterations, yielding 36 total unique models developed. Each model was developed and trained on PD training group labels while the PD test features and labels were held out from analysis. Critically, training and test features were sorted subject-wise so that feature sets for a given participant were sorted into either the training set or the test set. Training model coefficients were saved for model evaluation.
Model evaluation was implemented by inspecting how well each model performed in predicting PD test labels. To this end, we applied the training model coefficients to the independent test set features to generate predictions of PD test group labels. PD labels and model predictions were convolved into a 2x2 table from which model performance metrics of accuracy, sensitivity, and specificity were calculated. These procedures were implemented iteratively across all Monte Carlo simulations (n=100), generating a sample of independent model prediction metrics based on random assortment of features into training and test sets on each pass through the Monte Carlo simulation. Critically, all models were evaluated on the same training and test data across each Monte Carlo simulation. Models were trained and tested using the scikit-learn Python library.
Model selection was performed to identify the model most predictive of PD status. To this end, we used Wilcox sign-rank test to compare paired samples (n=100) of model performance metrics generated by each Monte Carlo simulation iteration. In cases where multiple high accuracy models were statistically indistinguishable from each other, the most parsimonious model was selected. Here, the most parsimonious model was defined as the least complex model requiring the fewest number of feature processing procedures.
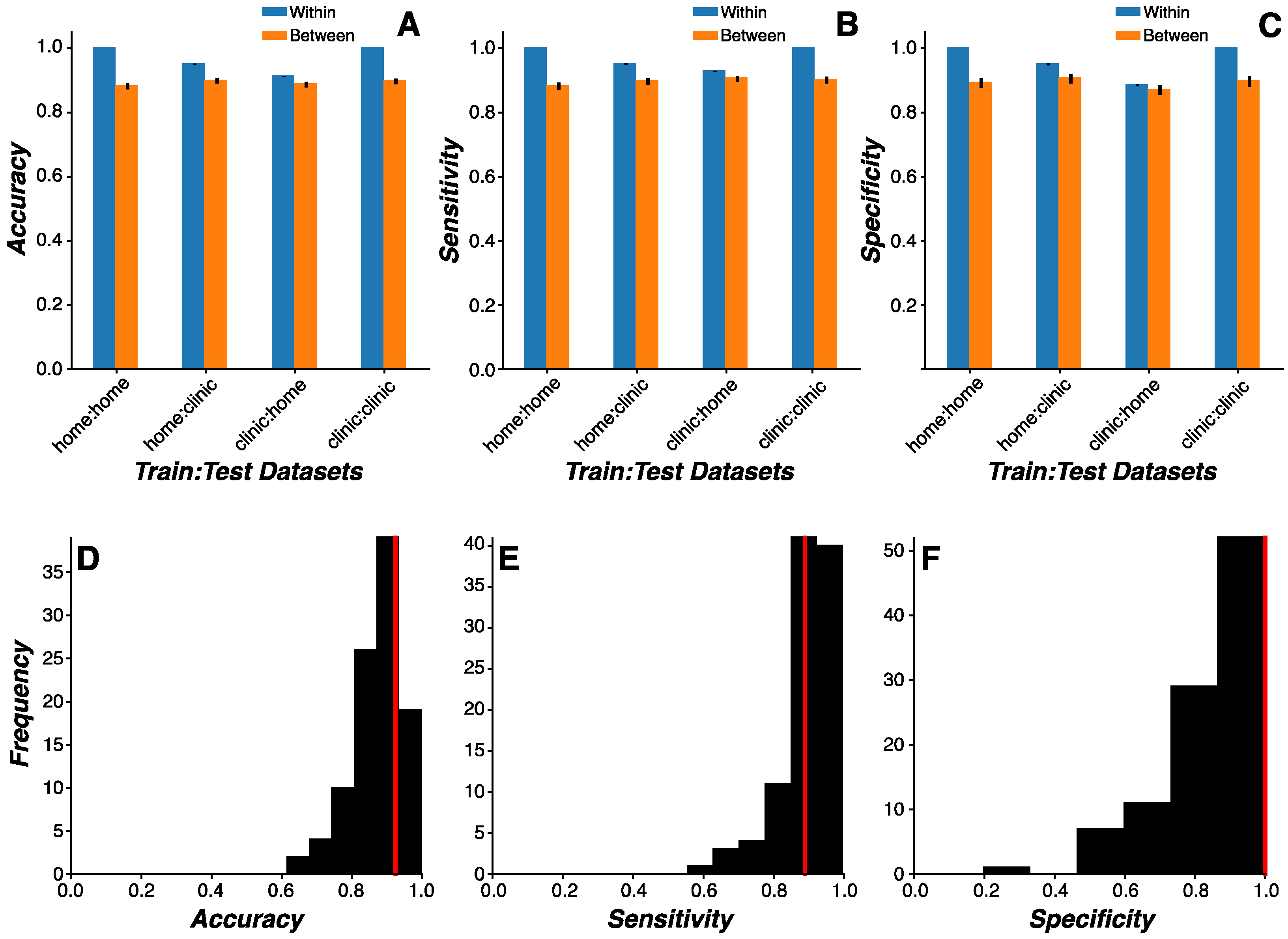
Cross-Environmental Predictions. We sought to determine whether performance and sensor data generated in clinic and home environments were of sufficient comparability to support remotely screening PD status based on home assessments alone. To this end, cross-environmental learning analyses trained models on data generated by sensors and assessments in one environment (e.g., clinic) and predicted PD status based on data generated by the same