Submitted:
22 August 2024
Posted:
22 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
- The trade-off between the enhancement of image contrast and details is set as a multi-objective problem. Unlike the traditional mono-objective approach, which only provides an optimal solution with a predefined priority, the current proposal offers the best solutions regarding the compromises between both criteria along the Pareto front.
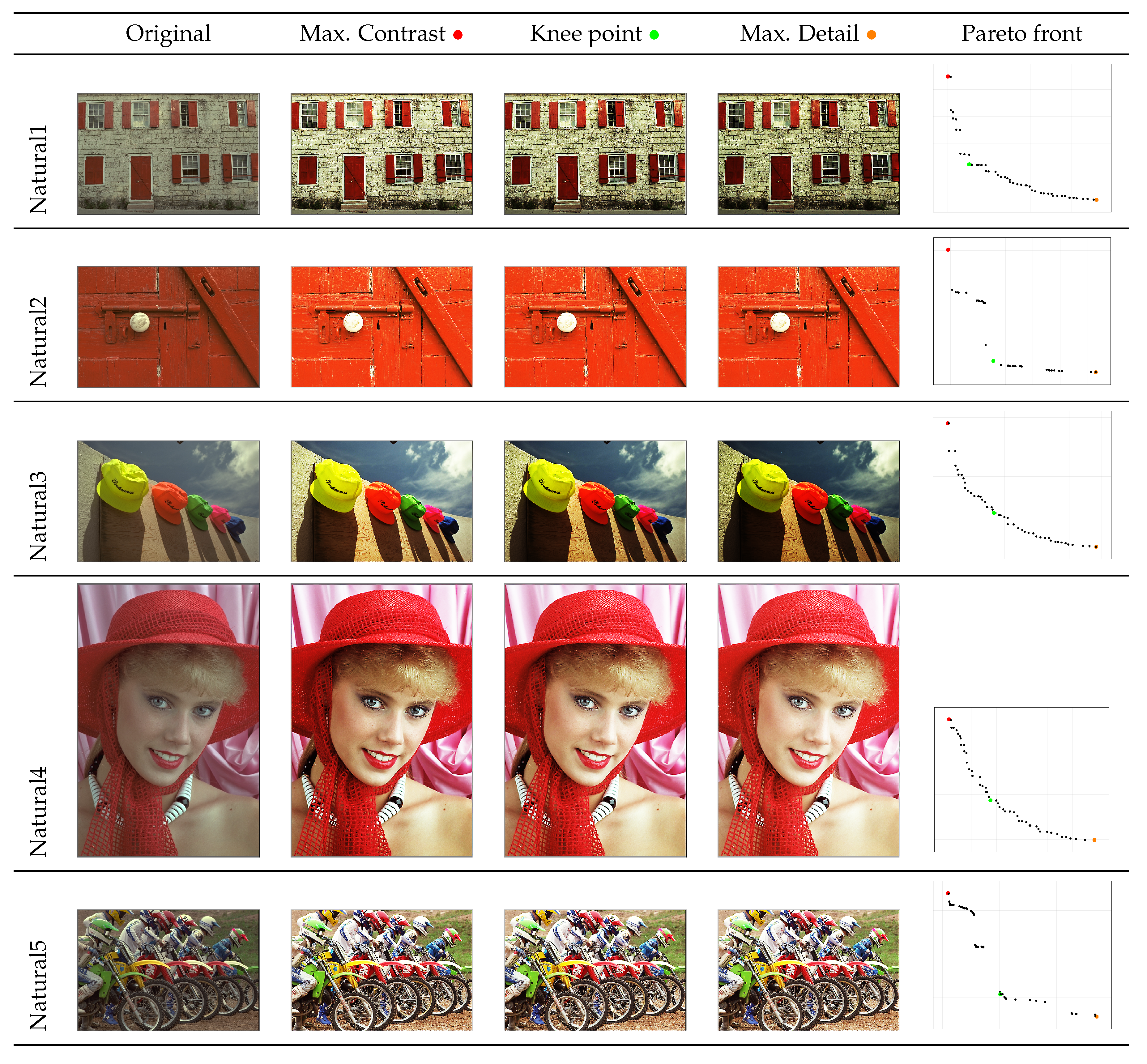
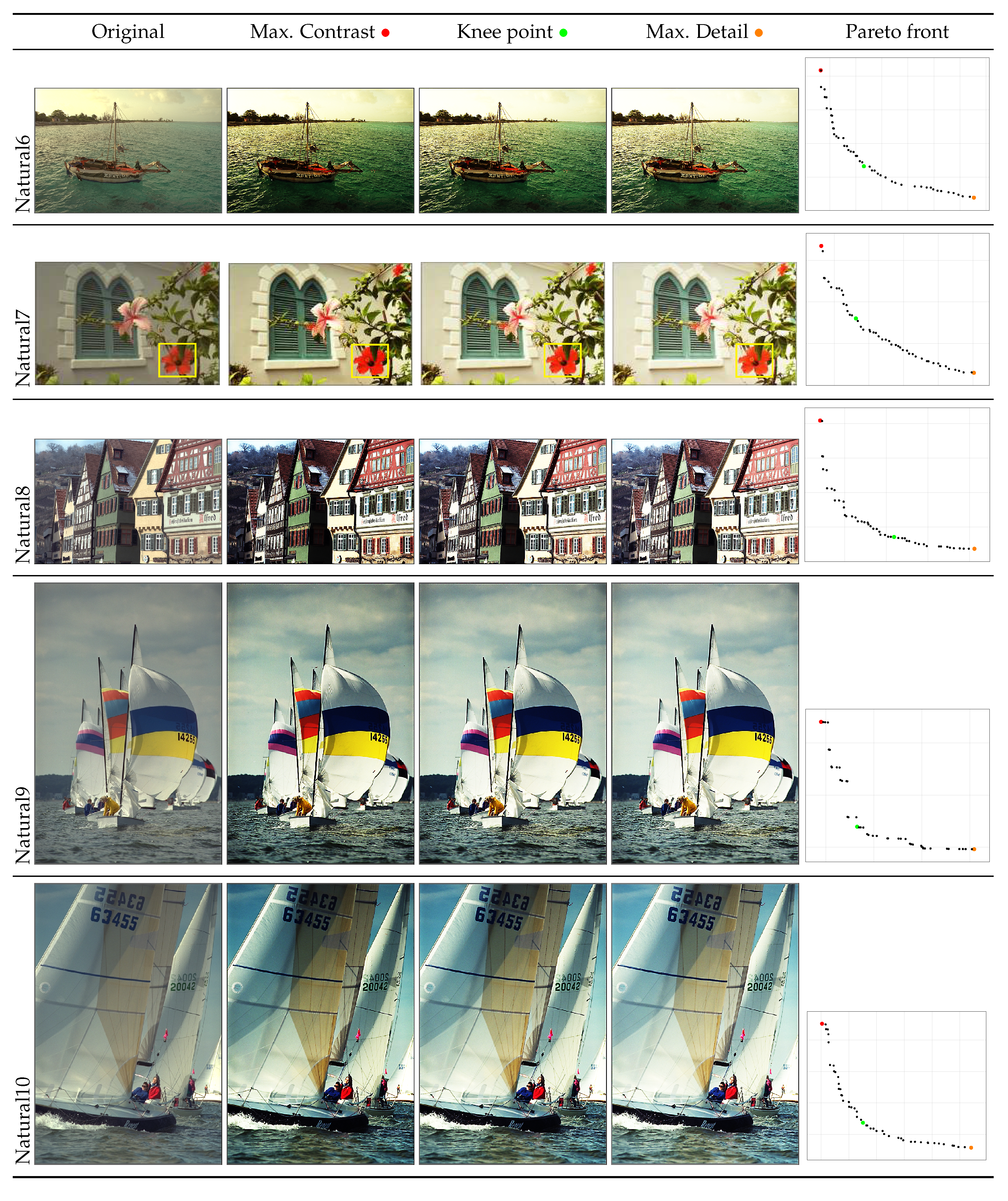
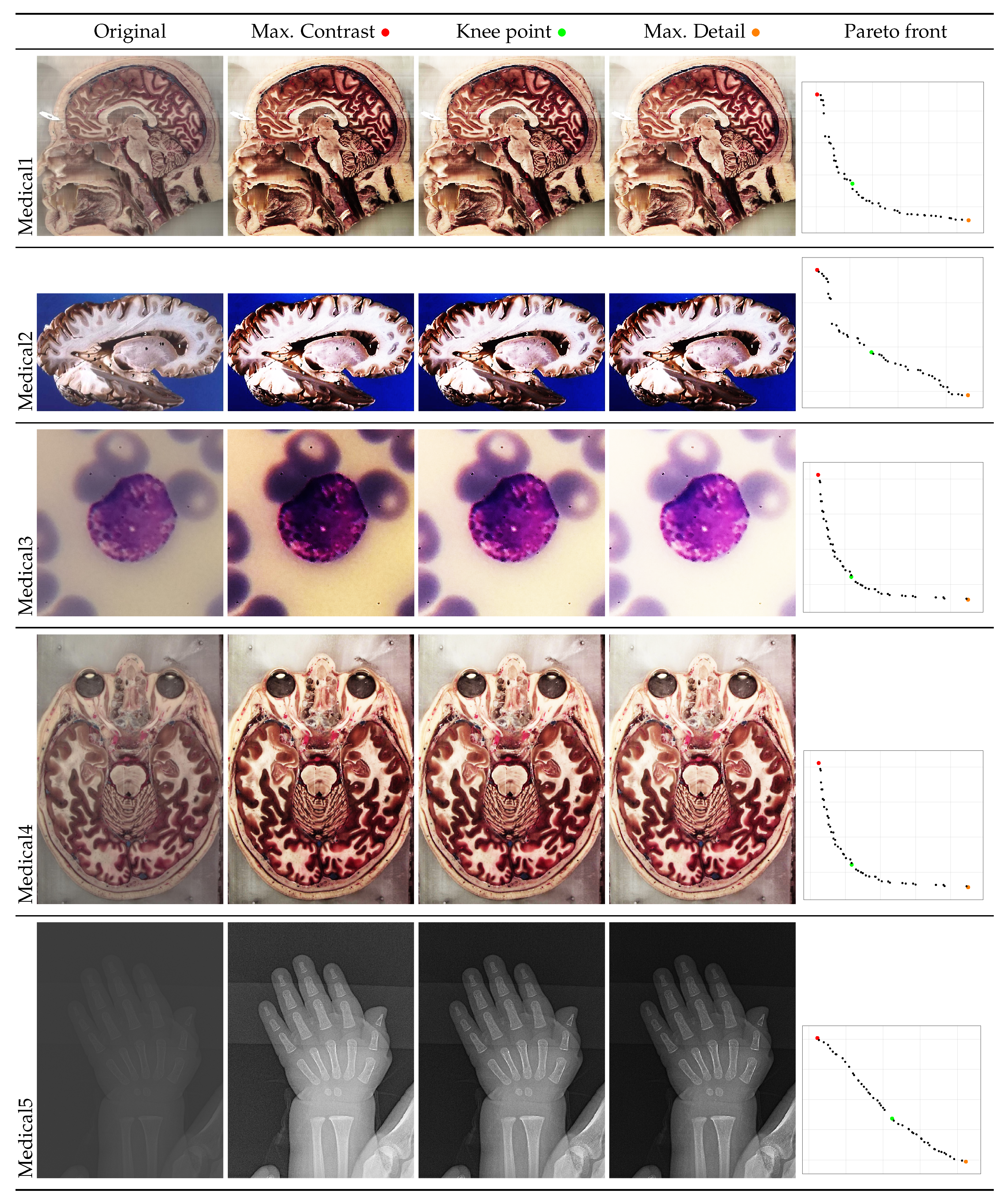
