Submitted:
17 August 2024
Posted:
19 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
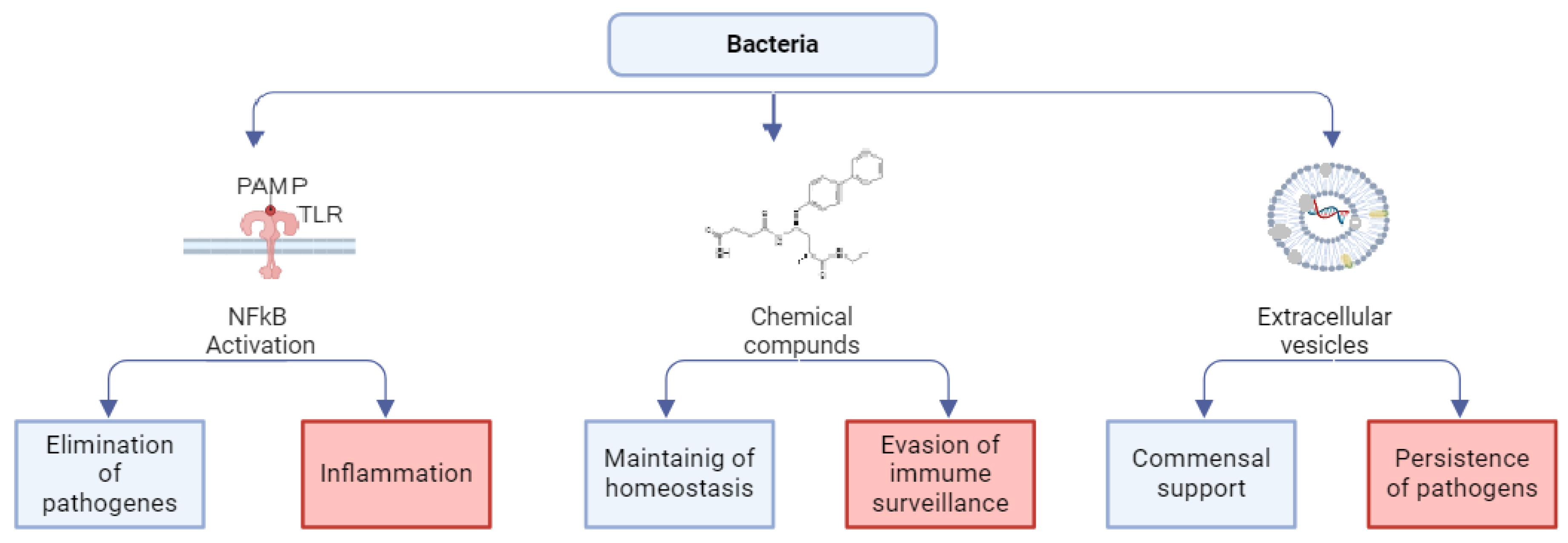
2. Bacteria Modes of Action on Host Cells
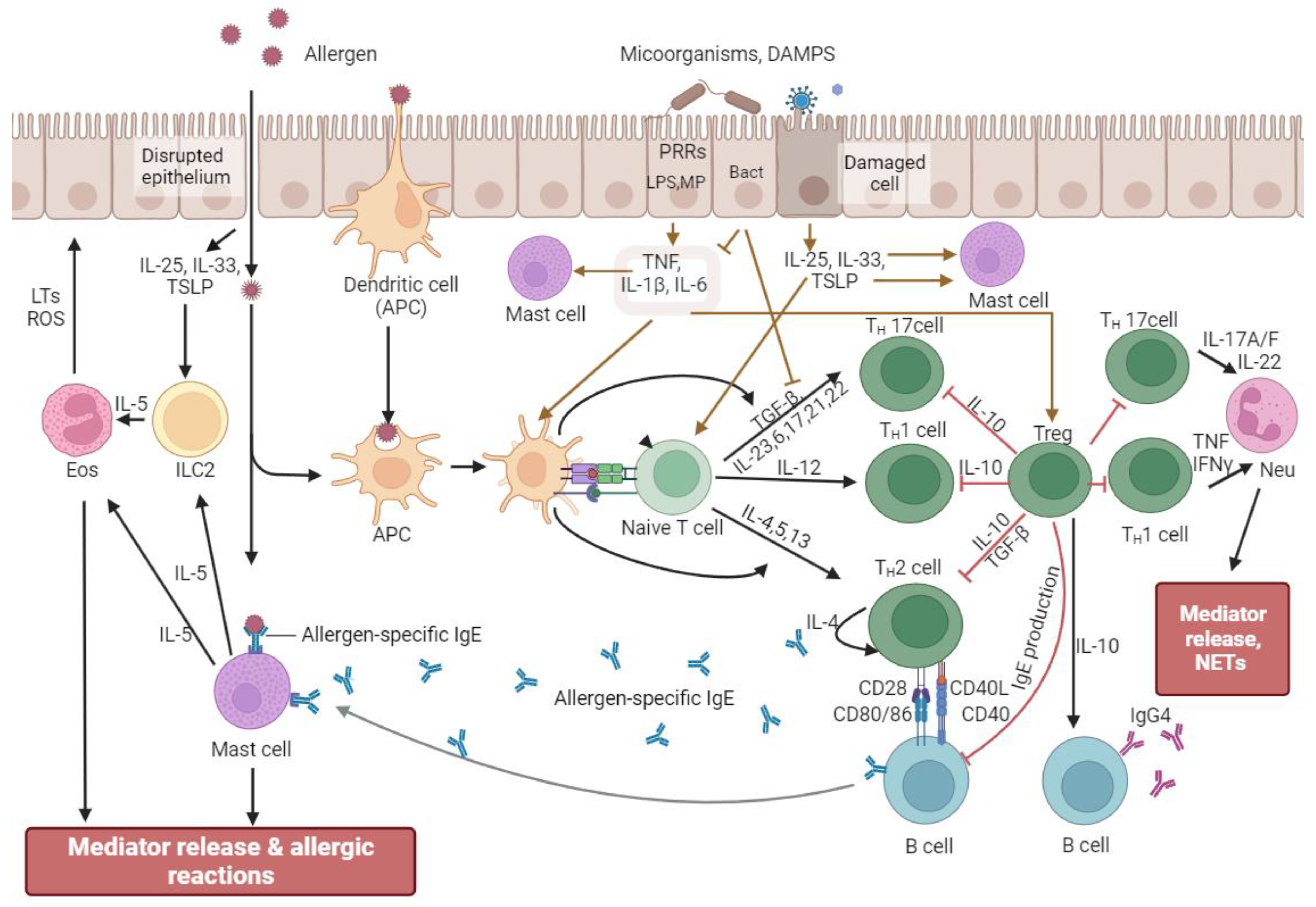
3. Modulation of Allergic Reactions by Bacterial Regulators
4. The Impact of Bacteria on Allergic Diseases
4.1. Gastrointestinal Tract
4.2. Upper and lower respiratory tract
4.3. Skin
5. Concluding Remarks
Funding
Data Availability Statement
Conflicts of Interest
References
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Zubeldia-Varela, E.; Barker-Tejeda, T.C.; Obeso, D.; Villaseñor, A.; Barber, D.; Pérez-Gordo, M. Microbiome and Allergy: New Insights and Perspectives. J. Investig. Allergol. Clin. Immunol. 2022, 32, 327–344. [Google Scholar] [CrossRef] [PubMed]
- Droste, J.H.; Wieringa, M.H.; Weyler, J.J.; Nelen, V.J.; Vermeire, P.A.; Van Bever, H.P. Does the use of antibiotics in early childhood increase the risk of asthma and allergic disease? Clin. Exp. Allergy. 2000, 30, 1547–53. [Google Scholar] [CrossRef]
- Yamamoto-Hanada, K.; Yang, L.; Narita, M.; Saito, H.; Ohya, Y. Influence of antibiotic use in early childhood on asthma and allergic diseases at age 5. Ann. Allergy Asthma Immunol. 2017, 119, 54–58. [Google Scholar] [CrossRef]
- Metzler, S.; Frei, R.; Schmaußer-Hechfellner, E.; von Mutius, E.; Pekkanen, J.; Karvonen, A.M.; Kirjavainen, P.V.; Dalphin, J.C.; Divaret-Chauveau, A.; Riedler, J.; et al. Association between antibiotic treatment during pregnancy and infancy and the development of allergic diseases. Pediatr. Allergy Immunol. 2019, 30, 423–433. [Google Scholar] [CrossRef]
- Zou, Z.; Liu, W.; Huang, C.; Sun, C.; Zhang, J. First-Year Antibiotics Exposure in Relation to Childhood Asthma, Allergies, and Airway Illnesses. Int. J. Environ. Res. Public Health 2020, 17, 5700. [Google Scholar] [CrossRef]
- Matricardi, P.M.; Ronchetti, R. Are infections protecting from atopy? Curr Opin Allergy Clin Immunol. 2001, 1, 413–9. [Google Scholar] [CrossRef] [PubMed]
- Holgate, S.T. The airway epithelium is central to the pathogenesis of asthma. Allergol. Int. 2008, 57, 1–10. [Google Scholar] [CrossRef]
- Kilpeläinen, M.; Terho, E.O.; Helenius, H.; Koskenvuo, M. Farm environment in childhood prevents the development of allergies. Clin. Exp. Allergy 2000, 30, 201–8. [Google Scholar] [CrossRef]
- Wills-Karp, M.; Santeliz, J.; Karp, C.L. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat. Rev. Immunol. 2001, 1, 69–75. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Hoskinson, C.; Dai, D.L.Y.; Del Bel, K.L.; Becker, A.B.; Moraes, T.J.; Mandhane, P.J.; Finlay, B.B.; Simons, E.; Kozyrskyj, A.L.; Azad, M.B.; et al. Delayed gut microbiota maturation in the first year of life is a hallmark of pediatric allergic disease. Nat. Commun. 2023, 14, 4785. [Google Scholar] [CrossRef] [PubMed]
- Hanski, I.; von Hertzen, L.; Fyhrquist, N.; Koskinen, K.; Torppa, K.; Laatikainen, T.; Karisola, P.; Auvinen, P.; Paulin, L.; Mäkelä, M.J.; et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc. Natl. Acad. Sci. USA. 2012, 109, 8334–9. [Google Scholar] [CrossRef]
- Lisik, D.; Ermis, S.S.Ö.; Ioannidou, A.; Milani, G.P.; Nyassi, S.; Spolidoro, G.C.I.; Kankaanranta, H.; Goksör, E.; Wennergren, G.; Nwaru, B.I. Siblings and risk of allergic rhinitis: A systematic review and meta-analysis. Pediatr. Allergy Immunol. 2023, 34, e13991. [Google Scholar] [CrossRef] [PubMed]
- Strachan, D.P.; Aït-Khaled, N.; Foliaki, S.; Mallol, J.; Odhiambo, J.; Pearce, N.; Williams, H.C.; ISAAC Phase Three Study Group. Siblings, asthma, rhinoconjunctivitis and eczema: a worldwide perspective from the International Study of Asthma and Allergies in Childhood. Clin. Exp. Allergy 2015, 45, 126–36. [Google Scholar] [CrossRef]
- Perez-Munoz, M.E.; Arrieta, M.C.; Ramer-Tait, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome 2017, 5, 48. [Google Scholar] [CrossRef]
- Jimenez, E.; Fernandez, L.; Marin, M.L.; Martin, R.; Odriozola, J.M.; Nueno-Palop, C.; et al. (). Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 2005, 51, 270–274. [Google Scholar] [CrossRef]
- Jimenez, E.; Marin, M.L.; Martin, R.; Odriozola, J.M.; Olivares, M.; Xaus, J. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, 187–193. [Google Scholar] [CrossRef]
- Moles, L.; Gómez, M.; Heilig, H.; Bustos, G.; Fuentes, S.; de Vos, W.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One 2013, 8, e66986. [Google Scholar] [CrossRef]
- Senn, V.; Bassler, D.; Choudhury, R.; Scholkmann, F.; Righini-Grunder, F.; Vuille-Dit-Bile, R.N.; Restin, T. Microbial Colonization From the Fetus to Early Childhood-A Comprehensive Review. Front. Cell. Infect. Microbiol. 2020, 10, 573735. [Google Scholar] [CrossRef]
- Machado, M.E.; Porto, L.C.; Alves Galvão, M.G.; Sant’Anna, C.C.; Lapa E Silva, J.R. SNPs, adipokynes and adiposity in children with asthma. J. Asthma 2023, 60, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Ishida-Yamamoto, A.; Kishibe, M.; Honma, M. Desmosomes and corneodesmosomes and their relevance to genetic skin diseases. G. Ital. Dermato.l Venereol. 2017, 152, 148–157. [Google Scholar] [CrossRef]
- Falcon, R.M.G.; Caoili, S.E.C. Immunologic, genetic, and ecological interplay of factors involved in allergic diseases. Front Allergy 2023, 4, 1215616. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.U.; Kim, J.D.; Park, C.S. Gene-Environment Interactions in Asthma: Genetic and Epigenetic Effects. Yonsei Med. J. 2015 56, 877–86. [CrossRef]
- Losol, P.; Sokolowska, M.; Hwang, Y.K.; Ogulur, I.; Mitamura, Y.; Yazici, D.; Pat, Y.; Radzikowska, U.; Ardicli, S.; Yoon, J.E.; et al. Epithelial Barrier Theory: The Role of Exposome, Microbiome, and Barrier Function in Allergic Diseases. Allergy Asthma Immunol. Res. 2023, 15, 705–724. [Google Scholar] [CrossRef]
- Ozdemir, C.; Kucuksezer, U.C.; Ogulur, I.; Pat, Y.; Yazici, D.; Ardicli, S.; Akdis, M.; Nadeau, K.; Akdis, C.A. Lifestyle Changes and Industrialization in the Development of Allergic Diseases. Curr. Allergy Asthma Rep. 2024, 24, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Diebold, Y.; Sahu, S.K.; Leonardi, A. Epithelial barrier dysfunction in ocular allergy. Allergy 2022, 77, 1360–1372. [Google Scholar] [CrossRef]
- Lu, H.F.; Zhou, Y.C.; Yang, L.T.; Zhou, Q.; Wang, X.J.; Qiu, S.Q.; Cheng, B.H.; Zeng, X.H. Involvement and repair of epithelial barrier dysfunction in allergic diseases. Front. Immunol. 2024, 15, 1348272. [Google Scholar] [CrossRef] [PubMed]
