1. Introduction
A precise and standardized classification system is essential in hematology. This system is based on detailed immunophenotypic and molecular characteristics of the diseases and ensures that patients receive personalized and effective treatment. Such classification is also important to advance research on the various aspects of hematologic diseases, including their causes, prevention strategies and therapeutic approaches.
The World Health Organization (WHO) classification of lymphoid tumors is an internationally recognized framework for the diagnosis of lymphoid neoplasms. The latest edition, the fifth, was published in 2022 and integrates the latest scientific discoveries and diagnostic advances. This comprehensive classification system ranks tumors based on their origin, morphological features and molecular biology. It was developed to increase diagnostic accuracy and improve the clinical management of these tumors [
1].
The latest edition of the WHO classification emphasizes the importance of the complete blood count (CBC) and the assessment of cell morphology in peripheral and bone marrow smears for the diagnosis of lymphoproliferative diseases. Microscopic examination of peripheral blood smears remains a cornerstone for the assessment of abnormal samples and the detection of pathologic cells. Despite its crucial role, this process is time-consuming, requires highly skilled personnel and a high level of expertise, and interpretation is highly dependent on the skills of the operator [
2].
In today's clinical laboratories, the workload is increasing, especially for CBC examinations, while the availability of specialized personnel is decreasing. This situation underscores the urgent need for effective methods to determine which blood values warrant further diagnostic testing. In addition, small clinical laboratories that focus on urgent testing and laboratories in developing countries with limited resources often do not have access to advanced diagnostic tests [
3]. In these situations, the CBC often serves as the primary tool for diagnosing suspected lymphoproliferative disorders.
Recent advances in hematology analyzers have greatly expanded their functionality beyond traditional leukocyte differential analysis. These state-of-the-art instruments utilize innovative chemical and physical principles for cell analysis, combined with advanced software management, to provide enriched data sets that include parameters for research use only (RUO) and detailed cell population data (CPD). This technological advance improves the detection of pathologic cells and aids in screening for hematologic neoplasms [
4,
5].
The clinical application of these parameters offers numerous advantages. They are easily accessible and are determined simultaneously with the conventional CBC parameters without the need for additional sampling. These parameters provide quantitative information on the morphological characteristics of leukocytes, including lymphocytes. The morphological characteristics can be accurately measured, making them very useful to identify dimensional, structural and morphological changes of lymphocytes under non-physiological conditions [
6,
7,
8,
9,
10,
11,
12].
This retrospective observational study assesses the diagnostic significance of the RUO parameters of the BC-6800 Plus hematology analyzer (Mindray, China) in differentiating the three major groups of lymphoid neoplasms: chronic lymphocytic leukemia (CLL), acute lymphoblastic leukemia (ALL) and other lymphoproliferative disorders. The aim is to propose new diagnostic algorithms that combine traditional parameters with RUO parameters to improve the sensitivity and specificity of the CBC in the differential diagnosis of lymphoid neoplasms.
2. Materials and Methods
2.1. Study Design
A retrospective observational study was conducted at the University Hospital “Maggiore della Carità”, Novara, Italy. The study protocol was approved by the Ethics Committee of the University Hospital “Maggiore della Carità” (CE162/2024), Novara, Italy, and was conducted according to the current revision of the Helsinki Declaration.
A total of 90 patients presenting at the emergency room with a pathologic blood count were included in the study. The blood counts were collected at the time of admission to the emergency department. This approach was chosen to analyse the traditional and research parameters of blood counts without possible interference from ongoing medications and/or chemotherapy. Inclusion criteria were confirmed follow-up diagnosis of lymphoid neoplasia, COVID negativity and age over 16 years.
The final diagnoses based on immunophenotyping, molecular markers and genotyping were ALL (n=14), CLL (n=47), and other lymphoproliferative disorders (n=29).
2.2. Data Collection
The complete blood count CBC was performed with EDTA-K2 anticoagulated whole blood samples using the BC-6800 Plus hematology analyzer (Mindray, China) at the Clinical Biochemistry Laboratory of the University Hospital “Maggiore della Carità”, Novara. The Mindray BC6800 Plus hematology analyzer features SF Cube technology, which provides quantitative parameters to describe the morphology of blood cells and detects atypical cells with warning signs. All these parameters can reflect the morphological changes in the peripheral blood cells [
13].
The quantitative RUO parameters NeuX, NeuY, NeuZ, MonX, MonY, MonZ, LymX, LymY and LymZ were evaluated. These RUO parameters are sophisticated metrics used to assess the morphology and activation state of different cell types in a blood sample and provide detailed insights into the physical and optical properties of neutrophils, monocytes, and lymphocytes [
13,
14]. NeuX measures the granularity and complexity of neutrophils. Elevated NeuX values indicate increased cytoplasmic granularity or abnormal neutrophil activation, which is often seen in inflammation or infection. MonX reflects the complexity of the cytoplasm and nucleus of monocytes. High MonX values indicate increased cytoplasmic and nuclear complexity, which can occur during monocyte activation or the conversion of monocytes into macrophages. LymX evaluates the internal complexity of lymphocytes. High LymX values may indicate activated lymphocytes, such as during immune reactions.