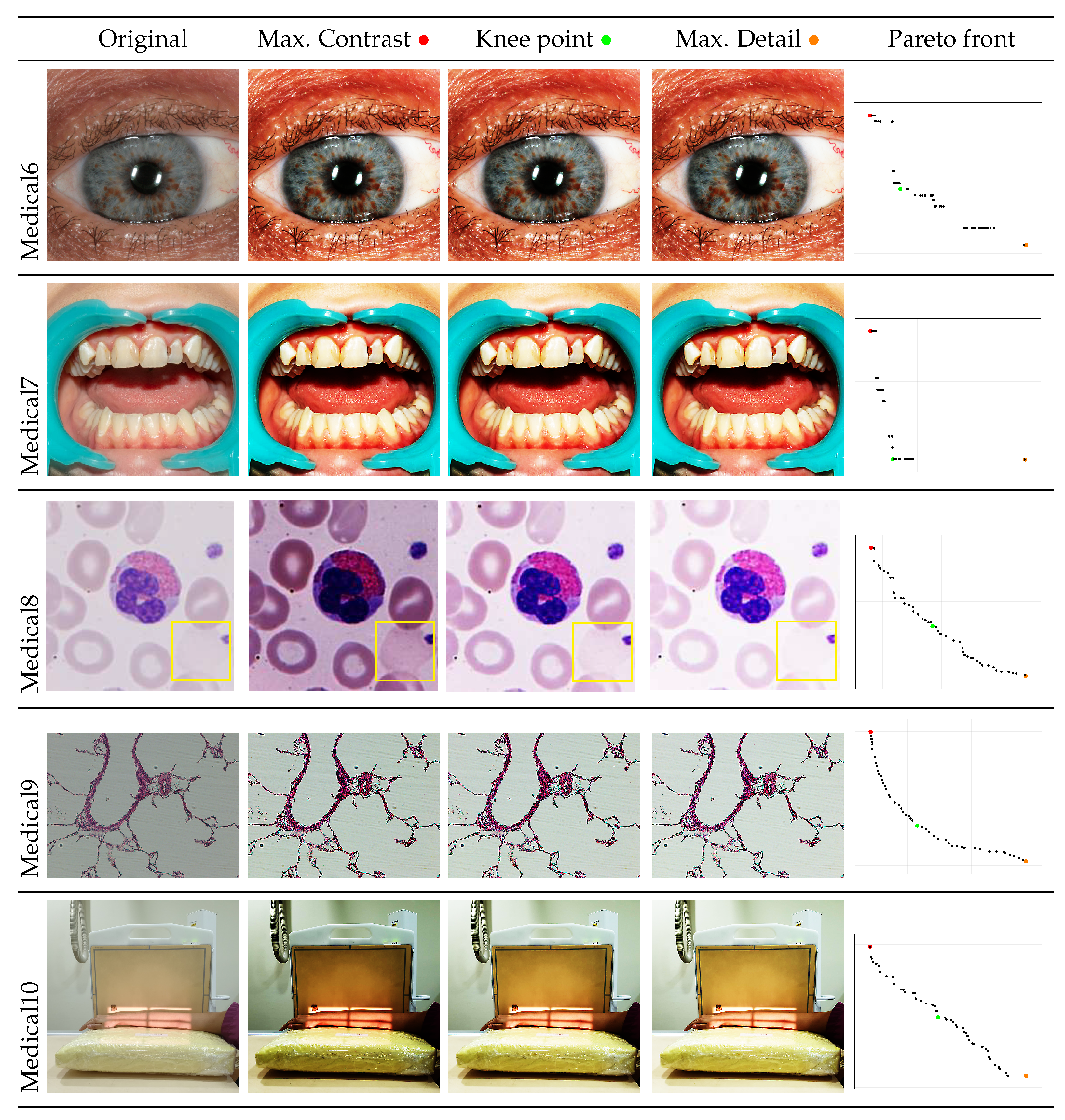
- A posterior preference operator is articulated, providing three key images from the Pareto front: the image with maximum contrast, the image with maximum detail, and the image at the knee of the front, which represents the image closest to the utopia point. This operator allows the user to select the most suitable solutions to their particular needs.
- An experiment is conducted with images of two categories: medical and natural scene images. Both categories represent research fields