1. Introduction
SARS-CoV-2, the Coronavirus causative of a severe acute respiratory syndrome, was identified in Wuhan, China in 2019. The SARS-CoV-2 spread quickly to several other countries and by 11th February 2020, the disease caused by this new Coronavirus, designated as COVID-19, was declared by the World Health Organization (WHO) as a Public Health Emergency of International Concern [
1]. Thereafter, it was formally declared as pandemic in March 2020. At the present time, SARS-CoV-2 has been responsible for more than 500 million infections and more than 6 million deaths worldwide [
2].
The laboratory diagnosis of SARS-CoV-2 infections has been primarily based on detection of viral RNA via Reverse Transcription Polymerase Chain Reaction (RT-PCR), which is considered the gold standard detection method for COVID-19. While the RT-PCR is highly sensitive and specific, the viral loads in upper respiratory tracks peak early during disease. These viral loads may quickly decline below the limit of detection for patients presenting later in the course of infection [
3], which can reduce the practical applicability of this method. Serological methods can be considered as a supplementary approach to fill this gap even after symptoms disappear or lack thereof. Moreover, serological tests could also provide useful epidemiological data such as background seroprevalence, persistence of class of antibodies to SAS-CoV-2 during and after the outbreak, and overall immunity status. In the past, we developed an in-house enzyme-linked immunosorbent assay (ELISA) for detecting specific IgG and IgG-isotypes antibodies to SARS-CoV-2 in a latin population that was first infected and then vaccinated with a mRNA vaccine [
4,
5]. In this study, we standardized an in-house ELISA using SARS-CoV-2 spike RBD as antigen to detect anti-SARS-CoV-2 IgM antibody. To standardize the assay, two panels of well-defined serum/plasma samples were used: one panel included SARS-CoV-2 confirmed positive samples collected during the first wave of the pandemic and the other panel included samples from healthy individuals or those carrying other viral/respiratory infections common in the latin population, collected before the 2019 outbreak.
2. Materials and Methods
2.1. Antigen and Reagents
We used commercially available recombinant SARS-CoV2 Spike-1-RBD from GenScript (No. Z03483-1), which can bind with Human ACE2 in a functional ELISA. This protein is produced in human cells with a predicted molecular weight of 30kDa and >90% purity as analyzed by SDS-PAGE (GenScript, Piscataway, NJ, USA). This assay also employed disposable, high-bind, clear, flat-bottomed, polystyrene 96-well plates (Costar, Corning Cat. No. CLS3361). For the IgM assay a mouse anti-human IgM-mu chain HRP conjugate (MyBiosource, Cat. No. MBS315374) was used as secondry antibody. Carbonate-bicarbonate buffer (Sigma-Aldrich, Cat. No. C3041-100CAP), and 3,3’, 5, 5’-Tetramethylbenzidine (TMB) (Sigma-Aldrich Cat. No. ES001) were used for coating and buffer substrate, respectively. For the neutralization assay, we used SARS-CoV-2 Receptor binding domain (RBD) horseradish peroxidase (HRP) conjugate (RBD-HRP) of the following variants: wild-type (GenScript, Cat. No. Z03594), Alpha (B.1.1.7) (GenScript, Cat. No. Z03595), Delta (B.1.617.2) (GenScript, Cat. No. Z03614) and Omicron (B.1.1.529) (GenScript, Cat. No. Z03730). The clinical performance comparison was performed using the FDA-EUA approved commercial kit sCoV-2 DetectTM IgM ELISA from InBios International, Inc. (Seattle, WA, USA) Cat. No. COVE-M.
2.2. Plasma/Serum Samples
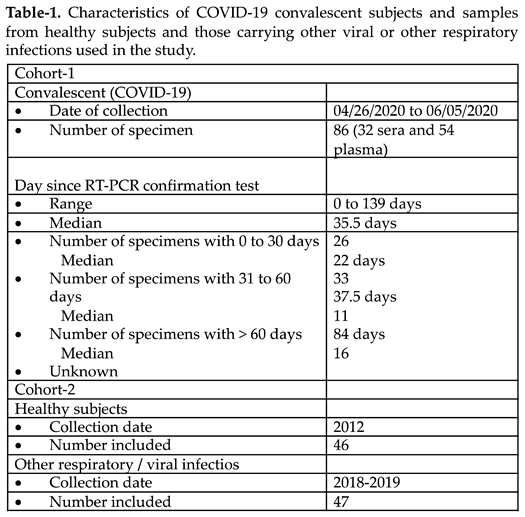
The study included a total of 178 samples from subjects of San Juan, Puerto Rico, from which eighty-six (86) were positive for SARS-CoV-2, and ninety-three (93) were negative (
Table 1). The SARS-CoV-2 positive specimens (32 serum samples and 54 plasma samples) were collected between December 2019 and March 2020, confirmed by RT-PCR and then banked at Clinical laboratories or blood banks affiliatted with the University of Puerto Rico-Medical Sciences Campus (UPR-MSC) network. Since the samples were not collected specifically for our study no personal identifiers were retained. Consequently, before the samples were received, all identifiers were removed to ensure the information could not be traced back to the individuals. The only details retrieved from 70 out of 86 subjects were the dates in which the donated specimen was collected, and the date in which the confirmatory RT-PCR for SARS-CoV-2 was performed. Hence, it was determined that 26 (30.23%) specimens had been collected between 1 to 30 days (median of 22 days) after the RT-PCR positive diagnosis; 33 (38.37%) specimens were collected between 31 to 60 days (median of 37.5 days) after the RT-PCR positive diagnosis and 11 (30.55%) specimens had been collected between 61 to 140 days (median of 84 days) following to the RT-PCR positive diagnosis. The date in which the RT-PCR was done was not available for 16 (18.60%) specimens. For this study, the time elapsed between the positive RT-PCR and the date of the collection of the sample was referred to as ‘infection time’.
Negative control group comprised 93 samples collected before the COVID-19 pandemic. Forty-six (46) samples were from healthy subjects and forty-seven (47) from subjects carrying the most common viral or respiratory allergies encountered by the Puerto Rican population. These samples had been banked at the Immunology and Molecular Parasitology Laboratory of UPR-MSC and the Virology Laboratory of UPR-MSC. The samples from subjects with viral infections were kindly donated by the Center for Disease Control and Prevention (CDC) Dengue Branch, San Juan, PR. These samples collected between 2018 to 2019 included 13 specimens from subjects with a history of respiratory allergies, 10 from subjects with Dengue virus-IgM positive diagnosis, 12 subjects with RT-qPCR Influenza A/B positive diagnosis, and 6 from subjects with Respiratory Syncytial Virus (RSV)-IgM positive diagnosis. Six (6) samples presented a positive diagnosis for Mycoplasma-IgM. All samples included in the present study were kept at -80oC and the aliquots tested had not been thawed since its collection.