NeuY evaluates the light scattering properties and fluorescence of neutrophils. Elevated NeuY levels signal increased cellular fluorescence, often due to less condensed chromatin and increased mRNA in the cytoplasm, indicating active protein synthesis and neutrophil activation. MonY measures the fluorescence of monocytes. High MonY values indicate increased light scattering, which may be due to active states or changes in chromatin structure and cytoplasm. LymY evaluates the fluorescence of lymphocytes. An increased LymY value reflects strong cellular fluorescence, typically indicating lymphocyte activation with changes in chromatin and increased mRNA levels.
Finally, NeuZ provides information about the size and volume of neutrophils. Deviations in NeuZ values may indicate changes in neutrophil size that may occur due to activation or pathological conditions. MonZ reflects the size of the monocytes. Variations in MonZ values indicate shifts in cell size that are often associated with monocyte activation or differentiation. LymZ measures the size and volume of lymphocytes. Changes in LymZ values may indicate changes in lymphocyte size that may be due to activation or other morphologic changes.
Neutrophil-to-monocyte ratio (NMR), lymphocyte-to-monocyte ratio (LMR) and neutrophil-to-lymphocyte ratio (NLR), which are suitable for routine use to visualize inflammation, have also been studied [
15,
16].
2.3. Statistical Analysis
Quantitative variables were expressed as median and interquartile range and qualitative variables as absolute frequencies and percentages. Differences between groups for continuous variables were determined using the Mann-Whitney test. The association between predictors and lymphoid neoplasms was analyzed using univariable and multivariable logistic regression. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated. Diagnostic accuracy in predicting CLL, ALL and other lymphoproliferative disorders was assessed by analyzing the receiver operating characteristic (ROC) curve and expressed as area under the curve (AUC) and 95% confidence interval. A p-value<0.05 was considered statistically significant. Statistical analysis was performed using STATA v.18 (StataCorp. 2023. Stata Statistical Software: Release 18. College Station, TX: StataCorp LLC).