- Loxham, M.; Davies, D.E. Phenotypic and genetic aspects of epithelial barrier function in asthmatic patients. J. Allergy Clin. Immunol. 2017, 139, 1736–1751. [Google Scholar] [CrossRef]
- Guryanova, S.V.; Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Bohle, B.; Ovchinnikova, T.V. How Do Pollen Allergens Sensitize? Front. Mol. Biosci. 2022, 9, 900533. [Google Scholar] [CrossRef]
- Yazici, D.; Ogulur, I.; Pat, Y.; Babayev, H.; Barletta, E.; Ardicli, S.; Bel Imam, M.; Huang, M.; Koch, J.; Li, M.; et al. The epithelial barrier: The gateway to allergic, autoimmune, and metabolic diseases and chronic neuropsychiatric conditions. Semin. Immunol. 2023, 70, 101846. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, R.; de Matos, M.R.; Cortes, L.; Nunes-Correia, I.; Todo-Bom, A.; Pires, E.; Veríssimo, P. Pollen Proteases Play Multiple Roles in Allergic Disorders. Int. J. Mol. Sci. 2020, 21, 3578. [Google Scholar] [CrossRef]
- Goleva, E.; Berdyshev, E.; Leung, D.Y. Epithelial barrier repair and prevention of allergy. J. Clin. Invest. 2019, 129, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Virgin, H.W. The virome in mammalian physiology and disease. Cell 2014, 157, 142–50. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–40. [Google Scholar] [CrossRef]
- Abrams, G.D.; Bishop, J.E. Effect of the normal microbial flora on gastrointestinal motility. Proc. Soc. Exp. Biol. Med. 1967, 126, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Dimidi, E.; Christodoulides, S.; Scott, S.M.; Whelan, K. Mechanisms of Action of Probiotics and the Gastrointestinal Microbiota on Gut Motility and Constipation. Adv. Nutr. 2017, 15, 484–494. [Google Scholar] [CrossRef]
- Høverstad, T.; Midtvedt, T. Short-chain fatty acids in germfree mice and rats. J. Nutr. 1986, 116, 1772–1776. [Google Scholar] [CrossRef]
- Jiménez, E.; Fernández, L.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Nueno-Palop, C.; Narbad, A.; Olivares, M.; Xaus, J.; Rodríguez, J.M. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 2005, 51, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, B.E.; Daft, F.S.; Mcdaniel, E.G.; Smith, J.C.; Fitzgerald, R.J. Effects of vitamin K-active compounds and intestinal microorganisms in vitamin K-deficient germfree rats. J. Nutr. 1962, 78, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Sun, E.W.; Rogers, G.B.; Keating, D.J. The Influence of the Gut Microbiome on Host Metabolism Through the Regulation of Gut Hormone Release. Front. Physiol. 2019, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Freter, R. The fatal enteric cholera infection in the guinea pig, achieved by inhibition of normal enteric flora. J. Infect. Dis. 1955, 97, 57–65. [Google Scholar] [CrossRef]
- Abt, M.C.; Pamer, E.G. Commensal bacteria mediated defenses against pathogens. Curr. Opin Immunol. 2014, 29, 16–22. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef]
- Hayes, C.L.; Dong, J.; Galipeau, H.J.; Jury, J.; McCarville, J.; Huang, X.; Wang, X.Y.; Naidoo, A.; Anbazhagan, A.N.; Libertucci, J.; et al. Commensal microbiota induces colonic barrier structure and functions that contribute to homeostasis. Sci. Rep. 2018, 8, 14184. [Google Scholar] [CrossRef]
- Sorbara, M.T.; Philpott, D.J. Peptidoglycan: a critical activator of the mammalian immune system during infection and homeostasis. Immunol. Rev. 2011, 243, 40–60. [Google Scholar] [CrossRef]
- Takeda, K.; Akira, S. Roles of Toll-like receptors in innate immune responses. Genes Cells 2001, 6, 733–42. [Google Scholar] [CrossRef]
- Krieg, A.M. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002, 20, 709–60. [Google Scholar] [CrossRef]
- Inohara, N.; Nuñez, G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 2003, 3, 371–82. [Google Scholar] [CrossRef] [PubMed]
- Girardin, S.E.; Boneca, I.G.; Viala, J.; Chamaillard, M.; Labigne, A.; Thomas, G.; Philpott, D.J.; Sansonetti, P.J. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 2003, 278, 8869–8872. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A. When signaling pathways collide: positive and negative regulation of toll-like receptor signal transduction. Immunity 2008, 29, 12–20. [Google Scholar] [CrossRef]
- Gorshkova, R.P.; Isakov, V.V.; Nazarenko, E.L.; Ovodov, Y.S.; Guryanova, S.V.; Dmitriev, B.A. Structure of the O-specific polysaccharide of the lipopolysaccharide from Yersinia kristensenii O:25.35. Carbohydr. Res. 1993, 241, 201–208. [Google Scholar] [CrossRef]
- L’vov, V.L.; Gur’ianova, S.V.; Rodionov, A.V.; Dmitriev, B.A.; Shashkov, A.S.; Ignatenko, A.V.; Gorshkova, R.P.; Ovodov, I.S. The structure of a repetitive unit of the glycerolphosphate- containing O-specific polysaccharide chain from Yersinia kristensenii strain 103 (0:12,26) lipopolysaccharide. Bioorg. Khim. 1990, 16, 379–389. [Google Scholar]
- L’vov, V.L.; Gur’yanova, S.V.; Rodionov, A.V.; Gorshkova, R.P. Structure of the repeating unit of the O-specific polysaccharide of the lipopolysaccharide of Yersinia kristensenii strain 490 (O:12,25). Carbohydr. Res. 1992, 228, 415–422. [Google Scholar] [CrossRef]
- Rohde, M. The Gram-Positive Bacterial Cell Wall. Microbiol Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Meshcheriakova, E.A.; Gur’ianova, S.V.; Makarov, E.A.; Andronova, T.M.; Ivanov, V.T. Structure-functional study of glycosaminylmuramoyl peptides. The effect of chemical modification of N-acetylglucosaminyl-N-acetylmuramoyldipeptide on its immunomodulating properties in vivo and in vitro. Bioorg. Chemistry 1991, 17, 1157–1165. [Google Scholar]
- Strober, W.; Murray, P.J.; Kitani, A.; Watanabe, T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat. Rev. Immunol. 2006, 6, 9–20. [Google Scholar] [CrossRef]
- Kufer, T.A.; Banks, D.J.; Philpott, D.J. Innate immune sensing of microbes by Nod proteins. Ann. N. Y. Acad. Sci. 2006, 1072, 19–27. [Google Scholar] [CrossRef]
- Guryanova, S.V. Regulation of Immune Homeostasis via Muramyl Peptides-Low Molecular Weight Bioregulators of Bacterial Origin. Microorganisms 2022, 10, 1526. [Google Scholar] [CrossRef]
- Tan, R.S.; Ho, B.; Leung, B.P.; Ding, J.L. TLR cross-talk confers specificity to innate immunity. Int Rev Immunol. 2014, 33, 443–53. [Google Scholar] [CrossRef]
- Becker, C.E.; O’Neill, L.A. Inflammasomes in inflammatory disorders: the role of TLRs and their interactions with NLRs. Semin. Immunopathol. 2007, 29, 239–48. [Google Scholar] [CrossRef]
- Merlo, A.; Calcaterra, C.; Mènard, S.; Balsari, A. Cross-talk between toll-like receptors 5 and 9 on activation of human immune responses. J. Leukoc. Biol. 2007, 82, 509–18. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef]