sensors and assessments in an independent environment (e.g., home). Prior to modeling, features generated across home and clinic visits were averaged together, yielding two vectors of data reflecting home and visit features for each participant. Only participants with both home and clinic vectors were submitted for analysis. During model development, participant data were partitioned into training and test sets (or cohorts) as described above.
We used the model selected from the Model Comparison analysis and implemented the same Monte Carlo simulation methods. Model training and testing routines were conducted using a 2 (training environment: home vs clinic visits) x 2 (testing environment: home vs clinic visits) x 2 (model test dataset: same vs independent cohort) study design. For example, one modeling condition would be to train on home data generated by the training cohort and test on clinic data generated by the training cohort (training environment=home, test environment=clinic, model test dataset=same cohort), while a separate modeling condition would be to train on home data generated by the training cohort and test on clinic data generated by the test cohort (training environment=home, test environment=clinic, model test dataset=independent cohort). Using this design, we were able to evaluate: (1) how the training environment affected classification accuracy; (2) how the testing environment affected classification accuracy; (3) how models trained and tested within the same environment performed relative to models trained and tested across environments; and (4) how cross-environment classification accuracy differed when models were trained and tested on the same or independent cohorts across independent environments.
Repeated-measures ANOVA was used to evaluate the omnibus effects of training environment, test environment, and model test dataset. Post-hoc analyses focused on: (1) effects of cohort to demonstrate proof-of-principle that training and testing on the same cohort inflates classification accuracy; (2) effects of environment on classification accuracy by evaluating same and independent cohort conditions separately; and (3) generalizability of classification results to the population by evaluating model performance metrics in the clinic training environment, home testing environment, and independent cohort condition. Importantly, classification results from model training and testing procedures performed on the same data were excluded from discussion to preclude obvious concerns of overfitting.