3. Results
The complete blood count of 90 patients admitted to the emergency room was examined. After admission, 47 (52.2%) were diagnosed with CLL, 29 (32.2%) with other lymphoproliferative disorders and 14 (15.6%) with ALL. The median age was 68 years (IQR 59-75). Males were 59 (65.5%) and females 32 (35.5%). In the present study, only blood counts were analyzed to assess the ability of blood count parameters alone, both traditional and RUO parameters, for early diagnosis of ALL, CLL and other lymphoproliferative disorders (
Table 1).
3.1. CLL vs ALL
On admission, many hematologic parameters differed significantly between patients with CLL and ALL: hemoglobin (Hb, p < 0.0001), white cell blood count (WBC, p=0.0002), neutrophil count (NE#, p= 0.0001), lymphocyte count (LY#, p=0.0001), NMR (p= 0.03), LMR (p=0.006), platelet count (PLT, p=0.0005), NeuX (p=0.0012), NeuY (p = 0.006), NeuZ (p<0.0001), LymY (p=0.0035), MonY (p=0.0001) (
Table 1).
In univariable logistic regression, Hb (p<0.0001), RDW-SD (p < 0.02), NE# (p=0.03), NLR (p=0.04), PLT (p=0.002), NeuX (p=0.006), NeuY (p=0.012), NeuZ (p =0.001), LymX (p=0.01), MonY (p=0.002), MonZ (p=0.03) were associated with CLL compared to ALL (
Table 2). Finally, in multivariable analysis, only Hb (p=0.02), NeuY (p=0.04), and MonY (p=0.01) were independent predictors of CLL compared to ALL (
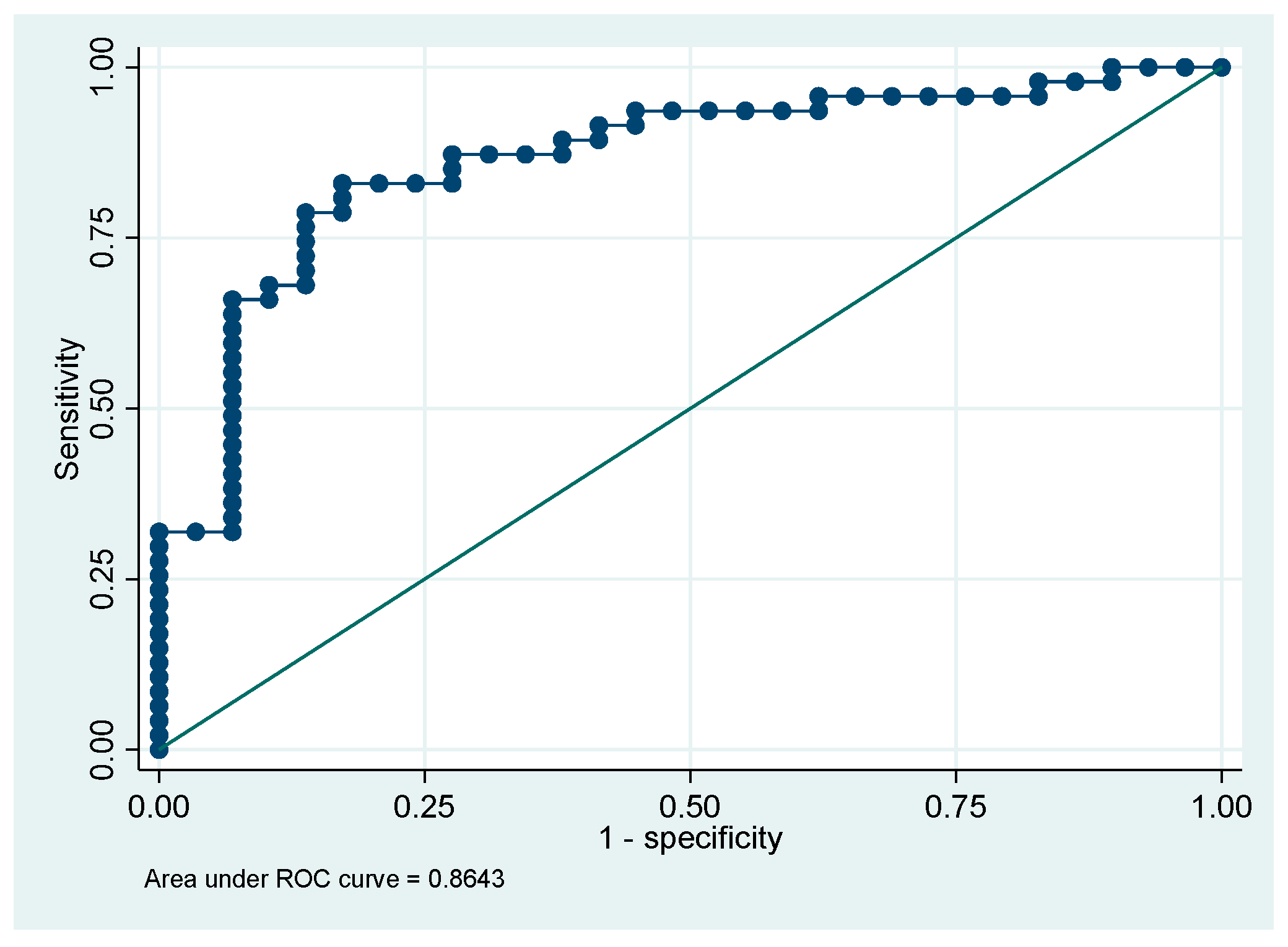
Table 3). The ROC curve for this model showed an area under the curve (AUC) of 0.91 (95%CI 0.81-1.0). The multivariable model correctly classified 93.4% of the cases (
Figure 1).
3.2. CLL and Other Lymphoproliferative Disorders
On admission, many hematologic parameters also differed significantly between patients with CLL and other lymphoproliferative disorders: Hb (p=0.02), NeuX (p=0.01), LymX (p=0.01), LymY (p=0.0001) and LymZ (p=0.01) (
Table 1). In univariable logistic regression, only Hb (p=0.01), NeuX (p=0.03), NeuY (p=0.03), NeuZ (p =0.04), LymX (p=0.01), LymY (p=0.001), LymZ (p= 0.017) were associated with CLL compared to other lymphoproliferative disorders (
Table 4). In multivariable analysis, only MO# (p=0.003), LymY (p<0.0001), and MonY (p=0.004), were independent predictors of CLL compared to other lymphoproliferative disorders (