- Garg, A.; Zhao, A.; Erickson, S.L.; Mukherjee, S.; Lau, A.J.; Alston, L.; Chang, T.K.; Mani, S.; Hirota, S.A. Pregnane X Receptor Activation Attenuates Inflammation-Associated Intestinal Epithelial Barrier Dysfunction by Inhibiting Cytokine-Induced Myosin Light-Chain Kinase Expression and c-Jun N-Terminal Kinase 1/2 Activation. J. Pharmacol. Exp. Ther. 2016, 359, 91–101. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Zhu, W. Crosstalk Between The Immune Receptors and Gut Microbiota. Curr. Protein. Pept. Sci. 2015, 16, 622–31. [Google Scholar] [CrossRef]
- Guryanova, S.V. Immunomodulation, Bioavailability and Safety of Bacteriocins. Life 2023, 13, 1521. [Google Scholar] [CrossRef] [PubMed]
- Gillor, O.; Ghazaryan, L. Recent advances in bacteriocin application as antimicrobials. Recent Pat. Antiinfect. Drug Discov. 2007, 2, 115–22. [Google Scholar] [CrossRef]
- Husted, A.S.; Trauelsen, M.; Rudenko, O.; Hjorth, S.A.; Schwartz, T.W. GPCR-Mediated Signaling of Metabolites. Cell. Metab. 2017, 25, 777–796. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Zhang, Y.; Song, M. Lactobacillus alleviates intestinal epithelial barrier function through GPR43-mediated M2 macrophage polarization. Animal Diseases 2024, 4, 20. [Google Scholar] [CrossRef]
- Kim, C.H. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell. Mol. Immunol. 2021, 18, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Seo, S.U.; Kweon, M.N. Gut microbiota-derived metabolites tune host homeostasis fate. Semin. Immunopathol. 2024, 11, 2. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. Microbiol Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Byndloss, M.X.; Rivera-Chávez, F.; Tsolis, R.M.; Bäumler, A.J. How bacterial pathogens use type III and type IV secretion systems to facilitate their transmission. Curr. Opin. Microbiol. 2017, 35, 1–7. [Google Scholar] [CrossRef]
- Galán, J.E.; Waksman, G. Protein-Injection Machines in Bacteria. Cell 2018, 172, 1306–1318. [Google Scholar] [CrossRef]
- Zhou, X.; Xie, F.; Wang, L.; Zhang, L.; Zhang, S.; Fang, M.; Zhou, F. The function and clinical application of extracellular vesicles in innate immune regulation. Cell. Mol. Immunol. 2020, 17, 323–334. [Google Scholar] [CrossRef]
- Charpentier, L.A.; Dolben, E.F.; Hendricks, M.R.; Hogan, D.A.; Bomberger, J.M.; Stanton, B.A. Bacterial Outer Membrane Vesicles and Immune Modulation of the Host. Membranes (Basel) 2023, 13, 752. [Google Scholar] [CrossRef]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–30. [Google Scholar] [CrossRef] [PubMed]
- Coelho, C.; Casadevall, A. Answers to naysayers regarding microbial extracellular vesicles. Biochem. Soc. Trans. 2019, 47, 1005–1012. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Jones, M.K. Role of Bacterial Extracellular Vesicles in Manipulating Infection. Infect Immun. 2023, 91, e0043922. [Google Scholar] [CrossRef] [PubMed]
- Peregrino, E.S.; Castañeda-Casimiro, J.; Vázquez-Flores, L.; Estrada-Parra, S.; Wong-Baeza, C.; Serafín-López, J.; Wong-Baeza, I. The Role of Bacterial Extracellular Vesicles in the Immune Response to Pathogens, and Therapeutic Opportunities. Int. J. Mol. Sci. 2024, 25, 6210. [Google Scholar] [CrossRef]
- Verbunt, J.; Jocken, J.; Blaak, E.; Savelkoul, P.; Stassen, F. Gut-bacteria derived membrane vesicles and host metabolic health: a narrative review. Gut Microbes 2024, 16, 2359515. [Google Scholar] [CrossRef]
- Díaz-Garrido, N.; Badia, J.; Baldomà, L. Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J. Extracell. Vesicles. 2021, 10, e12161. [Google Scholar] [CrossRef]
- Schorey, J.S.; Cheng, Y.; McManus, W.R. Bacteria- and host-derived extracellular vesicles - two sides of the same coin? J. Cell. Sci. 2021, 134, jcs256628. [Google Scholar] [CrossRef]
- Verbunt, J.; Jocken, J.; Blaak, E.; Savelkoul, P.; Stassen, F. Gut-bacteria derived membrane vesicles and host metabolic health: a narrative review. Gut Microbes 2024, 16, 2359515. [Google Scholar] [CrossRef]
- Hao, H.; Zhang, X.; Tong, L.; Liu, Q.; Liang, X.; Bu, Y.; Gong, P.; Liu, T.; Zhang, L.; Xia, Y.; et al. Effect of Extracellular Vesicles Derived From Lactobacillus plantarum Q7 on Gut Microbiota and Ulcerative Colitis in Mice. Front. Immunol. 2021, 12, 777147. [Google Scholar] [CrossRef] [PubMed]
- Gasaly, N.; de Vos, P.; Hermoso, M.A. Impact of Bacterial Metabolites on Gut Barrier Function and Host Immunity: A Focus on Bacterial Metabolism and Its Relevance for Intestinal Inflammation. Front. Immunol. 2021, 12, 658354. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.A.; Fu, J.; Chan, P.V. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U S A 2020, 117, 19376–19387. [Google Scholar] [CrossRef]
- Sun, M.; Ma, N.; He, T.; Johnston, L.J.; Ma, X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit. Rev. Food Sci. Nutr. 2020, 60, 1760–1768. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell. Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Kolesnikova, N.V.; Kozlov, I.G.; Guryanova, S.V.; Kokov, E.A.; Andronova, T.M. Clinical and immunological efficiency of muramyl dipeptide in the treatment of atopic diseases. Med. Immunol. 2016, 1, 15–20. [Google Scholar] [CrossRef]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Guryanova, S.; Khaitov, R. ; Glucosaminylmuramyldipeptide—GMDP: Effect on mucosal immunity (on the issue of immunotherapy and immunoprophylaxis). Immunologiya 2020, 41, 174–183. (In Russian) [Google Scholar] [CrossRef]
- Gholami, H.; Chmiel, J.A.; Burton, J.P.; Maleki Vareki, S. The Role of Microbiota-Derived Vitamins in Immune Homeostasis and Enhancing Cancer Immunotherapy. Cancers (Basel) 2023, 15, 1300. [Google Scholar] [CrossRef]
- Breiteneder, H.; Diamant, Z.; Eiwegger, T.; Fokkens, W.J.; Traidl-Hoffmann, C.; Nadeau, K.; O’Hehir, R.E.; O’Mahony, L.; Pfaar, O.; et al. Future research trends in understanding the mechanisms underlying allergic diseases for improved patient care. Allergy 2019, 74, 2293–2311. [Google Scholar] [CrossRef]
- Capucilli, P.; Hill, D.A. Allergic Comorbidity in Eosinophilic Esophagitis: Mechanistic Relevance and Clinical Implications. Clin. Rev. Allergy Immunol. 2019, 57, 111–127. [Google Scholar] [CrossRef]
- Letner, D.; Farris, A.; Khalili, H.; Garber, J. Pollen-food allergy syndrome is a common allergic comorbidity in adults with eosinophilic esophagitis. Dis. Esophagus. 2018, 31, 2. [Google Scholar] [CrossRef]
- Zysk, W.; Mesjasz, A.; Trzeciak, M.; Horvath, A.; Plata-Nazar, K. Gastrointestinal Comorbidities Associated with Atopic Dermatitis-A Narrative Review. Int. J. Mol. Sci. 2024, 25, 1194. [Google Scholar] [CrossRef]