2.3. In-house IgM ELISA (CovIgM-ELISA)
In-house CovIgM-ELISA is a semiquantitative assay that uses the indirect ELISA format to detect the presence of anti-SARS-CoV-2 IgM antibody in the sera and plasma. Briefly, Ninety-six-well plates were coated overnight with 100µl/well of spike RBD recombinant protein (GenScript, USA) at a concentration of 2g/ml. The unbound spike RBD was removed by washing three times the wells with 350µl/well of phosphate buffered saline (PBS) containing 0.5% Tween-20 (PBST). Non-specific binding was blocked by adding 300µl/well of 3% bovine serum albumin (BSA) diluted in PBST and incubated at 37oC for 30 minutes. Serum/plasma samples were diluted 1:100 in blocking solution and added to wells in duplicate (100µl /well). The blocking solution was used as blank. Plates were washed three times after incubation at 37oC for 30 min. Then, the secondary antibody, anti-human IgM-peroxidase conjugate, diluted 1:50,000 in PBST was added to each well (100µl/well) and incubated at 37oC at 30 min. The peroxidase reaction was visualized by adding the substrate solution 3,3’,5,5’-tetramethylbenzidine (TMB) and incubating in the dark at room temperature for 15 minutes. The reaction was stopped by adding 50µl/well of 1N HCl. Absorbances at 450 nm was determined with a spectrophotometer. The OD450 of blanks were subtracted from OD450 of each sample before data analysis.
2.4. Plasma/Serum Equivalence and Precision Study
To assess the equivalence of the IgM determinations made on plasma and serum, a comparison using both type of blood-derived samples, collected from the same individuals, was performed within the in-house CovIgM-ELISA. Five positive and five negative specimens were included in this study. To evaluate the precision of these determinations, specimens were tested in duplicate on three different days. Within-run and total analytical imprecision was calculated according to the Clinical and laboratory Standards Institute (CLSI) guideline EP5-A [
6].
2.5. Autoantibody Analysis
Human rheumatoid factor (RF) IgM was measured using a RF-IgM ELISA kit (Creative Diagnosis, Cat. No. DEIA 1697), which is a rapid test for the qualitative and semi-quantitative detection of RF-IgM class antibody in human serum.
2.6. cPass Neutralization Test
Having previously demonstrated that the results that provide the surrogate viral neutralization test (cPass
TM GenScript sVNT, Piscataway NJ) [
7] correlate perfectly with the traditional PRNT [
8] we used a cPass neutralization antibody test to determine the levels of neutralizing antibodies in the study population. cPass utilizes the recombinant RBD of the SARS-CoV2 spike protein to detect antibodies that block the RBD from binding to human ACE receptor. Briefly, specimens from negative and positive controls as well as the standards provided by the kit were diluted 1:10 in the sample dilution buffer according to the manufacturer’s instructions and pre-incubated with RBD-HRP for 30 min at 37
oC. This allows the interaction and binding of specific antibodies to RBD-HRP. Following incubation, each reaction mixture is then added to a 96 well capture plate coated with human ACE-2 protein. In this way, either free RBD-HRP or RBD-HRP bound to non-neutralizing antibodies can strongly interact with ACE2 being captured on the plate. RBD-HRP complexed with neutralizing antibodies remain in the supernatant and are removed in a subsequent wash step. Next, the reaction is developed by addition of TMB, followed by the addition of a stop solution, which allows for the visualization of bound RBD-HRP to the ACE2. Since this is an inhibition assay, color intensity is inversely proportional to the number of neutralizing antibodies present in the samples. Data is interpreted by calculating the percent of inhibition of RBD-HRP binding as follows: Percent inhibition = (1-OD value of sample/OD value of background) * 100%. Samples with neutralization activity ≥ 30% indicate the presence of SARS CoV-2 RBD-interacting antibodies capable of blocking the RBD-ACE2 interaction, thus, inhibiting the entrance of the virus into the cell.
2.7. Performance Comparison between the In-House CovIgM-ELISA and a Commercial EUA Approved IgM-ELISA Kit
The performance of the in-house CovIgM-ELISA was compared to that of a commercially available test by using 30 samples randomly selected from the COVID-19 confirmed cohort. The collected sera were additionally tested at double blind by two different operators using the FDA-EUA approved commercial kit SCoV-2 Detect
TM IgM ELISA, which uses S1 RBD as antigen. Serum samples for this test were processed and results interpreted according to the manufacturer’s instructions. Briefly, the immunological status ratios (ISR) of control and samples’ ODs over the calibrator were calculated. Results were interpreted by following InBios recommendations: ISR > 0.9 to 1.1 were defined as borderline and retested. If the average ISR from the repeated duplicate testing is >1.0, the sample is considered positive. If the average ISR from the duplicate testing is <1.0, the sample is considered negative. ISR >1.1 Positive, ISR <0.9 Negative. To complete the comparison between the in-house CovIgM-ELISA and the commercial InBios SCoV-2 Detect
TM IgM ELISA kit performance, a cost per sample analyzed at laboratory level was included. The latter evaluation only considered the cost of the reagents (in-house CovIgM-ELISA) or the commercial value of the kit (InBios SCoV-2 Detect
TM IgM ELISA). A list with the costs in USD of all reagents used for the CovIgM-ELISA has been included in
Table S1 & S2.
2.8. Data Analysis
All antibody determinations were done in duplicate and results reported as mean absorbance for each determination. A receiver operating characteristic (ROC) curve was generated to establish the cut-off value using the EpiTools epidemiological calculator (
http://epitools.ausvet.com.au). Arbitrary guidelines were followed when analyzing the area under curve (AUC) as follows: non-informative (AUC = 0.5); low accurate (0.5 < AUC < 0.7); moderately accurate (0.7 < AUC < 1); perfect (AUC = 1) [
9].
95% confidence intervals were calculated for sensitivity and specificity of different cut off values. Deming regression analysis [
10] was performed on paired plasma and serum specimens collected from the same individual to demonstrate the equivalence in the assays results for both specimens. To evaluate the agreement between the in-house CovIgM-ELISA and the RT-PCR, between in-house CovIgM-ELISA and a commercial kit as well as between in-house CovIgM-ELISA and the neutralization percentages determined by the cPass neutralization assay. The inter-rater agreement (kappa, κ) was applied according to the method described by Thrusfield [
11]. The κ values were considered as follows: slight agreement (κ = 0.01 to 0.2); fair agreement (κ = 0.21 to 0.40); moderate agreement (κ = 0.41 to 0.60); substantial agreement (κ = 0.61 to 0.80); almost perfect agreement (κ = 0.81 to 1.0) [
12,
13]. All statistical analyses were performed in GraphPad Prism 9.