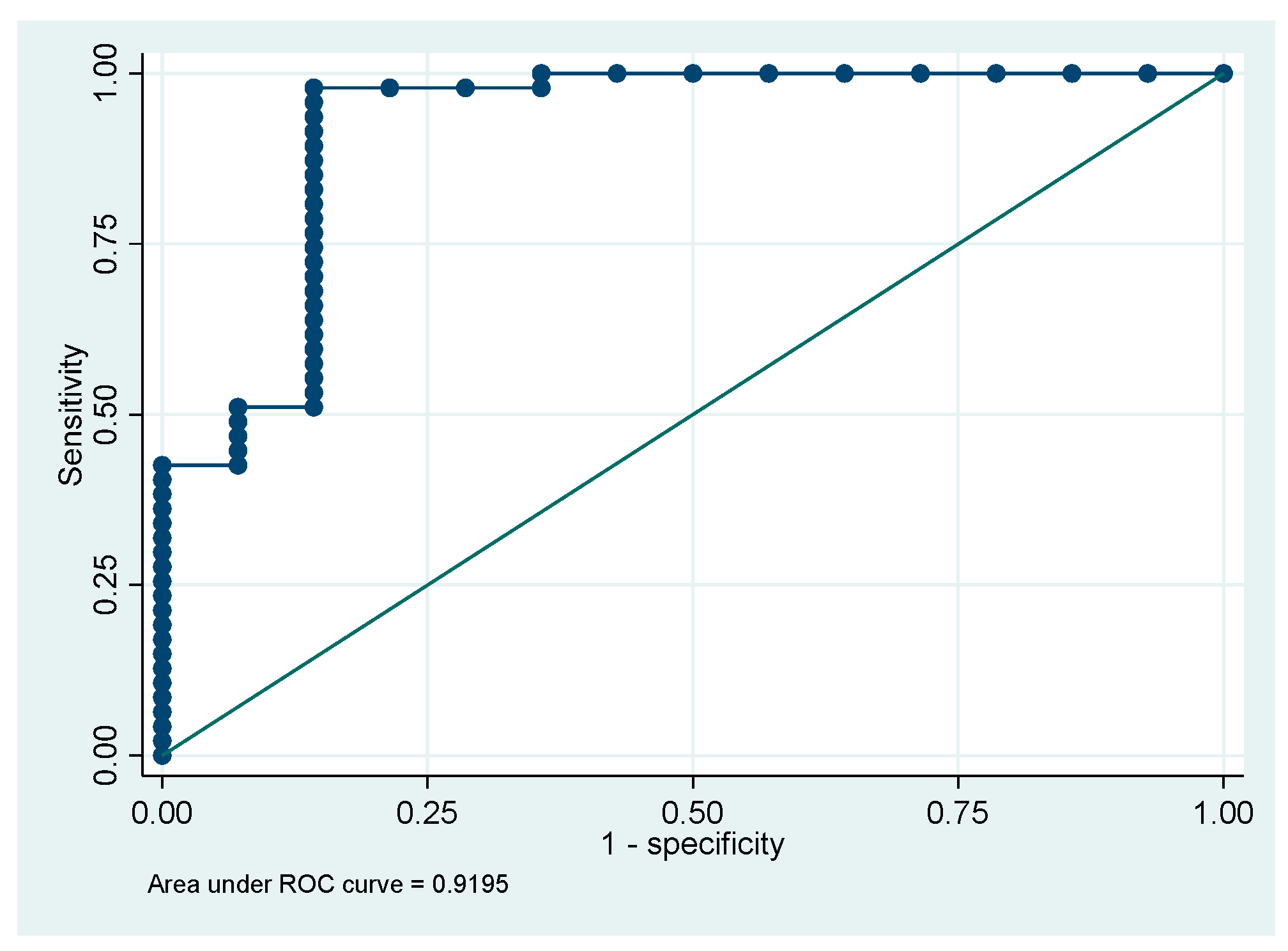
Table 5). The second multivariable model showed that the parameters absolute number of monocytes (MO#), LymY, and MonY were effective in distinguishing CLL patients from other lymphoproliferative disorders patients. This model achieved an AUC of 0.86 (95% CI 0.78-0.95). The multivariable model correctly classified 77.6% of the cases (
Figure 2).
3.3. ALL vs Other Lymphoproliferative Disorders
Finally, when comparing ALL and other lymphoproliferative disorders, the significant hematological parameters were: Hb (p=0.01), WBC (p=0.007), NE# (p=0.003), LY# (p=0.001), NMR (p=0.04), LMR (p=0.008), PLT (p=0.002), NeuY (p=0.03), NeuZ (p<0.0001), MonY (p=0.0002) (
Table 1). In univariable logistic regression, only Hb (p=0.02), PLT (p=0.01), NeuY (p=0.03), NeuZ (p =0.002), MonY (p=0.006), were associated with ALL compared to other lymphoproliferative disorders (
Table 6). In multivariable analysis, only NeuZ (p=0.01) and NeuY (p=0.04), were independent predictors of ALL compared to other lymphoproliferative disorders (
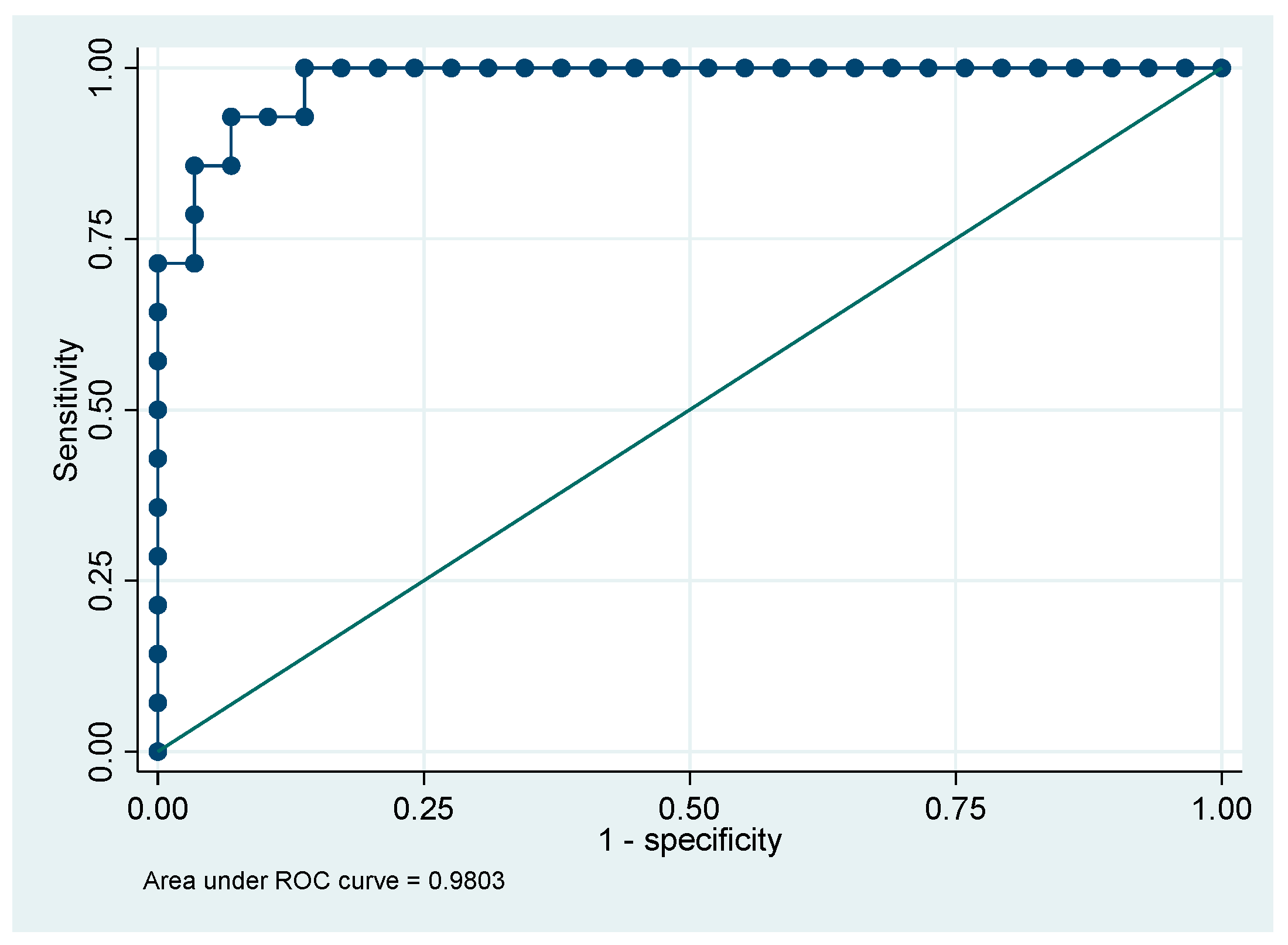
Table 7). The ROC curve for third model, based on the parameters NeuY and NeuZ, showed an excellent ability to discriminate patients with ALL from those with other lymphoproliferative disorders, with an AUC of 0.98 (95% CI 0.95-1.0)
The multivariable model correctly classified 93.0% of the cases (
Figure 3).
4. Conclusions
The new hematology systems offer numerous additional parameters to the conventional ones, which are useful for the differential diagnosis of hematologic neoplasms. The strength of these parameters, most of which have not yet been validated for clinical use and are therefore currently reserved for research purposes, lies in the fact that they are obtained in the DIFF channel without the use of dedicated reagents [
17,
18].
The RUO parameters are available within the time frame of a CBC, so they can be used immediately in the emergency room. This makes them extremely promising for the early and differential diagnosis of numerous pathologies, even in small hospitals where there are no specialized second-level tests and/or in hospitals in developing countries that do not have the sufficient resources to meet all diagnostic needs [