- Vasquez Ayala, A.; Hsu, C.Y.; Oles, R.E.; Matsuo, K.; Loomis, L.R.; Buzun, E.; Carrillo Terrazas, M.; Gerner, R.R.; Lu, H.H.; et al. Commensal bacteria promote type I interferon signaling to maintain immune tolerance in mice. J. Exp. Med. 2024, 221, e20230063. [Google Scholar] [CrossRef]
- Molloy, J.; Allen, K.; Collier, F.; Tang, M.L.; Ward, A.C.; Vuillermin, P. The potential link between gut microbiota and IgE-mediated food allergy in early life. Int. J. Environ. Res. Public Health 2013, 10, 7235–56. [Google Scholar] [CrossRef] [PubMed]
- Akdis, C.A. Does the epithelial barrier hypothesis explain the increase in allergy, autoimmunity and other chronic conditions? Nat. Rev. Immunol. 2021, 21, 739–751. [Google Scholar] [CrossRef]
- Kim, B.E.; Leung, D.Y.M. Significance of Skin Barrier Dysfunction in Atopic Dermatitis. Allergy Asthma Immunol. Res. 2018, 10, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Komlósi, Z.I.; van de Veen, W.; Kovács, N.; Szűcs, G.; Sokolowska, M.; O’Mahony, L.; Akdis, M.; Akdis, C.A. Cellular and molecular mechanisms of allergic asthma. Mol. Asp. Med. 2022, 85, 100995. [Google Scholar] [CrossRef]
- Turvey, S.E.; Broide, D.H. Innate immunity. J. Allergy Clin. Immunol. 2010, 125, S24–32. [Google Scholar] [CrossRef] [PubMed]
- Feili-Hariri, M.; Falkner, D.H.; Morel, P.A. Polarization of naive T cells into Th1 or Th2 by distinct cytokine-driven murine dendritic cell populations: implications for immunotherapy. J. Leukoc Biol. 2005, 78, 656–64. [Google Scholar] [CrossRef]
- Arango Duque, G.; Descoteaux, A. Macrophage cytokines: involvement in immunity and infectious diseases. Front. Immunol. 2014, 7, 491. [Google Scholar] [CrossRef]
- Creagh, E.M.; O’Neill, L.A. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006, 27, 352–7. [Google Scholar]
- Guryanova, S.V.; Kozlov, I.G.; Meshcheryakova, E.A.; Alekseeva, L.G.; Andronova, T.M. Glucosaminylmuramyl Dipeptide Normalizes Th1 / Th2 Balance in Atopic Bronchial Asthma. Immunology 2009, 5, 305–308. (In Russian) [Google Scholar]
- Trinchieri, G.; Pflanz, S.; Kastelein, R.A. The IL-12 family of heterodimeric cytokines: new players in the regulation of T cell responses, Immunity 2003, 19, 641–644, 2-s2.0-0345358584. [CrossRef]
- Delespesse, G.; Ohshima, Y.; Shu, U.; Yang, L.; Demeure, C.; Wu, C.; Byun, D.; Sarfati, M. Differentiation of naive human CD4 T cells into TH2/TH1 effectors. Allergology International 1997, 46, 63–72. [Google Scholar] [CrossRef]
- Luckheeram, R.V.; Zhou, R.; Verma, A.D.; Xia, B. CD4⁺T cells: differentiation and functions. Clin. Dev. Immunol. 2012, 925135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.M.; An, J. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef]
- Hong, H.; Liao, S.; Chen, F.; Yang, Q.; Wang, D.Y. Role of IL-25, IL-33, and TSLP in triggering united airway diseases toward type 2 inflammation. Allergy 2020, 75, 2794–2804. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.N.; Kohanski, M.A.; Maina, I.W.; Workman, A.D.; Herbert, D.R.; Cohen, N.A. Sentinels at the wall: epithelial-derived cytokines serve as triggers of upper airway type 2 inflammation. Int. Forum. Allergy Rhinol. 2019, 9, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Maggi, L.; Montaini, G.; Mazzoni, A.; Rossettini, B.; Capone, M.; Rossi, M.C.; Santarlasci, V.; Liotta, F.; Rossi, O.; et al. Human circulating group 2 innate lymphoid cells can express CD154 and promote IgE production. J. Allergy Clin. Immunol. 2017, 139, 964–976.e4. [Google Scholar] [CrossRef]
- Emami Fard, N.; Xiao, M.; Sehmi, R. Regulatory ILC2—Role of IL-10 Producing ILC2 in Asthma. Cells 2023, 12, 2556. [Google Scholar] [CrossRef]
- Gaudino, S.J.; Kumar, P. Cross-Talk Between Antigen Presenting Cells and T Cells Impacts Intestinal Homeostasis, Bacterial Infections, and Tumorigenesis. Front. Immunol. 2019, 10, 360. [Google Scholar] [CrossRef]
- Izquierdo, E.; Rodriguez-Coira, J.; Delgado-Dolset, M.I.; Gomez-Casado, C.; Barber, D.; Escribese, M.M. Epithelial Barrier: Protector and Trigger of Allergic Disorders. J. Investig. Allergol. Clin. Immunol. 2022, 32, 81–96. [Google Scholar] [CrossRef]
- Stadnyk, A.W. Cytokine production by epithelial cells. FASEB J. 1994, 8, 1041–7. [Google Scholar] [CrossRef]
- Koch, S.; Nusrat, A. The life and death of epithelia during inflammation: lessons learned from the gut. Annu. Rev. Pathol. 2012, 7, 35–60. [Google Scholar] [CrossRef]
- Onyiah, J.C.; Colgan, S.P. Cytokine responses and epithelial function in the intestinal mucosa. Cell. Mol. Life Sci. 2016, 73, 4203–4212. [Google Scholar] [CrossRef]
- Svanborg, C.; Agace, W.; Hedges, S.; Linder, H.; Svensson, M. Bacterial adherence and epithelial cell cytokine production. Zentralbl. Bakteriol. 1993, 278, 359–64. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Tsang, J.S.; Park, K. Systems immunology of regulatory T cells: Can one circuit explain it all? Trends Immunol. 2023, 44, 766–781. [Google Scholar] [CrossRef]
- Shao, Q.; Gu, J.; Zhou, J.; Wang, Q.; Li, X.; Deng, Z.; Lu, L. Tissue Tregs and Maintenance of Tissue Homeostasis. Front. Cell Dev. Biol. 2021, 9, 717903. [Google Scholar] [CrossRef]
- Sakaguchi, S.; Yamaguchi, T.; Nomura, T.; Ono, M. Regulatory T Cells and Immune Tolerance. Cell 2008, 133, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Ohue, Y.; Nishikawa, H. Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target? Cancer Sci. 2019, 110, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, A.; Carrier, Y.; Peron, J.P.; Bettelli, E.; Kamanaka, M.; Flavell, R.A.; Kuchroo, V.K.; Oukka, M.; Weiner, H.L. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat. Immunol. 2007, 8, 1380–9. [Google Scholar] [CrossRef]
- Pot, C.; Jin, H.; Awasthi, A.; Liu, S.M.; Lai, C.Y.; Madan, R.; Sharpe, A.H.; Karp, C.L.; Miaw, S.C.; Ho, I.C.; Kuchroo, V.K. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J. Immunol. 2009, 183, 797–801. [Google Scholar] [CrossRef]
- Kim, D.; Le, H.T.; Nguyen, Q.T.; Kim, S.; Lee, J.; Min, B. Cutting Edge: IL-27 Attenuates Autoimmune Neuroinflammation via Regulatory T Cell/Lag3-Dependent but IL-10-Independent Mechanisms In Vivo. J. Immunol. 2019, 202, 1680–1685. [Google Scholar] [CrossRef]
- Nguyen, Q.T.; Jang, E.; Le, H.T.; Kim, S.; Kim, D.; Dvorina, N.; Aronica, M.A.; Baldwin, W.M., 3rd; Asosingh, K.; Comhair, S.; et al. IL-27 targets Foxp3+ Tregs to mediate antiinflammatory functions during experimental allergic airway inflammation. JCI Insight 2019, 4, e123216. [Google Scholar] [CrossRef]