Reliability Analysis
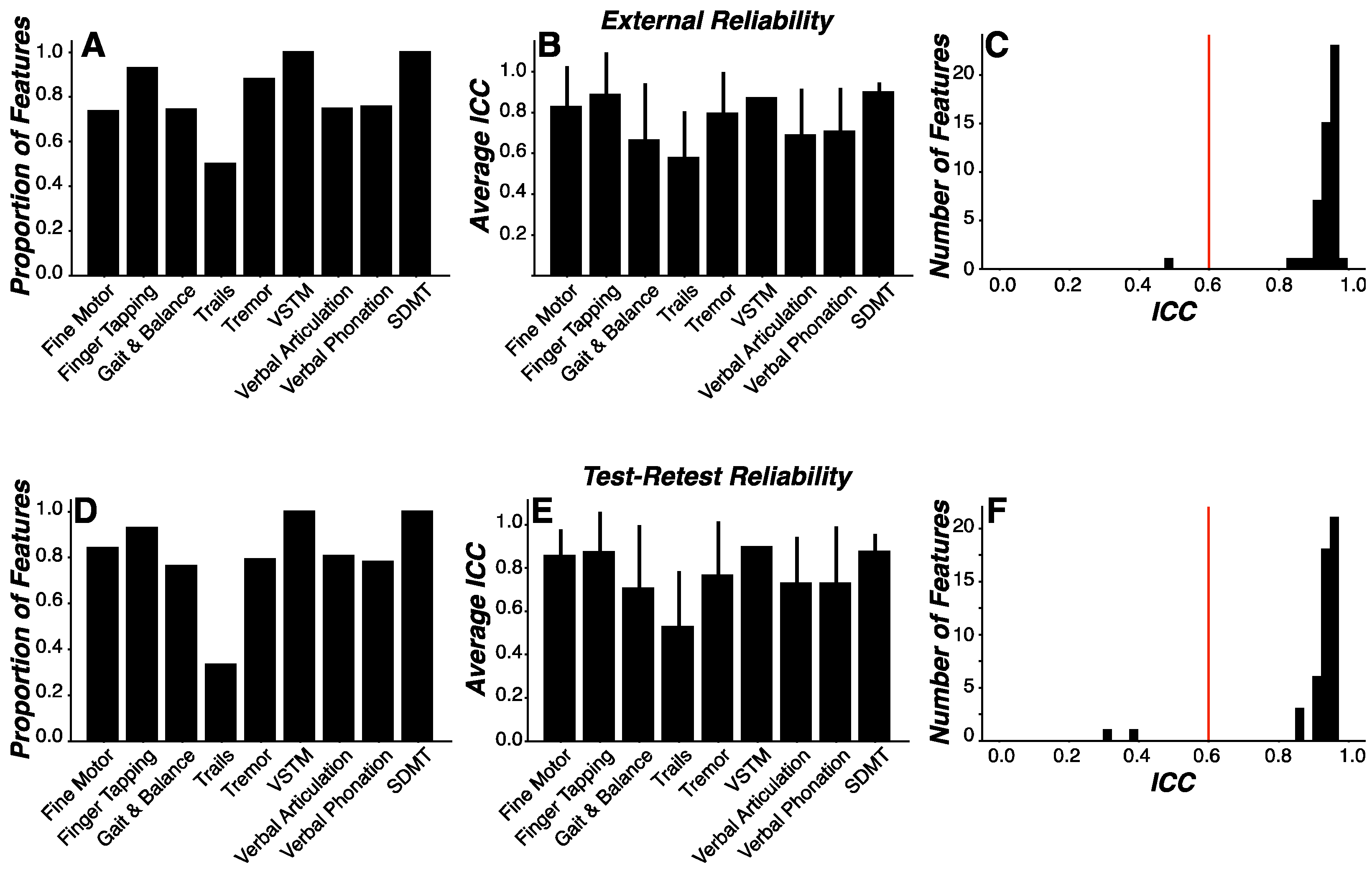
We sought to evaluate: (1) external reliability to assess measurement consistency across environments by comparing performance between clinic visits, conducted within a relatively controlled and supervised environment, and home visits, conducted within a relatively uncontrolled and unsupervised environment; and (2) test-retest reliability to assess measurement consistency across time. To this end, external and test-retest reliability was assessed for each engineered feature using intraclass correlations (ICC). Thresholds for acceptable reliability coefficients can range from 0.45 to 0.98.36 In the current work, we selected an ICC coefficient value of 0.6 as the threshold for acceptance as a reliable feature for both external and test-retest reliability analyses.
ICC coefficients were derived for each feature in both external and test-retest reliability analyses. Traditionally, reliability coefficients are reported individually for each feature or measurement assessed. Given the volume (n=3,621) of features engineered in this work, we evaluated the sample characteristics of our feature ICC coefficients relative to our reliability threshold. Specifically, we calculated the proportion of feature ICC coefficients exceeding the reliability threshold and whether the sample of feature ICC coefficients were statistically higher than the reliability threshold. To calculate the proportion of features exceeding the reliability threshold, we enumerated the number of feature ICC coefficients greater than 0.6 as a percentage of all features within a given assessment. To determine whether the feature ICC coefficients were statistically higher than the reliability threshold, we used one-sample t-tests to calculate whether the sample mean of ICC coefficients was greater than 0.6. Similar analyses were performed only on those features identified as important features during machine learning modeling procedures.