19].
There is currently only a very limited number of studies in the scientific literature investigating the diagnostic potential of Research Use Only (RUO) parameters [
20]. However, the prospects for the diagnostic application of RUOs are extremely promising and deserve further consideration and investigation, as they could open up new avenues for earlier and more accurate diagnosis and thus significantly improve the clinical approach and treatment of patients.
The BC 6800 PLUS analyzer (Mindray) is equipped with innovative analysis parameters categorized as Research Use Only (RUO) that could play an important role in improving the differential diagnosis of hematological neoplasms.
This study aimed to verify whether RUO parameters are useful for the differential diagnosis of three categories of lymphoid neoplasms: ALL, CLL, and other lymphoproliferative disorders at first admission in the emergency department.
As expected, many CBC parameters differed significantly between the three subgroups, but logistic regression analysis showed that only some RUO parameters were independent predictors of CLL, ALL, and other lymphoproliferative disorders. These RUO parameters may help to direct the diagnosis when circulating neoplastic cells are few or display an atypical morphology that can be hardly discriminated by the conventional parameters.
The first multivariable model showed that the parameters Hb, NeuY and MonY have a robust discriminatory ability to distinguish patients with CLL from those with ALL at the time of first admission to the emergency department with an AUC of 0.91. The second multivariable model showed that the absolute monocyte count (MO#), LymY and MonY parameters were effective in distinguishing CLL patients from other lymphoproliferative disorders patients, with an AUC of 0.86. The third model, based on the NeuY and NeuZ parameters, showed excellent ability to distinguish patients with CLL from those with other lymphoproliferative disorders, with an AUC of 0.98. These preliminary data are very promising but require further validation in larger patient cohorts to confirm their reliability and generalizability.