- Emami Fard, N.; Xiao, M.; Sehmi, R. Regulatory ILC2-Role of IL-10 Producing ILC2 in Asthma. Cells 2023, 12, 2556. [Google Scholar] [CrossRef]
- Iyer, S.S.; Cheng, G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 2012, 32, 23–63. [Google Scholar] [CrossRef]
- Kühn, R.; Löhler, J.; Rennick, D.; Rajewsky, K.; Müller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 1993, 75, 263–74. [Google Scholar] [CrossRef] [PubMed]
- Maynard, C.L. Contrasting roles for all-trans retinoic acid in TGF-[beta]-mediated induction of Foxp3 and Il10 genes in developing regulatory T cells. J. Exp. Med. 2009, 206, 343–57. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; Rutz, S.; Crellin, N.K.; Valdez, P.A.; Hymowitz, S.G. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 2011, 29, 71–109. [Google Scholar] [CrossRef] [PubMed]
- Maloy, K.J.; Powrie, F. Regulatory T cells in the control of immune pathology. Nature Immunol. 2001, 2, 816–22. [Google Scholar] [CrossRef]
- Jung, M.; Sabat, R.; Krätzschmar, J.; Seidel, H.; Wolk, K.; Schönbein, C.; Schutt, S.; Friedrich, M.; Docke, W.D.; Asadullah, K.; et al. Expression profiling of IL-10-regulated genes in human monocytes and peripheral blood mononuclear cells from psoriatic patients during IL-10 therapy. Eur. J. Immunol. 2004, 34, 481–93. [Google Scholar] [CrossRef]
- Kliem, C.V.; Schaub, B. The role of regulatory B cells in immune regulation and childhood allergic asthma. Mol. Cell. Pediatr. 2024, 11, 1. [Google Scholar] [CrossRef]
- Heinl, P.V.; Graulich, E.; Weigmann, B.; Wangorsch, A.; Ose, R.; Bellinghausen, I.; Khatri, R.; Raker, V.K.; Scheurer, S.; Vieths, S.; et al. IL-10-modulated dendritic cells from birch pollen- and hazelnut-allergic patients facilitate Treg-mediated allergen-specific and cross-reactive tolerance. Allergy 2024, 28. [Google Scholar] [CrossRef]
- Akdis, C.A.; Akdis, M. Mechanisms of immune tolerance to allergens: Role of IL-10 and Tregs. J. Clin. Investig. 2014, 124, 4678–4680. [Google Scholar] [CrossRef]
- Al-Qahtani, A.A.; Alhamlan, F.S.; Al-Qahtani, A.A. Pro-Inflammatory and Anti-Inflammatory Interleukins in Infectious Diseases: A Comprehensive Review. Trop. Med. Infect. Dis. 2024, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Wilson, E.B.; Brooks, D.G. The role of IL-10 in regulating immunity to persistent viral infections. Curr. Top. Microbiol. Immunol. 2011, 350, 39–65. [Google Scholar]
- Müller, A.; Oertli, M.; Arnold, I.C.H. pylori exploits and manipulates innate and adaptive immune cell signaling pathways to establish persistent infection. Cell Commun. Signal. 2011, 9, 25. [Google Scholar] [CrossRef]
- Yuan, C.; Qu, Z.-L.; Tang, X.-L.; Liu, Q.; Luo, W.; Huang, C.; Pan, Q.; Zhang, X.-L. Mycobacterium tuberculosis Mannose-Capped Lipoarabinomannan Induces IL-10-Producing B Cells and Hinders CD4+Th1 Immunity. iScience 2019, 11, 13–30. [Google Scholar] [CrossRef] [PubMed]
- González, L.A.; Melo-González, F.; Sebastián, V.P.; Vallejos, O.P.; Noguera, L.P.; Suazo, I.D.; Schultz, B.M.; Manosalva, A.H.; Peñaloza, H.F.; Soto, J.A.; et al. Characterization of the Anti-Inflammatory Capacity of IL-10-Producing Neutrophils in Response to Streptococcus pneumoniae Infection. Front. Immunol. 2021, 12, 638917. [Google Scholar] [CrossRef] [PubMed]
- Rechkina, E.A.; Denisova, G.F.; Masalova, O.V.; Lideman, L.F.; Denisov, D.A.; Lesnova, E.I.; Ataullakhanov, R.I.; Gur’ianova, S.V.; Kushch, A. Epitope mapping of antigenic determinants of hepatitis C virus proteins by phage display. Mol. Biologiia 2006, 40, 357–368. (In Russian) [Google Scholar]
- Guryanova, S.V.; Sigmatulin, I.; Gigani, O.O.; Lipkina, S.A. Mechanisms of regulation allergic and autoimmune reactions by bacterial origin bioregulators. RUDN Journal of Medicine. 2023, 27, 470–482. [Google Scholar] [CrossRef]
- Lathrop, S.K.; Bloom, S.M.; Rao, S.M.; Nutsch, K.; Lio, C.W.; Santacruz, N.; Peterson, D.A.; Stappenbeck, T.S.; Hsieh, C.S. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011, 478, 250–4. [Google Scholar] [CrossRef]
- Cebula, A.; Seweryn, M.; Rempala, G.A.; Pabla, S.S.; McIndoe, R.A.; Denning, T.L.; Bry, L.; Kraj, P.; Kisielow, P.; Ignatowicz, L. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 2013, 497, 258–62. [Google Scholar] [CrossRef]
- Krieg, A.M. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002, 20, 709–60. [Google Scholar] [CrossRef]
- Guryanova, S.V.; Gigani, O.B.; Gudima, G.O.; Kataeva, A.M.; Kolesnikova, N.V. Dual Effect of Low-Molecular-Weight Bioregulators of Bacterial Origin in Experimental Model of Asthma. Life 2022, 12, 192. [Google Scholar] [CrossRef] [PubMed]
- Prescott, D.; Maisonneuve, C.; Yadav, J.; Rubino, S.J.; Girardin, S.E.; Philpott, D.J. NOD2 modulates immune tolerance via the GM-CSF–dependent generation of CD103+ dendritic cells. Proc. Natl. Acad. Sci. USA 2020, 117, 10946–10957. [Google Scholar] [CrossRef] [PubMed]
- Pablo-Torres, C.; Garcia-Escribano, C.; Romeo, M.; Gomez-Casado, C.; Arroyo Solera, R.; Bueno-Cabrera, J.L.; Del Mar Reaño Martos, M.; Iglesias-Cadarso, A.; Tarín, C.; Agache, I.; et al. Transcriptomics reveals a distinct metabolic profile in T cells from severe allergic asthmatic patients. Front. Allergy 2023, 4, 1129248. [Google Scholar] [CrossRef]
- Zhang, Y.; Higashide, W.M.; McCormick, B.A.; Chen, J.; Zhou, D. The nflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol. Microbiol. 2006, 62, 786–793. [Google Scholar] [CrossRef]
- Piscatelli, H.; Kotkar, S.A.; McBee, M.E.; Muthupalani, S.; Schauer, D.B.; Mandrell, R.E.; Leong, J.M.; Zhou, D. The EHEC type III effector NleL is an E3 ubiquitin ligase that modulates pedestal formation. PLoS One 2011, 6, e19331. [Google Scholar] [CrossRef]
- Okuda, J.; Toyotome, T.; Kataoka, N.; Ohno, M.; Abe, H.; Shimura, Y.; Seyedarabi, A.; Pickersgill, R.; Sasakawa, C. Shigella effector IpaH9.8 binds to a splicing factor U2AF(35) to modulate host immune responses. Biochem. Biophys. Res. Commun. 2005, 333, 531–9. [Google Scholar] [CrossRef] [PubMed]
- Ashida, H.; Kim, M.; Schmidt-Supprian, M.; Ma, A.; Ogawa, M.; Sasakawa, C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat. Cel.l Biol. 2010, 12, 66–73, sup pp 1–9. [Google Scholar] [CrossRef]
- Ashida, H.; Nakano, H.; Sasakawa, C. Shigella IpaH0722 E3 ubiquitin ligase effector targets TRAF2 to inhibit PKC-NF-κB activity in invaded epithelial cells. PLoS Pathog. 2013, 9, e1003409. [Google Scholar] [CrossRef]
- Pawankar, R.; Canonica, G.W.; Holgate, S.T.; Lockey, RF, editors. WAO White Book on Allergy. Milwaukee, WI: World Allergy Organization; 2011. 20 p.