2.9. Ethics Statement
None of the samples analyzed in the present study were specifically collected for this study. COVID-19 samples were kindly donated by collaborators from local Clinical laboratories or Blood banks. The samples were stripped of all identifiers so that the information could not be traced back to the individuals. All samples donated to us were from subjects >21 years-old at the time in which the confirmatory positive RT-PCR was performed. The samples from negative population were all volunteers participating in the IRB-approved clinical protocol “Molecular Basis and Epidemiology of Viral Infections Circulating in Puerto Rico” (Pro0004333); approved by the Advarra IRB on 21 April 2020.
3. Results
3.1. Receiver Operating Characteristics (ROC), OD Distribution, and the Sensitivity and Specificity of the In-House CovIgM-ELISA
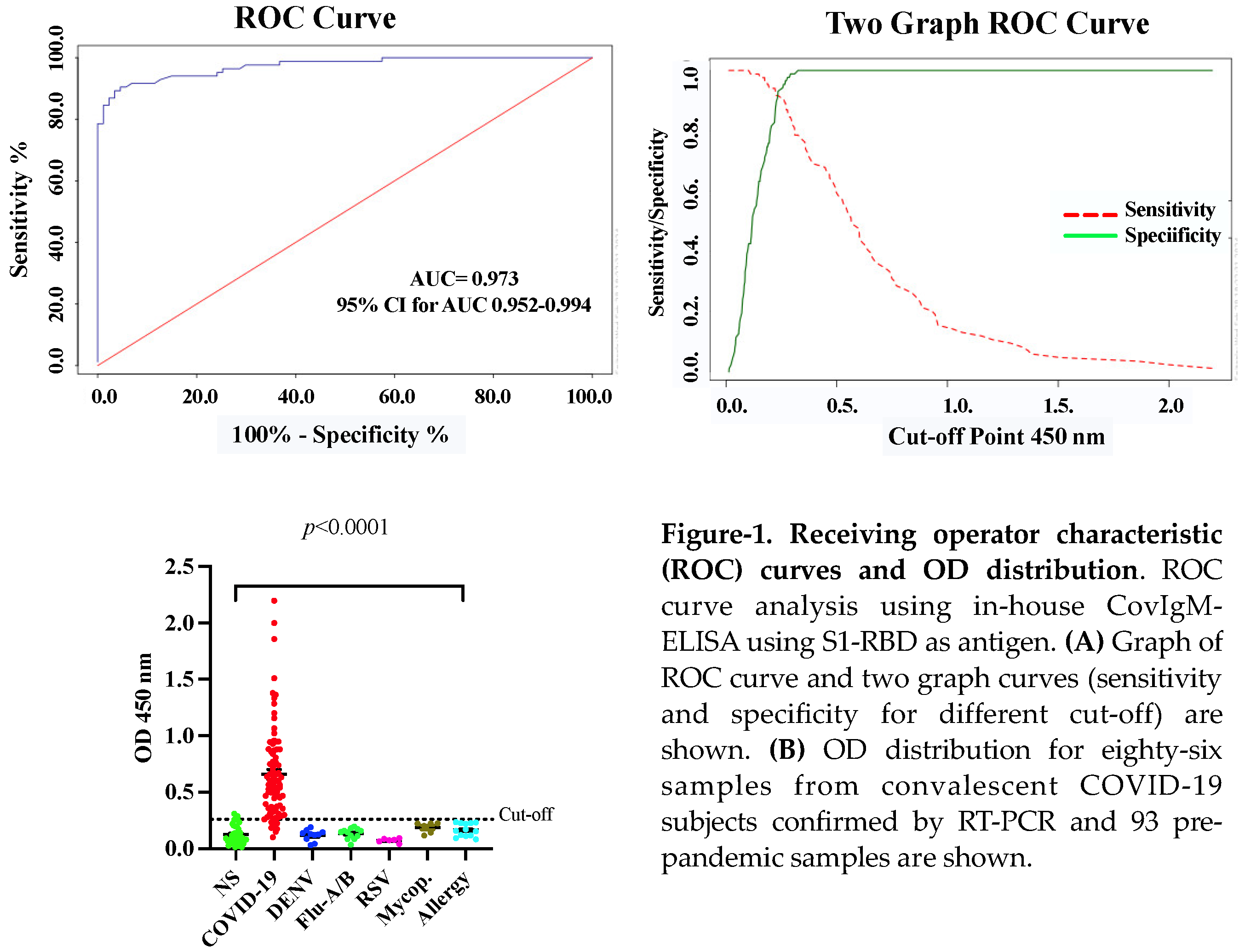
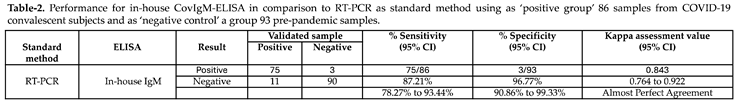
The in-house CovIgM-ELISA was standardized by measuring the background reading and using two types of controls: a pool of serum fromconvalescent patients as the positive control, and a pool from healthy subjects collected before the 2019 pandemic as negative control. Optimal concentrations of antigen, serum and conjugate for the in-house CovIgM-ELISA were defined by checkerboard titration to assess reproducibility and exclude nonspecific reactions. The background of the assay was defined as the reading measured from the detection system in the absence of any tested sera, which had to be lower than the reading of any serum ODs. Additionally, to confirm that the assay was working, the positive controls as well as the negative control at the optimized dilution was used as controls in each test. The in-house CovIgM-ELISA showed background readings that were close to zero and as expected, all sample readings had OD values lower than the positive controls. We created receiver operating characteristic (ROC) curves to establish the optimal positive cut-off for detecting IgM levels in SARS-CoV-2 infected samples. For this analysis we used 86 samples from subjects with confirmed SARS-CoV-2 infection, which had been confirmed by RT-PCR. Moreover, to provide increased reassurance of specificity a large panel of pre-pandemic samples collected prior to the 2019 pandemic from healthy donors as well as from subjects previously confirmed by RT-PCR or ELISA to have viral RNA or IgG/IgM antibodies for Dengue virus (DENV), Influenza A and B, Respiratory synctitial virus (RSV) and Mycoplasma, respectively, were used to further ensure specificity. ROC curve analysis indicated a mean sensitivity of 90.05% (95% CI: 82.3% to 95.10%), and a mean specificity of 96.77% (95% CI: 90.86% to 99.3%), with an AUC of 0.973 (95% CI: 0.952-0.994) (
Figure 1A) and an OD
450 positive cut-off value of 0.26, which represents 2-fold of mean OD of the negative samples. The cut-off represents the point in the ROC curves where the sensitivity and specificity were maximized for this set of samples. Thus, when the performance of the in-house CovIgM-ELISA was compared to the RT-PCR as standard method the agreement between both methods was assessed as almost perfect (kappa= 0.843, 95% CI: 0.764 to 0.922) (
Table 2).