In the multivariable model to distinguish between CLL and ALL, Hb level is expected to be included as a relevant variable. The reason for this is that ALL patients often have anemia, in contrast to CLL patients, where this is usually not the case. In addition, NeuY and MonY are significant predictors that allow effective differentiation between ALL and CLL. The increase in these parameters in patients with ALL might be ascribed to the bone marrow infiltration by leukemic blasts, which impairs the full maturation of neutrophils and monocytes. This leads to the release of immature cells into the bloodstream, a phenomenon that correlates with elevated NeuY and MonY levels [
21]. Furthermore, elevated NeuY and MonY levels may also reflect the morphological and functional changes of leukocytes during the immune response.
In the differential diagnosis between CLL and other lymphoproliferative disorders, multivariable logistic regression analysis underlines the importance of the absolute value of monocytes and the RUO parameters LymY and MonY as predictors, where monocytes were lower and LymY and MonY higher in other lymphoproliferative disorders than in CLL. In CLL, numerous studies have highlighted the role of monocytes and macrophages, in influencing CLL cell dynamics within the tumor microenvironment. These cells, derived from either peripheral blood inflammatory monocytes or tissue-resident macrophages [
23], play a critical role in modulating the viability, survival and resistance of CLL cells to chemotherapy-induced apoptosis [
24,
25,
26,
27]. There is evidence that circulating monocytes can infiltrate and colonize the macrophage compartments in CLL-affected lymph nodes, spleen and bone marrow. An increased presence of these monocytes in the bloodstream can potentially promote the survival and proliferation of CLL cells, ultimately leading to poorer clinical outcomes for patients with CLL [
28].
In addition, the increase of MonY and LymY in other lymphoproliferative disorders might be ascribed to misclassification of circulating other lymphoproliferative disorders cells or to activation of monocytes and lymphocytes [
29].
Finally, the multivariate logistic regression analysis in the context of differential diagnosis between ALL and other lymphoproliferative disorders showed decreased NeuZ and increased NeuY in ALL compared to other lymphoproliferative disorders and an excellent ability of these parameters to distinguish the two entities. In ALL, the accumulation of lymphoblasts in the bone marrow alters the hematopoietic microenvironment and impairs biochemical signals and cellular interactions important for myeloid cell differentiation, preventing normal neutrophil maturation.
In summary, our preliminary data indicate that RUO parameters, which provide additional insight into the cellular population and its properties, may facilitate the identification and characterization of abnormal cell populations associated with lymphoid neoplasms and contribute to their differentiation.
In the future, the introduction of RUO parameters could expand the diagnostic possibilities of blood counts and provide clinicians and haematologists with a more comprehensive tool for assessing hematologic malignancies. These parameters could improve the screening of hematologic malignancies and directly contribute to a more accurate and efficient differential diagnosis.
For diagnosis, RUO must be used in conjunction with established diagnostic methods such as morphology assessment, immunophenotyping and molecular features, but the integration of the new RUO parameters into validated and certified diagnostic algorithms could significantly revolutionise the early stages of haematological diagnostics. This development promises not only faster diagnosis, but also timely initiation of therapy, which could significantly improve patient outcomes. The introduction of innovative parameters could indeed make diagnostic processes faster and more accurate, so that hematologic diseases can be better detected at the first symptoms.
However, the present study is characterised by certain limitations that need to be taken into account. The most important one is the small size of the sample studied. This is due to the methodological decision to study only blood samples taken at the first admission to the emergency room, i.e. at the time of the first diagnosis of hematologic pathology. This approach was chosen to avoid the introduction of confounding factors that could bias the results, but it also limited the number of patients included in the study.
Another limitation concerns the subgroup of ALL that was further reduced by excluding patients younger than 16 years in order to maintain the homogeneity of the age of the three subgroups.
Therefore, larger future studies are needed to confirm these results. In particular, it is important to conduct large-scale retrospective studies and multicenter prospective studies with thousands of patients. Such studies may also provide large data sets that can be analysed using advanced artificial intelligence and machine learning techniques. These technologies could help to further improve the accuracy and speed of hematologic diagnosis, which could lead to more effective treatments and better management of hematologic diseases.
Author Contributions
Conceptualization, S.S., R.R., M.B.; methodology, S.S., V.Z., L.G.; validation, S.S, M.B., R.R. and D.F.; formal analysis, D.F.; investigation, S.S., V.Z and L.G; resources, S.S., R.R, G.G, A.P, V.Z., L.G., M.B., U.D.; data curation, S.S., M.B., V.Z, L.G AND R.R; Project administration, R.R., writing—original draft preparation, R.R., S.S., D.F., U.D., M.B., G.G., A.P.; writing—review and editing, R.R., S.S., U.D., M.B., G.G., A.P.; supervision, R.R., G.G. and U.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the University Hospital “Maggiore della Carità” (CE 162/2024), Novara, Italy.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO Classification of Tumours Online. Available online: https://tumourclassification.iarc.who.int/welcome/ (accessed on 18 July 2024).
- Assessment, U.E.N.C. for E. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Available online: https://hero.epa.gov/hero/index.cfm/reference/details/reference_id/786623 (accessed on 18 July 2024).