- GBD 2019 Diseases and Injuries Collaborators. Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Mattiuzzi, C.; Lippi, G. Worldwide asthma epidemiology: insights from the Global Health Data Exchange database. Int. Forum. Allergy Rhinol. 2020, 10, 75–80. [Google Scholar] [CrossRef]
- Pawankar, R. Allergic diseases and asthma: a global public health concern and a call to action. World Allergy Organ. J. 2014, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, N.G.; Agache, I.; Bavbek, S.; Bilo, B.M.; Braido, F.; Cardona, V.; Custovic, A.; Demonchy, J.; Demoly, P.; Eigenmann, P.; et al. Research needs in allergy: an EAACI position paper, in collaboration with EFA. Clin. Transl. Allergy 2012, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Lai, G.C.; Yao, L.J.; Aung, T.T.; Shental, N.; Rotter-Maskowitz, A.; Shepherdson, E.; Singh, G.S.N.; Pai, R.; Shanti, A. Microbial exposure during early human development primes fetal immune cells. Cell 2021, 184, 3394–3409.e20. [Google Scholar] [CrossRef]
- Kloepfer, K.M.; McCauley, K.E.; Kirjavainen, P.V. The Microbiome as a Gateway to Prevention of Allergic Disease Development. J. Allergy Clin. Immunol. Pract. 2022, 10, 2195–2204. [Google Scholar] [CrossRef]
- Rackaityte, E.; Halkias, J.; Fukui, E.M.; Mendoza, V.F; Hayzelden, C.; Crawford, E.D.; Fujimura, K.E.; Burt, T.D.; Lynch, S.V. Viable bacterial colonization is highly limited in the human intestine in utero. Nat. Med. 2020, 26, 599–607. [Google Scholar] [CrossRef]
- Durack, J.; Kimes, N.E.; Lin, D.L.; Rauch, M.; McKean, M.; McCauley, K.; Panzer, A.R.; Mar, J.S.; Cabana, M.D.; Lynch, S.V. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat. Commun. 2018, 16, 707. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 30, 307ra152. [Google Scholar] [CrossRef]
- Ismail, I.H.; Boyle, R.J.; Licciardi, P.V.; Oppedisano, F.; Lahtinen, S.; Robins-Browne, R.M.; Tang, M.L. Early gut colonization by Bifidobacterium breve and B. catenulatum differentially modulates eczema risk in children at high risk of developing allergic disease. Pediatr. Allergy Immunol. 2016, 27, 838–846. [Google Scholar] [CrossRef]
- Cukrowska, B.; Bierła, J.B.; Zakrzewska, M.; Klukowski, M.; Maciorkowska, E. The Relationship between the Infant Gut Microbiota and Allergy. The Role of Bifidobacterium breve and Prebiotic Oligosaccharides in the Activation of Anti-Allergic Mechanisms in Early Life. Nutrients 2020, 12, 946. [Google Scholar] [CrossRef]
- Fieten, K.B.; Totté, J.E.E.; Levin, E.; Reyman, M.; Meijer, Y.; Knulst, A.; Schuren, F.; Pasmans, S.G.M.A. Fecal Microbiome and Food Allergy in Pediatric Atopic Dermatitis: A Cross-Sectional Pilot Study. Int. Arch. Allergy Immunol. 2018, 175, 77–84. [Google Scholar] [CrossRef]
- Hattori, K.; Yamamoto, A.; Sasai, M.; Taniuchi, S.; Kojima, T.; Kobayashi, Y.; Iwamoto, H.; Namba, K.; Yaeshima, T. Effects of administration of bifidobacteria on fecal microflora and clinical symptoms in infants with atopic dermatitis. Arerugi 2003, 52, 20–30. [Google Scholar]
- Nylund, L.; Satokari, R.; Nikkila, J.; Rajilic-Stojanovic, M.; Kalliomaki, M.; Isolauri, E.; Salminen, S.; de Vos, W.M. Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic disease. BMC Microbiol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Hacini-Rachinel, F.; Gosoniu, M.L.; Bourdeau, T.; Holvoet, S.; Doucet-Ladeveze, R.; Beaumont, M.; Mercenier, A,.; Nutten, S. Immune-modulatory effect of probiotic Bifidobacterium lactis NCC2818 in individuals suffering from seasonal allergic rhinitis to grass pollen: an exploratory, randomized, placebo-controlled clinical trial. Eur. J. Clin. Nutr. 2013, 67. 161-7.
- Pessôa, R.; Clissa, P.B.; Sanabani, S.S. The Interaction between the Host Genome, Epigenome, and the Gut–Skin Axis Microbiome in Atopic Dermatitis. Int. J. Mol. Sci. 2023, 24, 14322. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Du, M.; Yang, H.; Shi, R.; Shi, Y.; Zhang, S.; Zhao, Y.; Lan, J. Short-chain fatty acid - A critical interfering factor for allergic diseases. Chem. Biol. Interact. 2023, 385, 110739. [Google Scholar] [CrossRef] [PubMed]
- Abbas, E.E.; Li, C.; Xie, A.; Lu, S.; Tang, L.; Liu, Y.H.; Elfadil, A.; Wen, S. Distinct Clinical Pathology and Microbiota in Chronic Rhinosinusitis With Nasal Polyps Endotypes. Laryngoscope 2021, 131, E34–E44. [Google Scholar] [CrossRef]
- Del Giudice, M.M.; Indolfi, C.; Capasso, M.; Maiello, N.; Decimo, F.; Ciprandi, G. Bifidobacterium mixture (B longum BB536, B infantis M-63, B breve M-16V) treatment in children with seasonal allergic rhinitis and intermittent asthma. Ital. J. Pediatr. 2017, 43, 25. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 29, 453–620. [Google Scholar] [CrossRef]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011, 332, 974–7. [Google Scholar] [CrossRef]
- Wang, R.X.; Lee, J.S.; Campbell, E.L.; Colgan, S.P. Microbiota-derived butyrate dynamically regulates intestinal homeostasis through regulation of actin-associated protein synaptopodin. Proc. Natl. Acad. Sci. USA 2020, 117, 11648–11657. [Google Scholar] [CrossRef]
- Grosheva, I.; Zheng, D.; Levy, M.; Polansky, O.; Lichtenstein, A. High-throughput screen identifies host and microbiota regulators of intestinal barrier function. Gastroenterology 2020, 159, 1807–1823. [Google Scholar] [CrossRef]
- Wong, C.B.; Iwabuchi, N.; Xiao, J.Z. Exploring the Science behind Bifidobacterium breve M-16V in infant health. Nutrients 2019, 11, 1724. [Google Scholar] [CrossRef]
- Inoue, Y.; Iwabuchi, N.; Xiao, J.Z.; Yaeshima, T.; Iwatsuki, K. Suppressive effects of Bifidobacterium breve strain M-16V on T-helper type 2 immune responses in a murine model. Biol. Pharm. Bull. 2009, 32, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; Marsland, B.J. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–66. [Google Scholar] [CrossRef]
- Stiemsma, L.T.; Arrieta, M.C.; Dimitriu, P.A.; Cheng, J.; Thorson, L.; Lefebvre, D.L.; Azad, M.B.; Subbarao, P.; Mandhane, P.; Becker, A.; et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin. Sci. (Lond). 2016, 130, 2199–2207. [Google Scholar] [CrossRef]
- Kaczynska, A.; Klosinska, M.; Chmiel, P.; Janeczek, K.; Emeryk, A. The Crosstalk between the Gut Microbiota Composition and the Clinical Course of Allergic Rhinitis: The Use of Probiotics, Prebiotics and Bacterial Lysates in the Treatment of Allergic Rhinitis. Nutrients 2022, 14, 4328. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Huang, J.L.; Chen, K.J.; Kong, M.S.; Hua, M.C.; Yeh, Y.M.; Chang, H.J. Comparison of 16S rRNA gene sequencing microbiota among children with serological IgE-mediated food hypersensitivity. Pediatr. Res. 2024, 95, 241–50. [Google Scholar] [CrossRef]
- Joseph, C.L.; Sitarik, A.R.; Kim, H.; Huffnagle, G.; Fujimura, K.; Yong, G.J.M.; Levin, A.M.; Zoratti, E.; Lynch, S.; Ownby, D.R.; et al. Infant gut bacterial community composition and food-related manifestation of atopy in early childhood. Pediatr. Allergy Immunol. 2022, 33, e13704. [Google Scholar] [CrossRef] [PubMed]
- Agache, I.; Laculiceanu, A.; Spanu, D.; Grigorescu, D. The Concept of One Health for Allergic Diseases and Asthma. Allergy Asthma Immunol. Res. 2023, 15, 290–302. [Google Scholar] [CrossRef]
- Brick, T.; Hettinga, K.; Kirchner, B.; Pfaffl, M.W.; Ege, M.J. The Beneficial Effect of Farm Milk Consumption on Asthma, Allergies, and Infections: From Meta-Analysis of Evidence to Clinical Trial. J. Allergy Clin. Immunol. Pract. 2020, 8, 878–889.e3. [Google Scholar] [CrossRef]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy. N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell. Host Microbe 2015, 17, 704–715. [CrossRef]
- Zeng, Y.; Liang, J.Q. Nasal Microbiome and Its Interaction with the Host in Childhood Asthma. Cells 2022, 11, 3155. [Google Scholar] [CrossRef]