To determine whether the results of our in-house CovIgM-ELISA could vary depending on the type of specimen (serum or plasma) used, we tested paired samples from the same subject and submitted the results to a Deming regression analysis. A high equivalence between sera and plasma samples was found, which was highly significant (
Figure S1). Moreover, at the comparison between duplicated runs on alternate days accomplished by different operators, CovIgM-ELISA showed an adequate consistency, and a low error levels, with intra-assay and inter-assay variations lower than 15%. The OD distribution of validated samples was shown in a scatter plot with cut-off lines (
Figure 1B). No cross-reactions were detected when sera from individuals with other respiratory or viral infection. However, three samples from healthy subjects resulted positive in the in-house CovIgM-ELISA with OD
450 values in the range between 0.26 to 0.35.
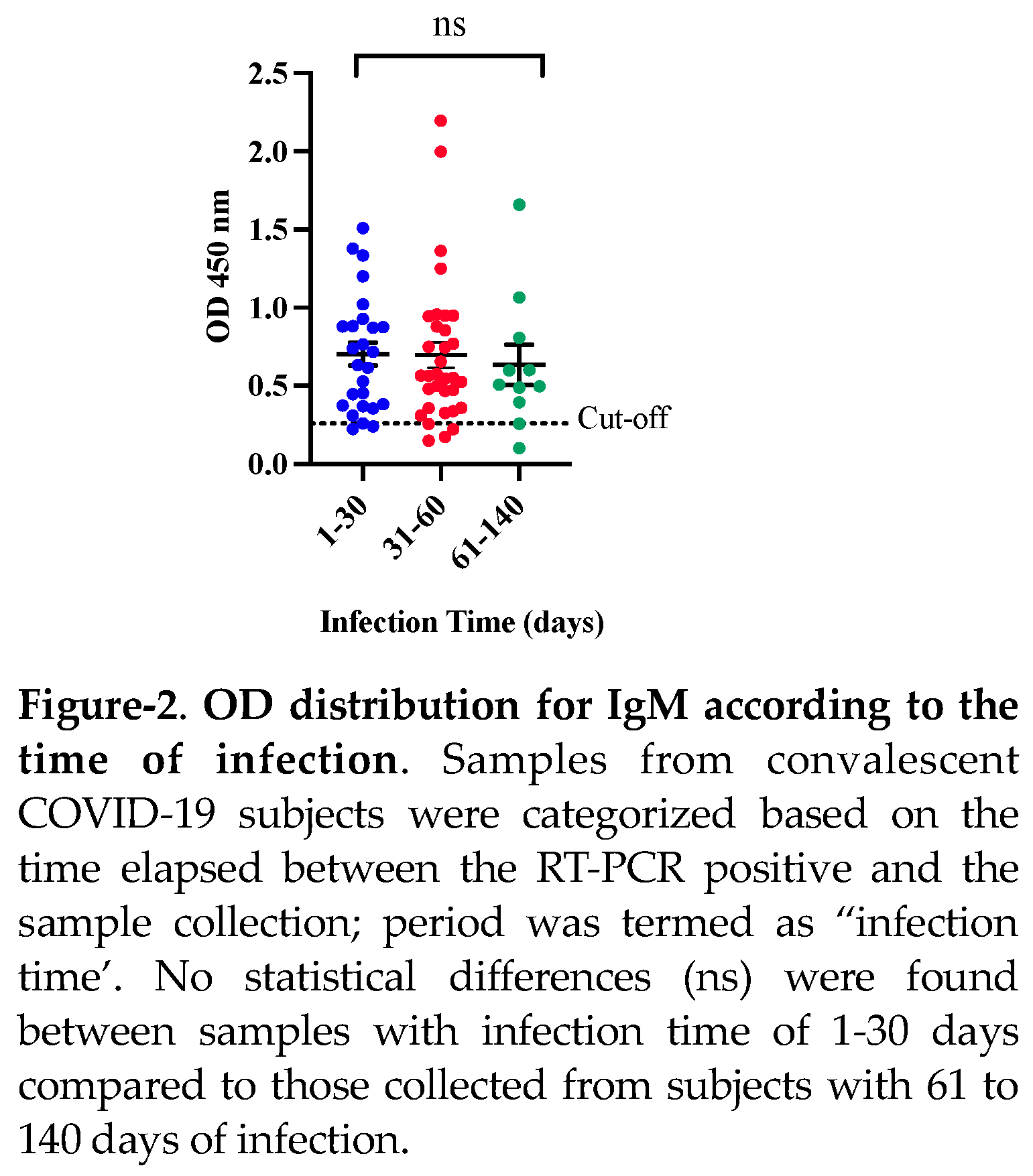
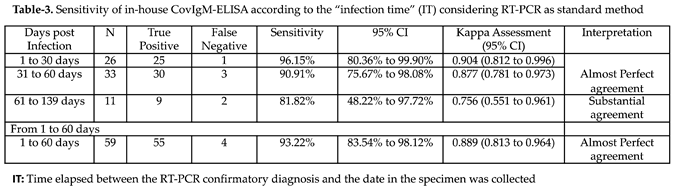
When the clinical performance of CovIgM-ELISA was evaluated by allotting the samples based on the estimated ‘infection time’ it was also found that 25 from 26 (96.15%) samples collected between 1 to 30 days following to the RT-PCR confirmatory diagnosis resulted IgM positive, whereas that 30 from 33 (90.91%) specimens collected between 31 to 60 days after the RT-PCR and 9 from 11 (81.8%) samples collected from subjects with more than 60 days after the RT-PCR were also found positive to IgM. Thus, in-house CovIgM-ELISA showed a sensitivity of 96.15% (95% CI: 80.36% to 99.90%) detecting anti-SARS-CoV-2 IgM antibody in samples from subjects with less of 30 days of infection and a sensitivity of 93.22% (95% CI: 83.84% to 98.12%) detecting IgM in samples from subjects with up to 60 days of infection (
Table 3). Paradoxically, although the sensitivity of in-house CovIgM-ELISA decreased to the extent that samples with longer infection times were included, the mean absorbance did not seem to decrease in the same proportion and no significant differences were observed between the IgM levels detected in the group of sera with l to 30 days, or 31 to 60 days with respect to those collected with more than 60 days of infection
(Figure 2).