where image processing is an essential endeavor. The results of this experiment demonstrate that the NSGA-II achieves images of superior quality compared to the original instances. Furthermore, a thorough analysis is conducted regarding the suitability of the obtained images according to the established preferences. For medical images, the evaluation focuses on how the selected solutions enhance the clarity and detail of relevant structures, which is crucial for diagnostics and analysis. For natural scene images, the analysis shows how the solutions improve contrast and detail, making the images more visually appealing and impactful.
2. Materials and Methods
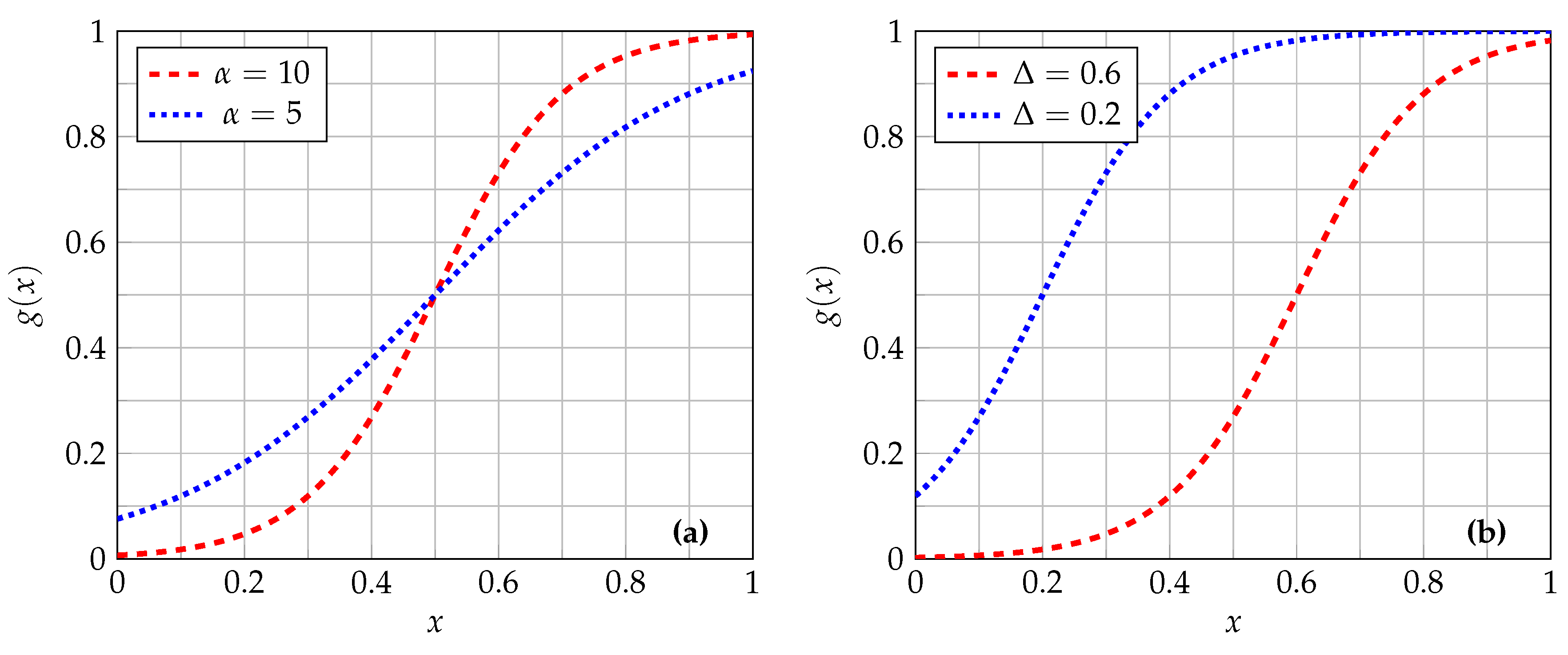
2.1. Sigmoid Correction
2.2. Unsharp Masking and Highboost Filtering
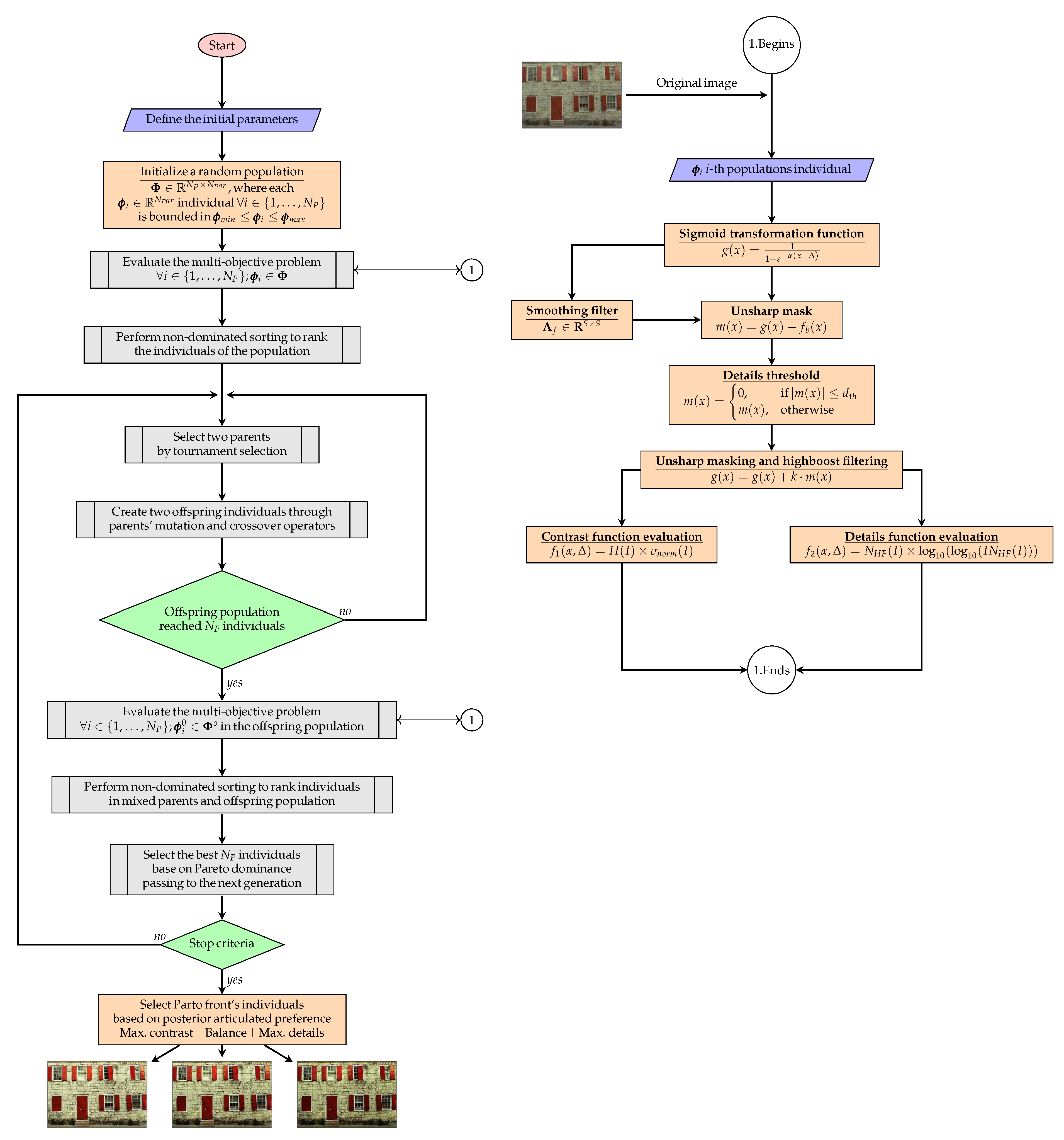
3. The Proposed Algorithm
3.1. Multi-Objective Optimization Problem
- Contrast Function: Defined as the product of entropy and the normalized standard deviation of the pixel intensities of the I image. The contrast function is expressed as:
- Details Function: Defined as the product of the number of pixels in high-frequency regions and the intensity of these high-frequency pixels of the I image. The details function is denoted as:
3.2. A Posterior Preference Articulation
4. Benchmark Results and Discussion
4.1. Experimental Design
4.2. Graphical Results
4.3. Quantitative Results
5. Conclusions
Author Contributions
Data Availability Statement
Acknowledgments
References
- Rafael, C.G.; Richard, E.W.; Steven, L.E.; Woods, R.; Eddins, S. Digital image processing using MATLAB; Tata McGraw-Hill, 2010.