- Rosas-Salazar, C.; Shilts, M.H.; Tovchigrechko, A.; Schobel, S.; Chappell, J.D.; Larkin, E.K.; Gebretsadik, T.; Halpin, R.A.; Nelson, K.E.; Moore, M.L.; et al. Nasopharyngeal Lactobacillus is associated with a reduced risk of childhood wheezing illnesses following acute respiratory syncytial virus infection in infancy. J. Allergy Clin. Immunol. 2018, 142, 1447–1456.e9. [Google Scholar] [CrossRef] [PubMed]
- Fazlollahi, M.; Lee, T.D.; Andrade, J.; Oguntuyo, K.; Chun, Y.; Grishina, G.; Grishin, A.; Bunyavanich, S. The nasal microbiome in asthma. J. Allergy Clin. Immunol. 2018, 142, 834–843.e2. [Google Scholar] [CrossRef]
- Tang, H.H.F.; Lang, A.; Teo, S.M.; Judd, L.M.; Gangnon, R.; Evans, M.D.; Lee, K.E.; Vrtis, R.; Holt, P.G.; Lemanske, R.F., Jr.; et al. Developmental patterns in the nasopharyngeal microbiome during infancy are associated with asthma risk. J. Allergy Clin. Immunol. 2021, 147, 1683–1691. [Google Scholar] [CrossRef]
- Toivonen, L.; Karppinen, S.; Schuez-Havupalo, L.; Waris, M.; He, Q.; Hoffman, K.L.; Petrosino, J.F.; Dumas, O.; Camargo, C.A., Jr.; Hasegawa, K.; et al. Longitudinal Changes in Early Nasal Microbiota and the Risk of Childhood Asthma. Pediatrics 2020, 146, e20200421. [Google Scholar] [CrossRef] [PubMed]
- McCauley, K.; Durack, J.; Valladares, R.; Fadrosh, D.W.; Lin, D.L.; Calatroni, A. Distinct Nasal Airway Bacterial Microbiota Differentially Relate to Exacerbation in Pediatric Asthma. Journal of Allergy and Clinical Immunology 2019, 144, 1187–97. [Google Scholar] [CrossRef]
- Depner, M.; Ege, M.J.; Cox, M.J.; Dwyer, S.; Walker, A.W.; Birzele, L.T. Bacterial microbiota of the upper respiratory tract and childhood asthma. J. Allergy Clin. Immunol. 2017, 139, 826–834.e13. [Google Scholar] [CrossRef] [PubMed]
- Bisgaard, H.; Hermansen, M.N.; Buchvald, F.; Loland, L.; Halkjaer, L.B.; Bønnelykke, K.; Brasholt, M.; Heltberg, A.; Vissing, N.H.; Thorsen, S.V.; et al. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med. 2007, 357, 1487–95. [Google Scholar] [CrossRef]
- Green, B.J.; Wiriyachaiporn, S.; Grainge, C.; Rogers, G.B.; Kehagia, V.; Lau, L.; Carroll, M.P.; Bruce, K.D.; Howarth, P.H. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One 2014, 9, e100645. [Google Scholar] [CrossRef]
- Yang, X.; Li, H.; Ma, Q.; Zhang, Q.; Wang, C. Neutrophilic Asthma Is Associated with Increased Airway Bacterial Burden and Disordered Community Composition. Biomed. Res. Int. 2018, 2018, 9230234. [Google Scholar] [CrossRef]
- Versi, A.; Ivan, F.X.; Abdel-Aziz, M.I.; Bates, S.; Riley, J.; Baribaud, F.; Kermani, N.Z.; Montuschi, P.; Dahlen, S.E.; Djukanovic, R.; et al. . Haemophilus influenzae and Moraxella catarrhalis in sputum of severe asthma with inflammasome and neutrophil activation. Allergy 2023, 78, 2906–2920. [Google Scholar] [CrossRef]
- Fraga-Silva, T.F.C.; Boko, M.M.M.; Martins, N.S.; Cetlin, A.A.; Russo, M.; Vianna, E.O.; Bonato, V.L.D. Asthma-associated bacterial infections: Are they protective or deleterious? J. Allergy Clin. Immunol. Glob. 2022, 2, 14–22. [Google Scholar] [CrossRef]
- Simpson, J.L.; Daly, J.; Baines, K.J.; Yang, I.A.; Upham, J.W.; Reynolds, P.N.; Hodge, S.; James, A.L.; Hugenholtz, P.; Willner, D.; et al. Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur. Respir. J. 2016, 47, 792–800. [Google Scholar] [CrossRef]
- Huang, Y.J.; Nariya, S.; Harris, J.M.; Lynch, S.V.; Choy, D.F.; Arron, J.R.; Boushey, H. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J. Allergy Clin. Immunol. 2015, 136, 874–884. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.B.; Hong, S.W.; Kim, D.K.; Jeon, S.G.; Kim, K.R.; Cho, S.H.; Gho, Y.S.; Jee, Y.K.; Kim, Y.K. Decreased diversity of nasal microbiota and their secreted extracellular vesicles in patients with chronic rhinosinusitis based on a metagenomic analysis. Allergy 2014, 69, 517–526. [Google Scholar] [CrossRef]
- Ruokolainen, L.; Fyhrquist, N.; Laatikainen, T.; Auvinen, P.; Fortino, V.; Scala, G.; Jousilahti, P.; Karisola, P.; Vendelin, J.; Karkman, A.; et al. Immune-microbiota interaction in Finnish and Russian Karelia young people with high and low allergy prevalence. Clin. Exp. Allergy 2020, 50, 1148–1158. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, J.H.; Lee, S.W. Application of Microbiome-Based Therapies in Chronic Respiratory Diseases. J. Microbiol. 2024, 62, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, J.; Zhou, X. Lung microbiome: new insights into the pathogenesis of respiratory diseases. Sig. Transduct. Target Ther. 2024, 9, 19. [Google Scholar] [CrossRef]
- Yagi, K.; Huffnagle, G.B.; Lukacs, N.W.; Asai, N. The Lung Microbiome during Health and Disease. Int. J. Mol. Sci. 2021, 22, 10872. [Google Scholar] [CrossRef]
- Taglialegna, A. Fibre-deprived Akkermansia worsens food allergy. Nat. Rev. Microbiol. 2023, 21, 769. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Liu, H.; Qiu, W.; Li, Z.; Ge, S.; Luo, Y.; Zeng, N.; Chen, M.; Zhou, Q.; Cai, S.; et al. The nasal microbiota is a potential diagnostic biomarker for sepsis in critical care units. Microbiol. Spectr. 2024, 12, e0344123. [Google Scholar] [CrossRef] [PubMed]
- Dvořák, Z.; Li, H.; Mani, S. Microbial Metabolites as Ligands to Xenobiotic Receptors: Chemical Mimicry as Potential Drugs of the Future. Drug Metab. Dispos. 2023, 51, 219–227. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U S A 2010, 107, 11971–5. [Google Scholar] [CrossRef]
- Chu, D.M.; Ma, J.; Prince, A.L.; Antony, K.M.; Seferovic, M.D.; Aagaard, K.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 2017, 23, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Meylan, P.; Lang, C.; Mermoud, S.; Johannsen, A.; Norrenberg, S.; Hohl, D.; Vial, Y.; Prod’hom, G.; Greub, G.; Kypriotou, M.; Christen-Zaech, S. Skin Colonization by Staphylococcus aureus Precedes the Clinical Diagnosis of Atopic Dermatitis in Infancy. J. Invest. Dermatol. 2017, 137, 2497–2504. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D. NISC Comparative Sequence Program; Murray, P.R.; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–9. [Google Scholar] [CrossRef]
- Paller, A.S.; Kong, H.H.; Seed, P.; Naik, S.; Scharschmidt, T.C.; Gallo, R.L.; Luger, T.; Irvine, A.D. The microbiome in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2019, 143, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Chen, T.H.; Narala, S. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, aah4680. [Google Scholar] [CrossRef] [PubMed]
- Demessant-Flavigny, A.L.; Connétable, S.; Kerob, D.; Moreau, M.; Aguilar, L.; Wollenberg, A. Skin microbiome dysbiosis and the role of Staphylococcus aureus in atopic dermatitis in adults and children: A narrative review. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 3–17. [Google Scholar] [CrossRef]
- Fox, C.; Nelson, D.; Wareham, J. The timing of skin acidification in very low birth weight infants. J. Perinatol. 1998, 18, 272–5. [Google Scholar]
- Zhou, C.; Fey, P.D. The acid response network of Staphylococcus aureus. Curr. Opin. Microbiol. 2020, 55, 67–73. [Google Scholar] [CrossRef]
- Fernández, L.; Gutiérrez, D.; García, P. Environmental pH is a key modulator of Staphylococcus aureus biofilm development under predation by the virulent phage phiIPLA-RODI. ISME J. 2021, 15, 245–259. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Costa, F.; Blake, K.J. S. S. aureus drives itch and scratch-induced skin damage through a V8 protease-PAR1 axis. Cell 2023, 186, 5375–5393.e5325. [Google Scholar] [CrossRef]