To rule out that the anti-SARS-CoV-2 IgM levels detected by the in-house CovIgM-ELISA were influenced by the rheumatoid factor (RF) as it has been reported elsewhere [
14], we proceed to determine the presence of IgM-RF in our cohort of samples from COVID-19 convalescent subjects. This possibility caanot be ruled out, as rheumatoid arthritis (RA) is one of the most frequent autoimmune diseases in the Puerto Rican population [
15], and the clinical status of the subjects used in the present study is unknown. Our results demonstrated that none of samples with anti-SARS-CoV-2 IgM antibodies resulted positive to IgM-RF, which confirm that the levels of IgM detected in these samples were selicited against the SARS-CoV-2 and specifically detected by our in-house CovIgM-ELISA. Detailed information regarding the average OD values for IgM, type of specimen, time of infection and result interpretation is provided in
Table S3.
3.2. Clinical Performance of the In-House CovIgM-ELISA in Comparison to the SCoV-2 DetectTM IgM ELISA (InBios International Inc)
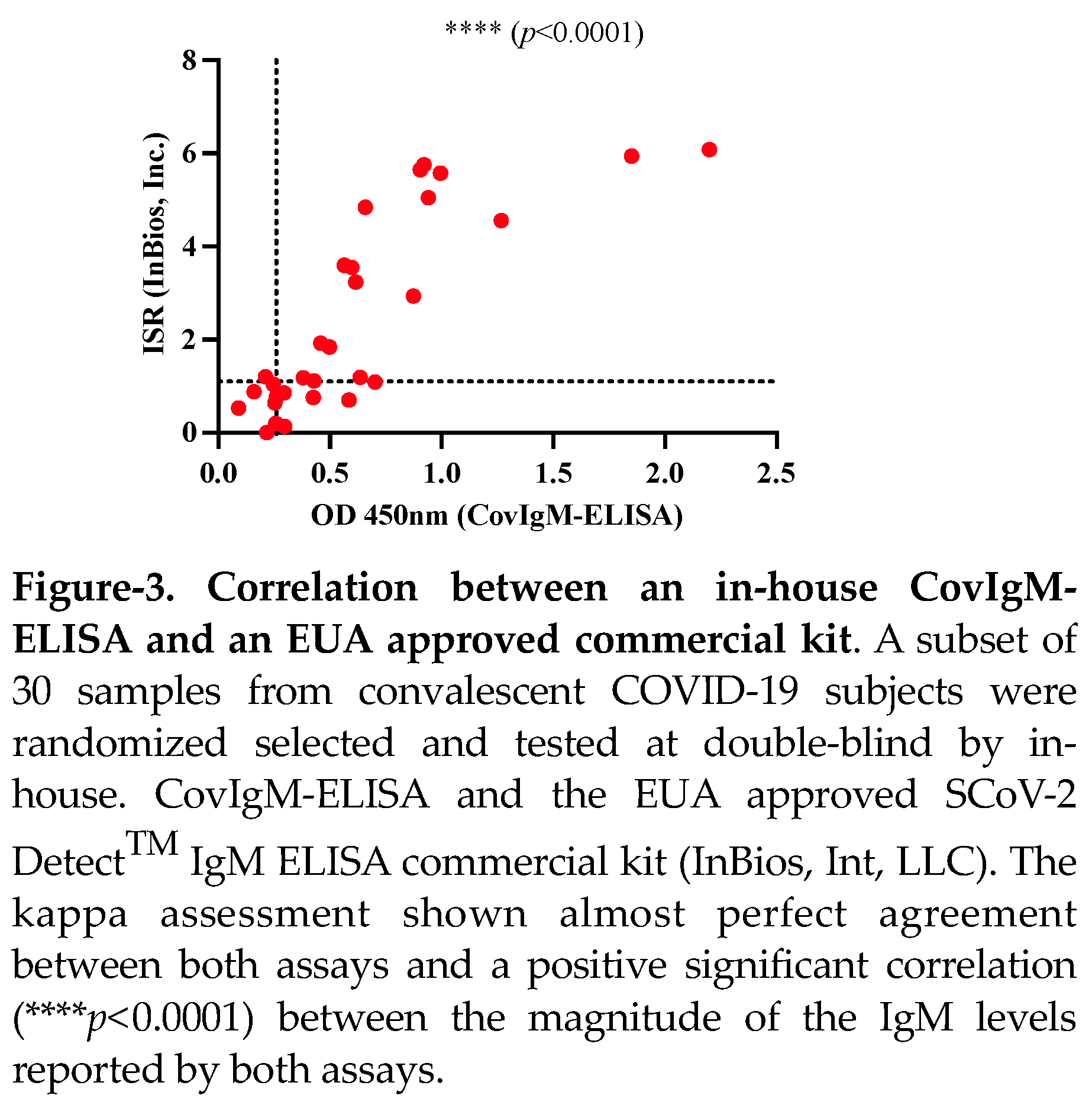
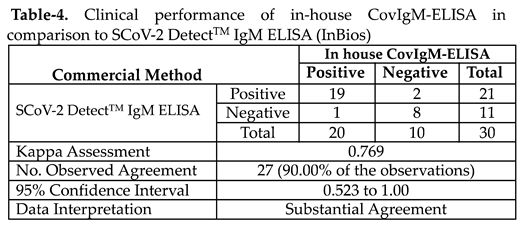
The clinical performance of our in-house CovIgM-ELISA was compared with a commercial ELISA kit (SCoV-2 Detect
TM IgM ELISA) using a sub-cohort of 30 samples randomly selected, which were tested at double blind by two different operators. We found that the in-house CovIgM-ELISA had substantial agreement with the SCoV-2 Detect
TM IgM ELISA kit (kappa= 0.769, 95CI: 0.523 to 1.00) (
Table 4, Table S4). Also, it was found a positive and significant correlation (
p<0.0001) in the levels of IgM detected by both assays (
Figure 3). The use of SCoV-2 Detect
TM IgM ELISA kit had a cost of
$7.50 per analyzed serum, whereas the in-house CovIgM-ELISA had a cost of
$2.45 per serum (
Table S2), which is 3.6 times cheaper than a commercial test, when labor is not considered.