- Russ, J.C.; Russ, J.C. Introduction to image processing and analysis; CRC press, 2017.
- Gonzalez, R.C.; Woods, R.E. Digital image processing 4th edition, global edition, 2018.
- Aşuroğlu, T.; Sümer, E. Performance analysis of spatial and frequency domain filtering in high resolution images. 2015 23nd Signal Processing and Communications Applications Conference (SIU). IEEE, 2015, pp. 935–938.
- Lepcha, D.C.; Goyal, B.; Dogra, A.; Sharma, K.P.; Gupta, D.N. A deep journey into image enhancement: A survey of current and emerging trends. Information Fusion 2023, 93, 36–76.
- Guo, X.; Li, Y.; Ling, H. LIME: Low-light image enhancement via illumination map estimation. IEEE Transactions on image processing 2016, 26, 982–993.
- Wang, W.; Wu, X.; Yuan, X.; Gao, Z. An experiment-based review of low-light image enhancement methods. Ieee Access 2020, 8, 87884–87917.
- Ullah, Z.; Farooq, M.U.; Lee, S.H.; An, D. A hybrid image enhancement based brain MRI images classification technique. Medical hypotheses 2020, 143, 109922.
- Dhal, K.G.; Ray, S.; Das, A.; Das, S. A survey on nature-inspired optimization algorithms and their application in image enhancement domain. Archives of Computational Methods in Engineering 2019, 26, 1607–1638.
- Bhandarkar, S.M.; Zhang, Y.; Potter, W.D. An edge detection technique using genetic algorithm-based optimization. Pattern Recognition 1994, 27, 1159–1180.
- Braik, M.; Sheta, A.F.; Ayesh, A. Image Enhancement Using Particle Swarm Optimization. World congress on engineering, 2007, Vol. 1, pp. 978–988.
- Behera, S.K.; Mishra, S.; Rana, D. Image enhancement using accelerated particle swarm optimization. Int J Eng Res Technol 2015, 4, 1049–1054.
- Nguyen-Thi, K.N.; Che-Ngoc, H.; Pham-Chau, A.T. An efficient image contrast enhancement method using sigmoid function and differential evolution. Journal of Advanced Engineering and Computation 2020, 4, 162–172.
- Bhandari, A.K.; Maurya, S. Cuckoo search algorithm-based brightness preserving histogram scheme for low-contrast image enhancement. Soft Computing 2020, 24, 1619–1645.
- Acharya, U.K.; Kumar, S. Genetic algorithm based adaptive histogram equalization (GAAHE) technique for medical image enhancement. Optik 2021, 230, 166273.
- Acharya, U.K.; Kumar, S. Directed searching optimized texture based adaptive gamma correction (DSOTAGC) technique for medical image enhancement. Multimedia Tools and Applications 2024, 83, 6943–6962.
- Braik, M. Hybrid enhanced whale optimization algorithm for contrast and detail enhancement of color images. Cluster Computing 2024, 27, 231–267.
- Rani, S.S. Colour image enhancement using weighted histogram equalization with improved monarch butterfly optimization. International Journal of Image and Data Fusion 2024, pp. 1–27.
- Du, N.; Luo, Q.; Du, Y.; Zhou, Y. Color image enhancement: a metaheuristic chimp optimization algorithm. Neural Processing Letters 2022, 54, 4769–4808.