- National Academies of Sciences, Engineering, and Medicine; Health and Medicine Division; Board on Health Care Services; Board on Global Health; Committee on Improving the Quality of Health Care Globally. Crossing the Global Quality Chasm: Improving Health Care Worldwide; The National Academies Collection: Reports funded by National Institutes of Health; National Academies Press (US): Washington (DC), 2018; ISBN 978-0-309-47789-5. [Google Scholar]
- Lecompte, T.P.; Bernimoulin, M.P. Novel Parameters in Blood Cell Counters. Clin Lab Med 2015, 35, 209–224. [Google Scholar] [CrossRef] [PubMed]
- Urrechaga, E. Reviewing the Value of Leukocytes Cell Population Data (CPD) in the Management of Sepsis. Ann Transl Med 2020, 8, 953. [Google Scholar] [CrossRef]
- Silva, M.; Fourcade, C.; Fartoukh, C.; Lenormand, B.; Buchonnet, G.; Callat, M.P.; Leclerc, C.; Basuyau, J.P.; Vasse, M. Lymphocyte Volume and Conductivity Indices of the Haematology Analyser Coulter GEN.S in Lymphoproliferative Disorders and Viral Diseases. Clin Lab Haematol 2006, 28, 1–8. [Google Scholar] [CrossRef]
- Haschke-Becher, E.; Vockenhuber, M.; Niedetzky, P.; Totzke, U.; Gabriel, C. A New High-Throughput Screening Method for the Detection of Chronic Lymphatic Leukemia and Myelodysplastic Syndrome. Clin Chem Lab Med 2008, 46, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Jean, A.; Boutet, C.; Lenormand, B.; Callat, M.-P.; Buchonnet, G.; Leclerc, C.; Vasse, M. Combination of Cellular Population Data and CytoDiff Analyses for the Diagnosis of Lymphocytosis. Clin Chem Lab Med 2011, 49, 1861–1868. [Google Scholar] [CrossRef] [PubMed]
- Furundarena, J.R.; Uranga, A.; Sainz, M.R.; González, C.; Uresandi, N.; Argoitia, N.; Araiz, M. Usefulness of the Lymphocyte Positional Parameters in the Sysmex XN Haematology Analyser in Lymphoproliferative Disorders and Mononucleosis Syndrome. Int J Lab Hematol 2018, 40, 41–48. [Google Scholar] [CrossRef]
- Poutakidou, D.; Ruth, I.; Gulbis, B. Differential Diagnosis of Lymphocytosis in Routine Laboratory Practice: Contribution of Lymphocyte Parameters Using the Sysmex-XN9000 Haematology Analyzer. Int J Lab Hematol 2023, 45, 685–690. [Google Scholar] [CrossRef]
- Seghezzi, M.; Buoro, S.; Previtali, G.; Moioli, V.; Manenti, B.; Simon-Lopez, R.; Ottomano, C.; Lippi, G. A Preliminary Proposal for Quality Control Assessment and Harmonization of Leukocytes Morphology-Structural Parameters (Cell Population Data Parameters). J Med Biochem 2018, 37, 486–498. [Google Scholar] [CrossRef]
- Pérez, I.; Redín, M.E. Stability of Leukocyte Research Parameters over Time on the Sysmex XN: How to Quantify the Changes in Cell Morphology. Int J Lab Hematol 2018, 40, 569–576. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, Z.; Pan, S.; Li, J.; Yang, Y.; Qi, H.; Xie, J.; Qu, J. The Clinical Value of Hematological Neutrophil and Monocyte Parameters in the Diagnosis and Identification of Sepsis. Ann Transl Med 2021, 9, 1680. [Google Scholar] [CrossRef]
- Sun, P.; Li, N.; Zhang, S.; Liu, S.; Zhang, H.; Yue, B. Combination of NeuX and NeuZ Can Predict Neutrophil Dysplasia Features of Myelodysplastic Neoplasms in Peripheral Blood. Int J Lab Hematol 2023, 45, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Buonacera, A.; Stancanelli, B.; Colaci, M.; Malatino, L. Neutrophil to Lymphocyte Ratio: An Emerging Marker of the Relationships between the Immune System and Diseases. Int J Mol Sci 2022, 23, 3636. [Google Scholar] [CrossRef]
- Mandaliya, H.; Jones, M.; Oldmeadow, C.; Nordman, I.I. Prognostic Biomarkers in Stage IV Non-Small Cell Lung Cancer (NSCLC): Neutrophil to Lymphocyte Ratio (NLR), Lymphocyte to Monocyte Ratio (LMR), Platelet to Lymphocyte Ratio (PLR) and Advanced Lung Cancer Inflammation Index (ALI). Transl Lung Cancer Res 2019, 8, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Crouser, E.D.; Parrillo, J.E.; Seymour, C.; Angus, D.C.; Bicking, K.; Tejidor, L.; Magari, R.; Careaga, D.; Williams, J.; Closser, D.R.; et al. Improved Early Detection of Sepsis in the ED With a Novel Monocyte Distribution Width Biomarker. Chest 2017, 152, 518–526. [Google Scholar] [CrossRef]