3.3. Association between Levels of IgM Detected by the In-House CovIgM-ELISA and the Neutralizing Activity
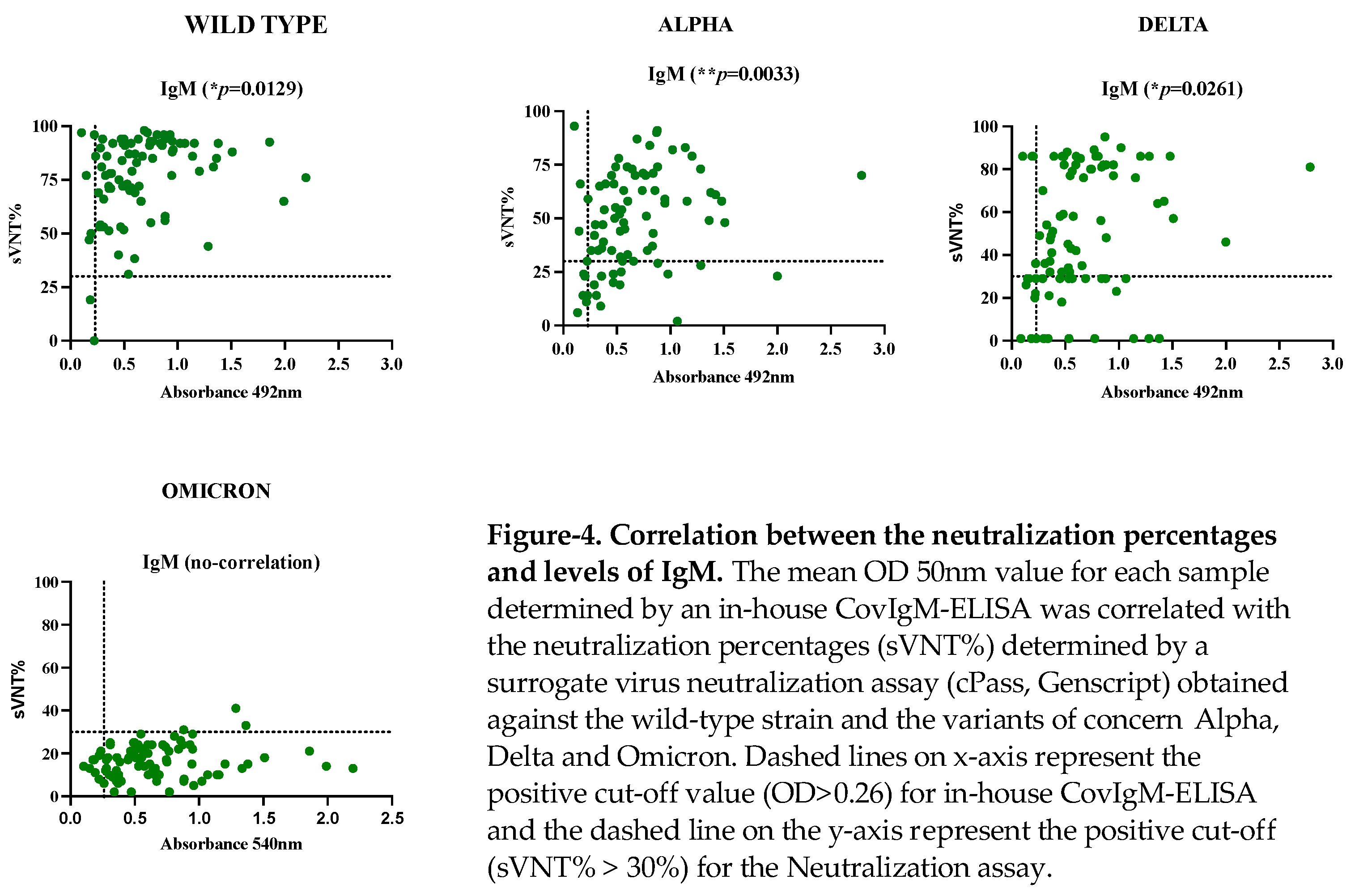
The samples from the SARS-CoV-2 confirmed cohort were tested by a surrogate virus neutralization assay (cPass) to quantify the neutralization percentages (sVNT%) against different variants of concern (VOCs). We found that the 98.82% of samples had detectable sVNT% above the positive threshold against the wildtype (WT) strain, 75.3% had detectable sVNT% against the ALPHA variant, 85.88% had detectable sVNT% against DELTA variant and only 3.5% of samples had detectable sVNT% against OMICRON. Consistent with these results we found that the IgM levels positively correlated with the sVNT% against the WT strain and the VOCs ALPHA and DELTA
(Figure 4, Table S5). 4. Discussion
We have developed an in-house ELISA method for measuring levels of IgM to SARS-CoV-2 in serum/plasma of patoents infected with SARS-CoV-2 using as antigen a recombinant variant of the domain S1 from spike protein and the receptor binding domain (S1-RBD). S1 and RBD were selected as target antigens among the SARS-CoV-2 proteins because these two proteins have been reported as the most suitable for diagnostic assays. The S1 protein is highly specific whereas the RBD exhibit greater sensitivity in the detection of subjects with mild infections [
16]. Other viral proteins, such as the S2 domain and nucleocapsid (N) exhibit cross-reactivity with the spike protein of MERS-CoV [
17], and even more so with SARS patient sera [
18]; therefore, they have been shown to be are less suitable for diagnosis. From our cut-off in-house CovIgM-ELISA, optimized the OD value of the IgM cut-off over 0.26, providing high sensitivity and specificity. This positive cut-off, which represents a 2-fold of the mean OD of the negative samples, is concordant with other ELISA assays that set up their cut-off values at three standard deviations (SD) above the mean of the negatives [
19].
IgM is a marker of acute infection, and it can be detected as early as 3 to 5 days after symptoms onset [
20]. IgM can reach its peak at 15 to 22 days, then it will gradually decline, disappearing after 61-90 days in all patients [
21,
22,
23], which could also reflex an antibody class switch to IgG [
24]. This dynamic is consistent with the observation that the in-house CovIgM-ELISA is more sensible (96.15%) when the analysis is made with samples collected between 1-30 days of infection, as seen in other studies [
20]. Only a single sample from this group, collected one day after RT-PCR resulted negative to IgM (see
Table S3). Since all COVID-19 samples were RT-PCR positive and were donated to us without identifiers, and no clinical data was available, it is possible to speculate that at the time of the sampling, the adaptive antibody response of this subject had not yet developed, which often occur few days after infection [
20].
We also found IgM positive in samples collected from subjects with up to 139 days of infection. Although this is an atypical finding it is consistent with some studies reporting the presence of anti-SARS-CoV-2 IgM antibody specific for S1-domain in subjects with up to 90 days after the symptom’s onset [
23]; whereas other studies report that IgM-antibodies specific to RBD could remain detectable 3 to 6 months after the disease onset [
25]. We have ruled-out that these atypical IgM levels can be cross-reactions with the RF or infection with other viruses because all these samples were negative to RF-IgM, and our assay did not showed cross-reaction with other viruses highly endemic in the island. Moreover, the samples were also positive by a commercial kit of high sensibility and specificity that received an Emergency Use Authorization (EUA) status from the US Food and Drug Administration (FDA) for COVID-19. Therefore, we infer that the atypical persistency of IgM in these samples could be a characteristic of the Latin population given that genetic background has shown to influence in the response to the SARS-CoV-2 virus [
26]. Alternatively, these subjects could have been reinfected with other Alpha or Delta subvariants. Although previous evidence suggest that antibodies from a primary SARS-CoV-2 infection can provide protection [
27], and convalescent plasma or vaccination programs are based on the assumption that humoral response can help prevent reinfection [
28,
29], it can still occur [30].