- Krishnan, S.N.; Yuvaraj, D.; Banerjee, K.; Josephson, P.J.; Kumar, T.C.A.; Ayoobkhan, M.U.A. Medical image enhancement in health care applications using modified sun flower optimization. Optik 2022, 271, 170051.
- Ma, G.; Yue, X.; Zhu, J.; Liu, Z.; Zhang, Z.; Zhou, Y.; Li, C. A novel slime mold algorithm for grayscale and color image contrast enhancement. Computer Vision and Image Understanding 2024, 240, 103933.
- More, L.G.; Brizuela, M.A.; Ayala, H.L.; Pinto-Roa, D.P.; Noguera, J.L.V. Parameter tuning of CLAHE based on multi-objective optimization to achieve different contrast levels in medical images. 2015 IEEE International Conference on Image Processing (ICIP). IEEE, 2015, pp. 4644–4648.
- Abouhawwash, M.; Alessio, A.M. Multi-objective evolutionary algorithm for PET image reconstruction: Concept. IEEE transactions on medical imaging 2021, 40, 2142–2151.
- Kuran, U.; Kuran, E.C. Parameter selection for CLAHE using multi-objective cuckoo search algorithm for image contrast enhancement. Intelligent Systems with Applications 2021, 12, 200051.
- Cuevas, E.; Zaldívar, D.; Pérez-Cisneros, M. Multi-objective Optimization of Anisotropic Diffusion Parameters for Enhanced Image Denoising. In New Metaheuristic Schemes: Mechanisms and Applications; Springer, 2023; pp. 241–268.
- Jaimes, A.L.; Martınez, S.Z.; Coello, C.A.C.; others. An introduction to multiobjective optimization techniques. Optimization in Polymer Processing 2009, 1, 29.
- Lee, D.H.; Kim, K.J.; Köksalan, M. A posterior preference articulation approach to multiresponse surface optimization. European Journal of Operational Research 2011, 210, 301–309.
- Wang, H.; Olhofer, M.; Jin, Y. A mini-review on preference modeling and articulation in multi-objective optimization: current status and challenges. Complex & Intelligent Systems 2017, 3, 233–245.
- Deb, K.; Pratap, A.; Agarwal, S.; Meyarivan, T. A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE transactions on evolutionary computation 2002, 6, 182–197.
- Imtiaz, M.S.; Wahid, K.A. Image enhancement and space-variant color reproduction method for endoscopic images using adaptive sigmoid function. 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE, 2014, pp. 3905–3908.
- Srinivas, K.; Bhandari, A.K. Low light image enhancement with adaptive sigmoid transfer function. IET Image Processing 2020, 14, 668–678.
- Eastman Kodak Company. Kodak Lossless True Color Image Suite. https://r0k.us/graphics/kodak/, 2013. Accessed: 2024-08-18.
- Hartung, M. Visible Human Project - Brain Images. Case study. https://doi.org/10.53347/rID-93598, 2024. Accessed on 10 Aug 2024.
- Al Kabbani, A. Human brain - lateral view. Case study. https://doi.org/10.53347/rID-72871, 2024. Accessed on 10 Aug 2024.
- Nickparvar, M. White Blood Cells Dataset: A Large Dataset of Five White Blood Cells Types. https://www.kaggle.com/datasets/masoudnickparvar/white-blood-cells-dataset/data, 2022. Accessed on 10 Aug 2024.
- Mooney, P. Blood Cell Image Dataset. https://www.kaggle.com/datasets/paultimothymooney/blood-cells/discussion/437393, note = Accessed on 10 Aug 2024,, 2018.
- Radiological Society of North America. Pediatric Bone Age Machine Learning Challenge Dataset. https://www.kaggle.com/code/plarmuseau/image-contrast-enhancement-techniques/input, 2017. Accessed on 10 Aug 2024.
- Thurston, M. Lisch nodules (photo). https://doi.org/10.53347/rID-59408, 2024. Accessed on 10 Aug 2024.
- Dhairya, S. Dental Condition Dataset. https://www.kaggle.com/datasets/sizlingdhairya1/oral-infection, 2024. Accessed on 10 Aug 2024.
- Sinitca, A.M.; Lyanova, A.I.; Kaplun, D.I.; Hassan, H.; Krasichkov, A.S.; Sanarova, K.E.; Shilenko, L.A.; Sidorova, E.E.; Akhmetova, A.A.; Vaulina, D.D.; others. Microscopy Image Dataset for Deep Learning-Based Quantitative Assessment of Pulmonary Vascular Changes. Scientific Data 2024, 11, 635.