Feature Comparison
We evaluated the predictive power of mPower features extracted using our pipeline using a voting classifier in sci-kit learn/python. This approach was modeled after the ensemble approach deployed in R and described in Sieberts et al 2021 [
33]. Specifically, a voting classifier was constructed from elastic net, random forest, support vector machine, k-nearest neighbor, and neural network-based approaches. The classifier was trained on 80% of the data (per-subject median of each feature) and tested on the other 20% over 50 bootstraps and area under the receiver operating characteristic (auROC) curve was used as an accuracy metric. For comparison, we repeated this approach on accelerometer feature sets published along with Sieberts at al. 2021.
4. Discussion
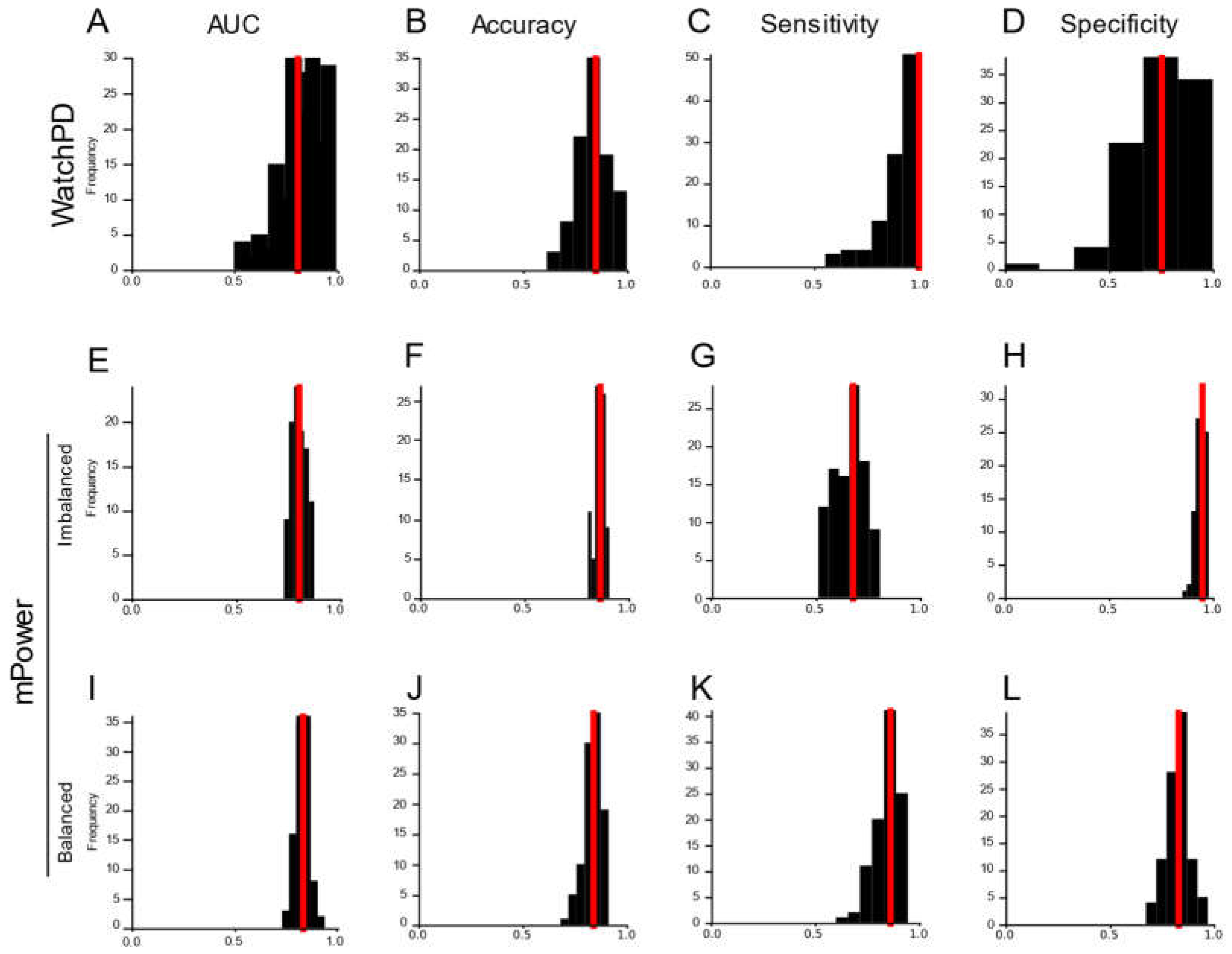
Consumer-grade wearable devices and sensors offer the potential for improved PD prevalence and surveillance metrics via broader access to screening tools. Here, we used a combination of feature engineering and multivariate machine learning modeling to develop a PD screening tool based on data generated from the large-scale multi-center WATCH-PD study. Our approach focused on clinically confirmed early-stage PD and HC participants generating high-dimensional data from multiple sensors while completing a multidomain (i.e., cognitive, psychomotor, speech, and mobility) battery of assessments on consumer-grade smartwatch and smartphone devices over a one-year period. We engineered a library of low-level signal-based disease- and behavior-relevant features from wearable device and sensor data generated by participants during study participation. Our machine learning model comparison approach revealed a Random Forest model that predicted early-stage PD status with high accuracy, sensitivity, and specificity that persisted across changes in environmental contexts. Moreover, the same feature engineering and model selection approach also accurately classified later stage PD status in an independent dataset generated on a different platform. Together, these results demonstrate the potential of consumer-grade technologies in screening early-stage PD status.