Despite no cross-reaction with DENV, Mycoplasma, RSV or Influenza were detected in our study, a small number of pre-pandemic samples (6.5%) from healthy donors tested positive for IgM, despite having very low OD values. We hypothesize that these false positives could be cross-reactions with seasonal human coronaviruses such as NL63, 229E and NL63, which have circulated within the Puerto Rican population for a long time and are responsible of a high percentage of common colds [
19]. A more robust cross-reactive analysis to rule-out these possibilities could be needed prior to accomplish large epidemiological studies. Despite this limitation, our in-house CovIgM-ELISA has several advantages that should be highlighted. This assay is not only easy to perform but also a low-cost alternative when compared to the one offered by InBios, which is one of the cheapest commercially available kits that uses S1-RBD as target antigen. The manufacturer of ScoV-2 Detect
TM IgM ELISA reported a sensitivity of 96.7% when testing human serum collected between 7-60 days post onset of symptoms (
http://inbios.com/product/scov-2-detect-igm-elisa-kit/). A similar sensitivity could be seen by our in-house CovIgM-ELISA when testing samples with 30 or up-to 60 days of infection.
Another important observation of this study is that the IgM levels determined by the in-house CovIgM-ELISA correlated with the neutralization percentages (sVNT%) against the VOCs of the Alpha and Delta variants determined by the a surrogate virus neutralization assay. This is consistent with previous studies, which had demonstrated that IgM is one of the antibody classes that contribute most to SARS-CoV-2 neutralization [31]. By the time that all the samples were collected, Alpha and Delta were the dominant VOCs circulating in Puerto Rico [
23], which emerged from the original Wuhan strain, and they exhibit a relatively small number of mutations in their genome. The observation that IgM antibodies in these samples were completely infective against Omicron was expected because this variant developed more than 50 mutations [
24].
5. Conclusions
In conclusion, the high sensitivity, specificity and accuracy demonstrated by the in-house CovIgM-ELISA indicate that our assay could not only detect anti-SARS-CoV-2 IgM antibody, but also could performs at a level of performance comparable to that of the RT-PCR and SCoV-2 Detect
TM IgM ELISA kits. From a diagnostic point of view, detection IgM is highly advantageous because it is the first antibody class elicited shortly after infection with SARS-CoV-2. Indeed, IgM detection could be more sensitive than RT-PCR for more than five days of duration [32], even if the viral load is under RT-PCR detection limits. However, IgM detection alone should not be used as the only method for screening COVID-19 cases. It should also be combined with IgG detection for a more accurate and sensitive diagnostic approach. In this context, we believe that in-house CovIgM-ELISA along with the CovIgG-ELISA developed by our research group for detecting anti-SARS-CoV-2 IgG antibody [
4,
8], can be used for the diagnosing suspected COVID-19 patients or for conducting large epidemiological studies in a resources-limited setting, such as Puerto Rico, at a fraction of the cost, when qualified personnel are available. The in-house CovIgM-ELISA is no time-consuming; it can be completed in a maximum of 2.5 hours, and 96 single samples or 48 samples in duplicate, could be tested per each single plate in the same period. Our university laboratories are well-equipped and counts with full-time skilled technicians. The in-house-CovIgM-ELISA is an available alternative that can make feasible to conduct COVID-19 surveillance programs at academic institutions or similar facilities with limited access to diagnostic and research resources.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Deming analysis for serum/plasma equivalence and reproducibility of the in-house IgM ELISA; Table S1: In-house ConvIgM-ELISA reagents and instruments; Table S2: Bulk cost in USA dollars of all reagents used to run in-house CovIgM-ELISA; Table S3: IgM levels measured by the in-house CovIgM-ELISA in samples from COVID-19 convalescent subjects. Cut-off >0.26; Table S4: Results obtained with the in-house CovIgM-ELISA compared to an EUA approved commercial kit (SCoV-2 DetectTM IgM ELISA); Tabl S5: Levels of IgM and neutralization percentages determined within samples from no-vaccinated, COVID-19 convalescent subjects.
Author Contributions
Conceptualization, A.M.E. and C.A.S; methodology, A.M.E, C.A.S., A.A.R, validation, A.M.E., C.O.M, P.C., formal analysis, A.M.E.; C.A.S., investigation, A.M.E., C.A.S., A.A.R, L.A., and C.O. M. resources, C.A.S. and A.M.E.; data curation, A.M.E; writing—original draft preparation, A.M.E.; writing—review and editing, C.A.S, R., A.A.R, L.A., C.O.M, P.C.; supervision, A.M.E, C.A.S.; funding acquisition, C.A.S. , A.M.E. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by 1U01CA260541-01 to CAS (NCI/NIAID). The Puerto Rico Science, Technology and Research Trust and Department of Economic Development of Puerto Rico also supported the research reported in this work under agreement number 2020-00272 to CAS and AME and agrement number 2024-000450 to AME, respectively.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the University of Puerto Rico Medical Sciences Campus IRB (protocol: #1250121) and the Advarra IRB-approved clinical protocol “Molecular Basis and Epidemiology of Viral Infections Circulating in Puerto Rico” (Pro0004333). Continuing review approval March 10, 2023.
Acknowledgments
Authors acknowledge Dr. Jorge Muñoz from CDC-San Juan for kindly donate the samples from subjects carrying viral diseases.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Organization WH. WHO DirectorGeneral’s opening remarks at the media briefting on COVID-19: 11 March 2020. https://wwwwhoint/dg/speeches/detail/who-directorgeneral-s-opening-remarks-atthe-media-briefingon-covid-19. 2020.
- Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, Bruggen MC, et al. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75(7):1564-81. [CrossRef] [PubMed] [PubMed Central]
- Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465-9. [CrossRef] [PubMed]
- Espino AM, Armina-Rodriguez A, Alvarez L, Ocasio-Malave C, Ramos-Nieves R, Rodriguez Martino EI, et al. The Anti-SARS-CoV-2 IgG1 and IgG3 Antibody Isotypes with Limited Neutralizing Capacity against Omicron Elicited in a Latin Population a Switch toward IgG4 after Multiple Doses with the mRNA Pfizer-BioNTech Vaccine. Viruses. 2024;16(2). [CrossRef] [PubMed] [PubMed Central]
- Sariol CA, Pantoja P, Serrano-Collazo C, Rosa-Arocho T, Armina A, Cruz L, et al. Function is more reliable than quantity to follow up the humoral response to the Receptor Binding Domain of SARS- CoV-2 Spike protein after natural infection or COVID-19 vaccination. medRxiv : the preprint server for health sciences. 2021. [CrossRef] [PubMed] [PubMed Central]
- protocol CE-A. Evaluating of precision performance of quantitative measurements methods. Apprved guidelines. 2004;24, 2nd ed. Wayne, PA: CLSI.