- Cork, M.J.; Danby, S.G.; Vasilopoulos, Y.; Hadgraft, J.; Lane, M.E.; Moustafa, M.; Guy, R.H.; Macgowan, A.L.; Tazi-Ahnini, R.; Ward, S.J. Epidermal barrier dysfunction in atopic dermatitis. J. Invest. Dermatol. 2009, 129, 1892–908. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.L.; Curran-Everett, D.; Leung, D.Y.M. Food allergy is associated with Staphylococcus aureus colonization in children with atopic dermatitis. J.Allergy Clin. Immunol. 2016, 137, 1247–1248.e1243. [Google Scholar] [CrossRef]
- Tsilochristou, O.; du Toit, G.; Sayre, P.H.; Roberts, G.; Lawson, K.; Sever, M.L.; Bahnson, H.T.; Radulovic, S.; Basting, M.; et al. Immune Tolerance Network Learning Early About Peanut Allergy Study Team. Association of Staphylococcus aureus colonization with food allergy occurs independently of eczema severity. J. Allergy Clin. Immunol. 2019, 144, 494–503. [Google Scholar] [CrossRef]
- Du Toit, G.; Roberts, G.; Sayre, P.H.; Bahnson, H.T.; Radulovic, S.; Santos, A.F.; Brough, H.A.; Phippard, D.; Basting, M.; Feeney, M.; et al. Randomized trial of peanut consumption in infants at risk for peanut allergy. N. Engl. J. Med. 2015, 26, 803–13. [Google Scholar] [CrossRef]
- Muluk, N.B.; Altın, F.; Cingi, C. Role of Superantigens in Allergic Inflammation: Their Relationship to Allergic Rhinitis, Chronic Rhinosinusitis, Asthma, and Atopic Dermatitis. Am. J. Rhinol Allergy 2018, 32, 502–517. [Google Scholar] [CrossRef] [PubMed]
- Boguniewicz, M.; Leung, D.Y. Atopic dermatitis: a disease of altered skin barrier and immune dysregulation. Immunol. Rev. 2011, 242, 233–46. [Google Scholar] [CrossRef]
- Clowry, J.; Dempsey, D.J.; Claxton, T.J.; Towell, A.M.; Turley, M.B.; Sutton, M.; Geoghegan, J.A.; Kezic, S.; Jakasa, I.; White, A.; et al. Distinct T cell signatures are associated with Staphylococcus aureus skin infection in pediatric atopic dermatitis. JCI Insight 2024, 9, e178789. [Google Scholar] [CrossRef]
- Zollner, T.M.; Wichelhaus, T.A.; Hartung, A.; Von Mallinckrodt, C.; Wagner, T.O.; Brade, V.; Kaufmann, R. Colonization with superantigen-producing Staphylococcus aureus is associated with increased severity of atopic dermatitis. Clin. Exp. Allergy. 2000, 30, 994–1000. [Google Scholar] [CrossRef]
- Czarnowicki, T.; Gonzalez, J.; Shemer, A.; Malajian, D.; Xu, H.; Zheng, X.; Khattri, S.; Gilleaudeau, P.; Sullivan-Whalen, M.; Suárez-Fariñas, M.; et al. Severe atopic dermatitis is characterized by selective expansion of circulating TH2/TC2 and TH22/TC22, but not TH17/TC17, cells within the skin-homing T-cell population. J. Allergy Clin. Immunol. 2015 136, 104-115.e7. [CrossRef]
- DeVore, S.B.; Gonzalez, T.; Sherenian, M.G.; Herr, A.B.; Khurana Hershey, G.K. On the surface: Skin microbial exposure contributes to allergic disease. Ann. Allergy Asthma Immunol. 2020, 125, 628–638. [Google Scholar] [CrossRef]
- Ménard, G.; Bonnaure-Mallet, M.; Donnio, P.Y. Adhesion of Staphylococcus aureus to epithelial cells: an in vitro approach to study interactions within the nasal microbiota. J. Med. Microbiol. 2020, 69, 1253–1261. [Google Scholar] [CrossRef]
- Guryanova, S.; Guryanova, A. sbv IMPROVER: Modern Approach to Systems Biology. Methods Mol Biol. 2017, 1613, 21–29. [Google Scholar] [CrossRef]
- Pirker, A.-L.; Vogl, T. Development of systemic and mucosal immune responses against gut microbiota in early life and implications for the onset of allergies. Front. Allergy 2024, 5, 1439303. [Google Scholar] [CrossRef] [PubMed]
- Namasivayam, A.A.; Morales, A.F.; Lacave, Á.M.; Tallam, A.; Simovic, B.; Alfaro, D.G.; Bobbili, D.R.; Martin, F.; Androsova, G.; Shvydchenko, I.; et al. Community-Reviewed Biological Network Models for Toxicology and Drug Discovery Applications. Gene Regul. Syst. Bio. 2016, 10, 51–66. [Google Scholar] [CrossRef]
- Cereta, A.D.; Oliveira, V.R.; Costa, I.P.; Afonso, J.P.R.; Fonseca, A.L.; de Souza, A.R.T.; Silva, G.A.M.; Mello, D.A.C.P.G.; de Oliveira, L.V.F.; da Palma, R.K. Emerging Cell-Based Therapies in Chronic Lung Diseases: What About Asthma? Front Pharmacol. 2021, 12, 648506. [Google Scholar] [CrossRef] [PubMed]
- Tap, J.; Mondot, S.; Levenez, F.; Pelletier, E.; Caron, C.; Furet, J.P.; Ugarte, E.; Muñoz-Tamayo, R.; Paslier, D.L.; Nalin, R.; et al. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 2009, 11, 2574–84. [Google Scholar] [CrossRef]
- Vuillermin, P.J.; Macia, L.; Nanan, R.; Tang, M.L.; Collier, F.; Brix, S. The maternal microbiome during pregnancy and allergic disease in the offspring. Semin. Immunopathol. 2017, 39, 669–675. [Google Scholar] [CrossRef] [PubMed]
- McCoy, K.D.; Köller, Y. New developments providing mechanistic insight into the impact of the microbiota on allergic disease. Clin. Immunol. 2015, 159, 170–6. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Kou, F.; Men, X.; Liu, Y.; Tang, L.; Wen, S. The Changes in Bacterial Microbiome Associated with Immune Disorder in Allergic Respiratory Disease. Microorganisms 2022, 10, 2066. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, F.; Balas, I.; Robinson, M.J.; Bakdash, G. Border Control: The Role of the Microbiome in Regulating Epithelial Barrier Function. Cells 2024, 13, 477. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Liang, S.; Jin, F. Gut microbiota and healthy longevity. Sci. China Life Sci. 2024, 2. [Google Scholar] [CrossRef]
- Ragonnaud, E.; Biragyn, A. Gut microbiota as the key controllers of “healthy” aging of elderly people. Immun. Ageing 2021, 18, 2. [Google Scholar] [CrossRef]
| Bacteria or Substances | Disease | Reference | |
|---|---|---|---|
| Gastrointestinal tract | Deficiency of Lachnospira, Veillonella, Rothia and Faecalibacterium | Asthma | [169] |
| An increase in the Lachnospira / Clostridium neonatale ratio | [187] | ||
| Deficiency of odd-chain fatty acids, 10-nonadecenoate and 10-hepatadecenoate Enriched for the aconitate |
[168] | ||
| Deficiency of short-chain fatty acids | [176,186] | ||
| Presence of Bifidobacterium catenulatum Deficiency of Bifidobacterium breve An increase of the species Firmicutes such as Clostridium and deficiency of Bacteroides Deficiency of oligosaccharides and short-chain fatty acids |
Atopic dermatitis | [170,171,172] [173] [174] [176] |
|
| Deficiency of Bifidobacterium breve Deficiency of Bifidobacterium lactis Deficiency of short-chain fatty acids |
Allergic rhinitis | [179] [175] [177] |
|
| Presence of Enterobacter and deficiency of Bifidobacterium | Chronic rhinosinusitis | [178] | |
| Presence of Clostridiaceae, Ruminococcaceae, Lachnospiraceae and Erysipelotrichaceae Deficiency of Lactobacillales, Bacteroidales |
Food allergy | [189] [190] |
|
| Upper and lower respiratory tract |
Streptococcus in nasopharynx Predominance of Moraxella in nasopharynx Low abundance of Lactobacillus in nasopharynx Low abundance of Firmicutes, Actinobacteria and Saccharibacteria in nasopharynx Haemophilus influenza, Moraxella catarrhalis and Tropheryma whipplei in sputum Moraxella catarrhalis, Haemophilus and Streptococcus in sputum Enriched with taxa from Bacteroidetes and Proteobacteria in nasopharynx Klebsiella in bronchial swab samples |
Asthma |
[193] [197,198,199] [195] [203] [204,206] [202] [196] [207] |
| Low abundance of Bacteroidetes and predominance of Staphylococcus aureus in nasal lavage | Chronic rhinosinusitis | [209] | |
| Low abundance of Acinetobacter | Allergic rhinitis | [210] | |
| Skin | Predominance of Staphylococcus aureus Deficiency of Staphylococcus epidermidis, Streptococcus, Propionibacterium and Corynebacterium Predominance of Staphylococcus aureus and deficiency of Staphylococcus epidermidis and Staphylococcus hominis |
Atopic dermatitis | [219,220,221] [220] [222] |
| Predominance of Staphylococcus aureus | Food allergy | [230,231,232] | |
| Low abundance of Acinetobacter | Allergic rhinitis | [210] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