Numerous studies have evaluated wearable devices and sensors in PD symptom monitoring [
16,
17,
18,
19,
20,
21,
22,
23,
24], though these studies focused on capturing data from a singular device or sensor modality to the exclusion of capturing the multi-domain sequelae of PD. Furthermore, few among these studies have evaluated whether wearable devices and sensors can generate predictive models of PD status with direct applications to patient screening and surveillance [
17,
24,
33]. Two studies evaluated separate cohorts of early-stage PD patients that completed voice, finger tapping, gait, balance, and memory assessments on a smartphone [
17,
24]. Arora and colleagues (2015)[
24] evaluated a small cohort of PD patients (n=10) and non-demographically-matched HC (n=10) participants. Using their features, they found a Random Forest model of PD status achieved sensitivity and specificity of 96.2% and 96.9%, respectively. Critically, however, investigators performed record-wise rather than subject-wise cross-validation, which overestimates classification performance due to non-independence of training and test datasets [
38]. Thus, this study was limited by the small sample size, non-demographically matched study groups, and overestimated classification performance. Omberg and colleagues (2021) [
17] evaluated population-level cohorts of self-reported PD patients (n=1,414) and self-reported non-PD participants (n=8,432) participating in the unsupervised remotely completed mPower study [
36]. Using their features, they found a Random Forest model of self-reported PD status achieved an AUC of 0.80. In a subsequent crowdsourcing analysis of the same study data [
33], the best crowdsourced model was a deep learning neural network that achieved an AUC of 0.87. This study was limited, however, by reduced control over enrollment screening and data quality, non-demographically matched study groups, and non-clinically confirmed PD status. In the current work, by contrast, we evaluated a larger demographically-matched and clinically confirmed cohort of PD (n=82) and HC (n=50) participants who completed visits both in clinic and remotely at home. Participants completed a battery of multidomain assessments that more comprehensively captured PD symptoms, including upper extremity tremor and bradykinesia using the smartwatch, relative to previous studies. Using more appropriate machine learning methodology, including subject-wise cross-validation, our Random Forest model achieved superior classification performance. Indeed, the same feature engineering and modeling approach selected a Gradient Boosted Tree model that accurately classified participants in the mPower study. Furthermore, we sought to understand the boundary conditions of our model by evaluating its performance across environmental contexts and found that our model performs equally well when tested on clinic data collected in a controlled environment and home data remotely collected in an uncontrolled environment. Taken together, we have instrumentally extended our understanding of how device and sensor selection, assessment-to-disease mapping, and analytic methodologies are critical to tracking PD status in real-world settings.