- Agnello, L.; Ciaccio, A.M.; Vidali, M.; Cortegiani, A.; Biundo, G.; Gambino, C.M.; Scazzone, C.; Lo Sasso, B.; Ciaccio, M. Monocyte Distribution Width (MDW) in Sepsis. Clin Chim Acta 2023, 548, 117511. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, E.M.A.; Formenti, P.; Pastori, S.; Roccaforte, V.; Gotti, M.; Panella, R.; Galimberti, A.; Costagliola, R.; Vetrone, F.; Umbrello, M.; et al. The Potential Role of Neutrophil-Reactive Intensity (NEUT-RI) in the Diagnosis of Sepsis in Critically Ill Patients: A Retrospective Cohort Study. Diagnostics (Basel) 2023, 13, 1781. [Google Scholar] [CrossRef]
- Sacchetti, S.; Vidali, M.; Esposito, T.; Zorzi, S.; Burgener, A.; Ciccarello, L.; Cammarota, G.; Zanotti, V.; Giacomini, L.; Bellan, M.; et al. The Role of New Morphological Parameters Provided by the BC 6800 Plus Analyzer in the Early Diagnosis of Sepsis. Diagnostics (Basel) 2024, 14, 340. [Google Scholar] [CrossRef]
- Silva, R.A.M.; de Mendonça, R.M.H.; Dos Santos Aguiar, S.; Yajima, J.C.; Marson, F.A.L.; Brandalise, S.R.; Levy, C.E. Induction Therapy for Acute Lymphoblastic Leukemia: Incidence and Risk Factors for Bloodstream Infections. Support Care Cancer 2022, 30, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Stewart, B.J.; Fergie, M.; Young, M.D.; Jones, C.; Sachdeva, A.; Blain, A.; Bacon, C.M.; Rand, V.; Ferdinand, J.R.; James, K.R.; et al. Spatial and Molecular Profiling of the Mononuclear Phagocyte Network in Classic Hodgkin Other lymphoproliferative disorders. Blood 2023, 141, 2343–2358. [Google Scholar] [CrossRef]
- Perdiguero, E.G.; Geissmann, F. The Development and Maintenance of Resident Macrophages. Nat Immunol 2016, 17, 2–8. [Google Scholar] [CrossRef]
- Burger, J.A.; Tsukada, N.; Burger, M.; Zvaifler, N.J.; Dell’Aquila, M.; Kipps, T.J. Blood-Derived Nurse-like Cells Protect Chronic Lymphocytic Leukemia B Cells from Spontaneous Apoptosis through Stromal Cell-Derived Factor-1. Blood 2000, 96, 2655–2663. [Google Scholar]
- Kurtova, A.V.; Balakrishnan, K.; Chen, R.; Ding, W.; Schnabl, S.; Quiroga, M.P.; Sivina, M.; Wierda, W.G.; Estrov, Z.; Keating, M.J.; et al. Diverse Marrow Stromal Cells Protect CLL Cells from Spontaneous and Drug-Induced Apoptosis: Development of a Reliable and Reproducible System to Assess Stromal Cell Adhesion-Mediated Drug Resistance. Blood 2009, 114, 4441–4450. [Google Scholar] [CrossRef]
- Nishio, M.; Endo, T.; Tsukada, N.; Ohata, J.; Kitada, S.; Reed, J.C.; Zvaifler, N.J.; Kipps, T.J. Nurselike Cells Express BAFF and APRIL, Which Can Promote Survival of Chronic Lymphocytic Leukemia Cells via a Paracrine Pathway Distinct from That of SDF-1alpha. Blood 2005, 106, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Seiffert, M.; Schulz, A.; Ohl, S.; Döhner, H.; Stilgenbauer, S.; Lichter, P. Soluble CD14 Is a Novel Monocyte-Derived Survival Factor for Chronic Lymphocytic Leukemia Cells, Which Is Induced by CLL Cells in Vitro and Present at Abnormally High Levels in Vivo. Blood 2010, 116, 4223–4230. [Google Scholar] [CrossRef]
- Friedman, D.R.; Sibley, A.B.; Owzar, K.; Chaffee, K.G.; Slager, S.; Kay, N.E.; Hanson, C.A.; Ding, W.; Shanafelt, T.D.; Weinberg, J.B.; et al. Relationship of Blood Monocytes with Chronic Lymphocytic Leukemia Aggressiveness and Outcomes: A Multi-Institutional Study. Am J Hematol 2016, 91, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Aladily, T.N.; Alhelou, Z.; Hamdan, N. Circulating Hodgkin Cells: Extending the Spectrum of Atypical Lymphocytes. Int J Lab Hematol 2022, 44, e253–e254. [Google Scholar] [CrossRef]
- Robak, T.; Krawczyńska, A.; Cebula-Obrzut, B.; Urbaniak, M.; Iskierka-Jażdżewska, E.; Robak, P. Atypical Chronic Lymphocytic Leukemia-The Current Status. Cancers (Basel) 2023, 15, 4427. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).