- Taylor SC, Hurst B, Charlton CL, Bailey A, Kanji JN, McCarthy MK, et al. A New SARS-CoV-2 Dual-Purpose Serology Test: Highly Accurate Infection Tracing and Neutralizing Antibody Response Detection. Journal of clinical microbiology. 2021;59(4). [CrossRef] [PubMed] [PubMed Central]
- Sariol CAA, Pantoja P, Serrano-Collazo C, Rosa-Arocho T, Armina-Rodriguez A, Cruz L, et al. Function Is More Reliable than Quantity to Follow Up the Humoral Response to the Receptor-Binding Domain of SARS-CoV-2-Spike Protein after Natural Infection or COVID-19 Vaccination. Viruses. 2021;13(10). [CrossRef] [PubMed] [PubMed Central]
- Hosmer DW, Jr.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression;. John Wiley & Sons: Hoboken, NJ, USA,. 2013.
- Linnet K BJ. Selection and analytical evaluation of methods - with statistical techniques. . In Burtis CA, Ashwood ER, Bruns DE (eds) Tietz Textbook of Clinical Chemistry and Molecular Diagnostics (5th edn) Elsevier Saunders, St Louis, MO,. 20012:201-28.
- Thrusfield M. Veterinary Epidemiology, 2end. Balckewell Science Ltd., 25 John Street, London, WC1N 2 BL, UK. 1995.
- Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37-46. [CrossRef]
- Wieckowska BK, K.B.; Jozwiak, P.; Moryson, W.; Stawinska-Witoszynska, B. Cohen’s Kappa Coefficient as a Measure to Assess Classification Improvement following the Addition of a New Marker to a Regression Model. Int J Environ Res Public Health. 2022;19:10213. [CrossRef]
- Oka S, Higuchi T, Furukawa H, Shimada K, Hashimoto A, Matsui T, et al. False-positive detection of IgM anti-severe acute respiratory syndrome coronavirus 2 antibodies in patients with rheumatoid arthritis: Possible effects of IgM or IgG rheumatoid factors on immunochromatographic assay results. SAGE Open Med. 2022;10:20503121221088090. [CrossRef] [PubMed] [PubMed Central]
- Miranda EM, Han X, Park SH, Suri S, Suryavanshi M. Treatment Patterns Among Patients with Rheumatoid Arthritis in Puerto Rico. Rheumatol Ther. 2022;9(2):609-19. [CrossRef] [PubMed] [PubMed Central]
- Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerging infectious diseases. 2020;26(7):1478-88. [CrossRef] [PubMed] [PubMed Central]
- Oliveira BA, Oliveira LC, Sabino EC, Okay TS. SARS-CoV-2 and the COVID-19 disease: a mini review on diagnostic methods. Revista do Instituto de Medicina Tropical de Sao Paulo. 2020;62:e44. [CrossRef] [PubMed] [PubMed Central]
- Chia WN, Tan CW, Foo R, Kang AEZ, Peng Y, Sivalingam V, et al. Serological differentiation between COVID-19 and SARS infections. Emerging microbes & infections. 2020;9(1):1497-505. [CrossRef] [PubMed] [PubMed Central]
- Mahallawi WH. A serological assay to detect human SARS-CoV-2 antibodies. J Taibah Univ Med Sci. 2021;16(1):57-62. [CrossRef] [PubMed] [PubMed Central]
- Al-Tamimi M, Tarifi AA, Qaqish A, Abbas MM, Albalawi H, Abu-Raideh J, et al. Immunoglobulins response of COVID-19 patients, COVID-19 vaccine recipients, and random individuals. PLoS One. 2023;18(2):e0281689. [CrossRef] [PubMed] [PubMed Central]
- Amellal H, Assaid N, Charoute H, Akarid K, Maaroufi A, Ezzikouri S, et al. Kinetics of specific anti-SARS-CoV-2 IgM, IgA, and IgG responses during the first 12 months after SARS-CoV-2 infection: A prospective longitudinal study. PLoS One. 2023;18(7):e0288557. [CrossRef] [PubMed] [PubMed Central]
- Kaduskar O, Gurav YK, Deshpande K, Desphande GR, Yadav P, Rakhe A, et al. Understanding the dynamics of IgM & IgG antibodies in COVID-19-positive patients. Indian J Med Res. 2022;155(5&6):565-9. [CrossRef] [PubMed] [PubMed Central]
- Ha B, Jadhao S, Hussaini L, Gibson T, Stephens K, Salazar L, et al. Evaluation of a SARS-CoV-2 Capture IgM Antibody Assay in Convalescent Sera. Microbiol Spectr. 2021;9(2):e0045821. [CrossRef] [PubMed] [PubMed Central]
- Valdes-Fernandez BN, Duconge J, Espino AM, Ruano G. Personalized health and the coronavirus vaccines-Do individual genetics matter? Bioessays. 2021;43(9):e2100087. [CrossRef] [PubMed] [PubMed Central]
- Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang ML, et al. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. Journal of clinical microbiology. 2020;58(11). [CrossRef] [PubMed] [PubMed Central]
- Corey L, Mascola JR, Fauci AS, Collins FS. A strategic approach to COVID-19 vaccine R&D. Science (New York, NY. 2020;368(6494):948-50. [CrossRef] [PubMed]
- Joyner MJ, Senefeld JW, Klassen SA, Mills JR, Johnson PW, Theel ES, et al. Effect of Convalescent Plasma on Mortality among Hospitalized Patients with COVID-19: Initial Three-Month Experience. medRxiv : the preprint server for health sciences. 2020. [CrossRef] [PubMed] [PubMed Central]
- Goldman JD, Wang K, Roltgen K, Nielsen SCA, Roach JC, Naccache SN, et al. Reinfection with SARS-CoV-2 and Waning Humoral Immunity: A Case Report. Vaccines (Basel). 2022;11(1). [CrossRef] [PubMed] [PubMed Central]
- Klingler J, Weiss S, Itri V, Liu X, Oguntuyo KY, Stevens C, et al. Role of IgM and IgA Antibodies in the Neutralization of SARS-CoV-2. medRxiv : the preprint server for health sciences. 2020. [CrossRef] [PubMed] [PubMed Central]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).