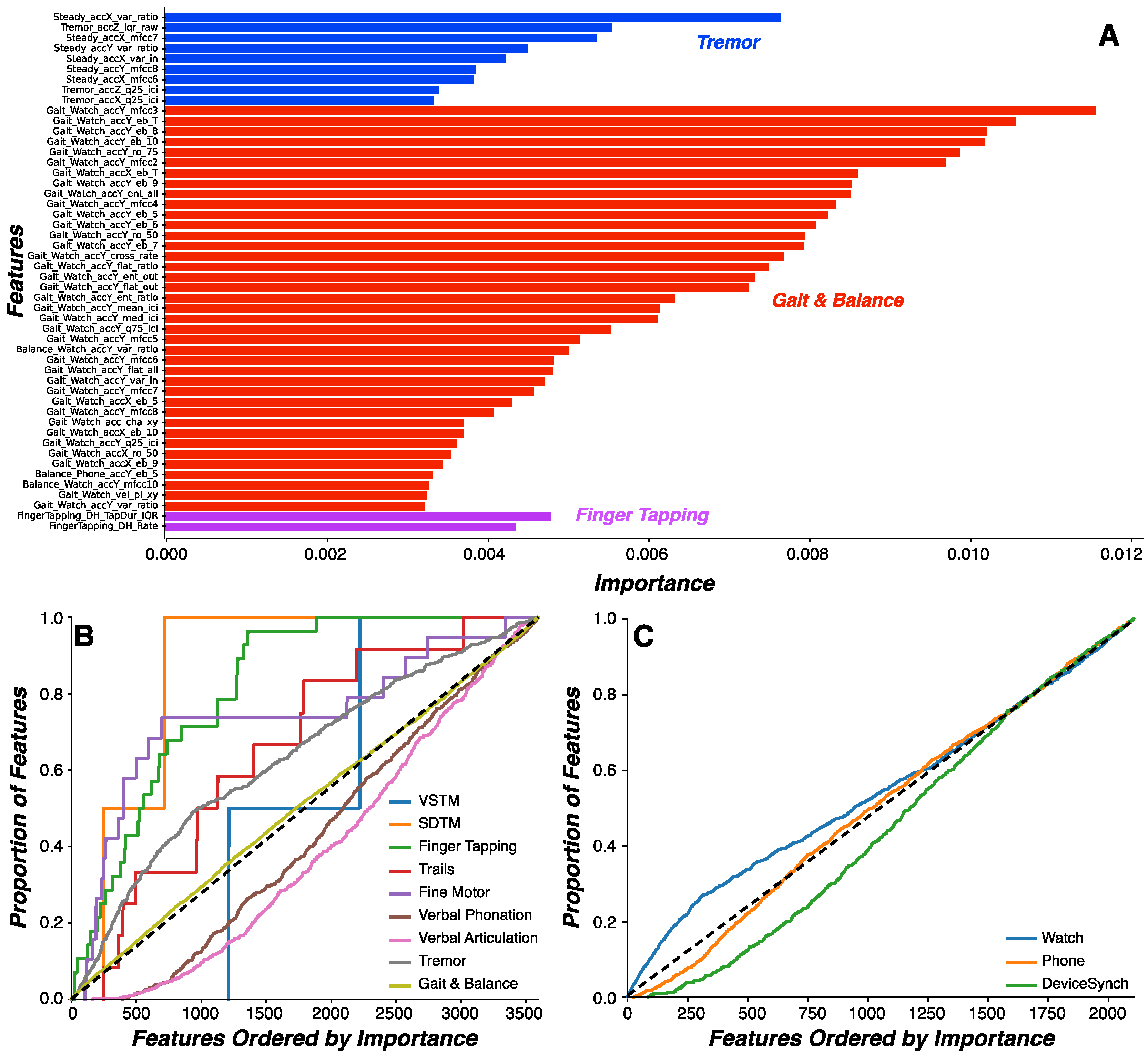
We sought to directly compare features generated by the watch and phone to understand whether either device is more effective at predicting PD status. To this end, our analysis focused on the same set of features engineered from acceleration-based signals generated concurrently from the watch and phone during the gait task. No other task generated similar data streams or features from both devices. We found that watch-derived features were probabilistically more important than phone-derived features, indicating features related to arm swing from the watch are more important than features related to gait parameters from the phone in predicting PD status. This finding raises important considerations regarding minimization of patient burden and the minimal number of devices and sensors required to capture and classify disease-relevant signals. Both devices used in the current work captured disease-relevant information. Indeed, the exercise of mapping disease-relevant features onto multidimensional clinical scales, such as the MDS-UPDRS, demands multiple sensors and devices. Here, the watch captured measurements relevant to tremor and arm swing, whereas the phone captured measurements relevant to gait, speech, and finger tapping measurements. In other studies, for example, the Roche PD Mobile Application v1 used only a smartphone to evaluate Parkinsonian symptoms [
20], whereas the subsequent Roche PD Mobile Application v2 added a smartwatch to better capture bradykinesia symptoms and passive mobility measurements [
21]. Despite the apparent advantage of multiple devices, better resolution of patient symptom burden is not achieved with numerous sensors. Lonini and colleagues, for example, developed models of bradykinesia and tremor and demonstrated a model including features from a single hand acceleration sensor performed just as well as a model including features from multiple sensors distributed across both hands, arms, and thighs [
18]. Thus, a single smartphone and smartwatch set is sufficient for minimizing patient burden while maximizing Parkinsonian symptom sensitivity.
Our approach has strong applicability to developing population-wide brief screening tools for early-stage PD. Using a combination of feature engineering and machine learning model comparison routines on sensor data streams generated from a fit-for-purpose mobile PD application, we were able to construct a Random Forest model that predicted early-stage PD status with 92% accuracy. Clinical diagnostic accuracy, by comparison, ranges between 58 to 85% [
8,
9]. Our approach may therefore support improvements in diagnostic accuracy and reductions in the prevalence of symptomatic undiagnosed PD cases [
11]. Critically, our approach aims to provide a platform for remotely screening individuals for PD and is therefore not intended to be a diagnostic tool. Indeed, the current platform could be a complementary tool to the MDS-UPDRS, whereby individuals identified as PD based on platform readout would require clinical evaluation to evaluate, diagnose, and stage disease. Using this remote technology platform, both physicians and patients may be better equipped to screen and monitor transition to early-stage PD, thereby minimizing clinic and patient burden, remotely generating accumulating diagnostic evidence over time, facilitating earlier diagnosis and access to treatment, and improving long-term quality of life.
Our feature engineering and machine learning approach performed well on the mPower study, an independent dataset collected from later stage PD patients on a different platform. This study shared Gait & Balance, Verbal Phonation, and Finger Tapping assessments with WATCH-PD. Using a reduced feature set derived from the shared assessments across platforms, we showed that a Random Forest model produced classification of WATCH-PD status in the upper range of clinical diagnostic accuracy (84.6%). Critically, another tree-based classifier performed well on the same feature set derived from the mPower dataset (Gradient Boosted Tree, 86.2% for imbalanced dataset and 84.1% accuracy for balanced dataset). While demonstrate accuracy metrics commensurate with clinical accuracy, models derived from the reduced feature set still underperform compared to the Random Forest classifier trained on the full WATCH-PD dataset (92.3%). One potential explanation for reduced accuracy is that the mPower study collected sensor data only from a smartphone, excluding measurements from smart watches that produced features putatively more important than phone-based features in the WATCH-PD data as described above. Regardless, these results suggest that our approach to feature engineering and model selection is platform-agnostic and thus may be applicable to a variety of existing and future studies. Standards will need to be developed, however, to account for inter-study differences in assessment implementation, sampling rate, and device type [
39].
Several study limitations demand further consideration and research to better understand the potential of remote digital health technologies in supporting population-wide early-stage PD screening. (1) Sensitivities reported here may have been affected by the fact that our study was comprised of an enriched sample of formally diagnosed early-stage PD patients who may have more severe symptoms relative to non-diagnosed individuals living with PD. Indeed, we developed models of PD status using clinically confirmed patients to ensure our model labels mirrored the ground truth diagnosis. Developing our models against traditional clinician ratings, which are demonstrably less accurate than biomarker confirmation, would have resulted in a less accurate model and a subsequently less useful tool for remotely screening PD status. To ensure the current screening tool can detect non-diagnosed individuals living with lower symptom burden, our approach and model must be validated in an independent study comprised of a larger sample with more heterogenous Parkinsonian symptoms. (2) Low-level signal-based features (e.g., acoustic audio features) were prioritized over high-level model-based heuristic features (e.g., speech lexical features). In PD, for example, both low-level signal-based [
40] and high-level model-based heuristic [
41] features have been developed for bradykinesia. Here, we prioritized low-level signal-based features to fully characterize the rhythmic activity embedded within device- and sensor-generated signals produced by neurotypical and Parkinsonian patterns of speech and movement. Further work focused on directly comparing low-level against high-level features will be necessary from perspectives of model predictability and explainability. (3) There are potential concerns over model overfitting given the large volume of features (n=3,621) and relatively small number of subjects (n=132). These concerns, however, are mitigated by our analysis design. First, we parametrically introduced feature selection and feature reduction routines, ensuring that models constructed from saturated feature sets were compared against models constructed from feature sets with reduced dimensionality. Second, we implemented cross-validation procedures to ensure that model training and testing were performed on independent datasets, preventing any influence of overfitting during model development on our evaluation of model performance. While true that some models demonstrated relatively low cross-validation accuracy, these models were equally represented across feature selection and feature reduction imputations. Thus, the selection of our best performing model – a Random Forest model constructed from a saturated feature set – was not due to overfitting. Validating our model in a larger study sample with greater heterogeneity in Parkinsonian symptomatology will further seek to address these concerns. (4) Binary classification of early-stage PD was prioritized over predicting MDS-UPDRS scores because we focused on evaluating the utility of the current platform as a screening tool. Consequently, we were unable to evaluate our features and model against the clinical gold-standard MDS-UPDRS, including comparing our model against MDS-UPDRS scores, understanding the additional diagnostic value of our platform relative to clinical gold-standards, and developing predictive models of PD severity and progression. (5) Our analysis was agnostic to the longitudinal design of this study. Indeed, the current work aimed to assess the potential of our approach and platform to be used as a screening tool without requirements to track change over time. Future analyses will focus on furthering our approach by evaluating feature sensitivity and digital phenotype progression over time.
PD is the fastest growing neurological disorder [
1], impairing multiple functional domains, including cognition, motor coordination, speech, and mobility [
7]. Contributors to underestimated global PD burden [
2,
3] include low diagnostic accuracy with clinical standard measures [
8,
9], symptomatic undiagnosed cases [
10], and challenges in identifying prodromal patients transitioning to PD [
11]. Broader access to objective, repeatable, and validated remote screening assessments that capture the multidomain features of PD symptomatology stands to improve our understanding of the global PD burden and to facilitate time to treatment and care. Here, we extend our understanding of how remotely monitored consumer-grade wearable devices and sensors can contribute to better global surveillance and screening of PD screening. Using our comprehensive platform approach, we demonstrate that PD status can be remotely evaluated population-wide across environmental conditions with high accuracy, sensitivity, and specificity. Further validation in an independent study cohort and subsequent regulatory approval will be necessary to align with the roadmap recommended by the Movement Disorders Society Task Force on Technology [
42].
Conflicts of Interest
D. Anderson, J. Severson., A. Best, M. Merickel., D. Amato, B. Severson, S. Jezewski, S. Polyak, A. Keil, M. Kantartjis, and S. Johnson are employees of Clinical ink, who acquired the BrainBaseline Movement Disorders platform in 2021. E.R. Dorsey has received compensation for consulting services from Abbott, Abbvie, Acadia, Acorda, Bial-Biotech Investments, Inc., Biogen, Boehringer Ingelheim, California Pacific Medical Center, Caraway Therapeutics, Curasen Therapeutics, Denali Therapeutics, Eli Lilly, Genentech/Roche, Grand Rounds, Huntington Study Group, Informa Pharma Consulting, Karger Publications, LifeSciences Consultants, MCM Education, Mediflix, Medopad, Medrhythms, Merck, Michael J. Fox Foundation, NACCME, Neurocrine, NeuroDerm, NIH, Novartis, Origent Data Sciences, Otsuka, Physician’s Education Resource, Praxis, PRIME Education, Roach, Brown, McCarthy & Gruber, Sanofi, Seminal Healthcare, Spark, Springer Healthcare, Sunovion Pharma, Theravance, Voyager and WebMD; research support from Biosensics, Burroughs Wellcome Fund, CuraSen, Greater Rochester Health Foundation, Huntington Study Group, Michael J. Fox Foundation, National Institutes of Health, Patient-Centered Outcomes Research Institute, Pfizer, PhotoPharmics, Safra Foundation, and Wave Life Sciences; editorial services for Karger Publications; stock in Included Health and in Mediflix, and ownership interests in SemCap. J.L. Adams has received compensation for consulting services from VisualDx and the Huntington Study Group; and research support from Biogen, Biosensics, Huntington Study Group, Michael J. Fox Foundation, National Institutes of Health/National Institute of Neurological Disorders and Stroke, NeuroNext Network, and Safra Foundation. M.A. Kostrzebski holds stock in Apple, Inc. T. Kangarloo is employees of and own stock in Takeda Pharmaceuticals, Inc. J. Cosman Cosman is an employee of and owns stock in AbbVie Pharmaceuticals.