- A, E. Trauma forearm positioning (photo). https://doi.org/10.53347/rID-76100, 2020. Accessed on 10 Aug 2024.
 |
 |
 |
 |
| Image | Solution | SSIM | ||||||
|---|---|---|---|---|---|---|---|---|
| Natural1 | Max. Contrast | 7.5531 | 0.5957 | 136328 | 28907.9607 | 4.4997 | 100511.1934 | 0.6658 |
| Knee | 7.5593 | 0.5919 | 140554 | 29866.6697 | 4.4741 | 103785.0868 | 0.6434 | |
| Max. Detail | 7.4855 | 0.5771 | 142301 | 30043.5271 | 4.3198 | 105104.0062 | 0.6153 | |
| Natural2 | Max. Contrast | 6.3958 | 0.2890 | 42665 | 7256.3766 | 1.8482 | 29298.3087 | 0.6388 |
| Knee | 6.3867 | 0.2889 | 43005 | 7323.3415 | 1.8449 | 29547.1093 | 0.6484 | |
| Max. Detail | 6.3633 | 0.2888 | 43039 | 7331.7722 | 1.8374 | 29572.3889 | 0.6509 | |
| Natural3 | Max. Contrast | 7.5080 | 0.5562 | 27263 | 4478.1528 | 4.1760 | 18199.8145 | 0.8264 |
| Knee | 7.4174 | 0.5437 | 31497 | 5253.1676 | 4.0331 | 21228.6115 | 0.7635 | |
| Max. Detail | 7.2310 | 0.5140 | 33082 | 5554.9158 | 3.7168 | 22370.5067 | 0.6750 | |
| Natural4 | Max. Contrast | 7.6028 | 0.5582 | 38563 | 6506.9944 | 4.2437 | 26317.5588 | 0.7603 |
| Knee | 7.5705 | 0.5498 | 41095 | 6840.4895 | 4.1620 | 28125.8294 | 0.7430 | |
| Max. Detail | 7.4589 | 0.5305 | 42358 | 6941.6262 | 3.9571 | 29014.4705 | 0.7154 | |
| Natural5 | Max. Contrast | 7.6397 | 0.6084 | 123121 | 25158.0587 | 4.6481 | 90179.9275 | 0.6053 |
| Knee | 7.6277 | 0.6070 | 123275 | 25177.7133 | 4.6297 | 90296.0866 | 0.5938 | |
| Max. Detail | 7.5816 | 0.6062 | 123311 | 25173.0702 | 4.5959 | 90321.6616 | 0.5894 | |
| Natural6 | Max. Contrast | 7.2965 | 0.7151 | 92537 | 18795.8519 | 5.2174 | 66825.3294 | 0.6586 |
| Knee | 7.3302 | 0.7005 | 96356 | 19321.6818 | 5.1345 | 69678.1741 | 0.6931 | |
| Max. Detail | 7.2851 | 0.6758 | 97675 | 19209.1750 | 4.9234 | 70611.6297 | 0.7175 | |
| Natural7 | Max. Contrast | 7.4601 | 0.5546 | 45798 | 8129.0078 | 4.1374 | 31650.5187 | 0.7839 |
| Knee | 7.3807 | 0.5442 | 48599 | 8487.5329 | 4.0163 | 33666.6691 | 0.7652 | |
| Max. Detail | 7.1648 | 0.5160 | 50088 | 8529.1422 | 3.6970 | 34707.5355 | 0.7388 | |
| Natural8 | Max. Contrast | 7.2975 | 0.7166 | 135043 | 30609.3702 | 5.2293 | 99829.9165 | 0.6525 |
| Knee | 7.3574 | 0.7065 | 142322 | 31648.0378 | 5.1979 | 105373.9500 | 0.6738 | |
| Max. Detail | 7.3067 | 0.6903 | 144811 | 31899.5143 | 5.0441 | 107256.0717 | 0.6770 | |
| Natural9 | Max. Contrast | 7.4927 | 0.5278 | 41161 | 7399.7872 | 3.9550 | 28296.7289 | 0.7953 |
| Knee | 7.4455 | 0.5261 | 42537 | 7682.5907 | 3.9174 | 29304.3657 | 0.8037 | |
| Max. Detail | 7.3050 | 0.5196 | 42823 | 7772.1429 | 3.7954 | 29520.5423 | 0.8067 | |
| Natural10 | Max. Contrast | 7.5721 | 0.5280 | 32834 | 5719.8726 | 3.9983 | 22240.9423 | 0.7855 |
| Knee | 7.5501 | 0.5198 | 35058 | 6002.2180 | 3.9246 | 23814.2353 | 0.7937 | |
| Max. Detail | 7.4591 | 0.5000 | 35621 | 6062.6948 | 3.7295 | 24210.7577 | 0.7868 |
| Image | Solution | SSIM | ||||||
|---|---|---|---|---|---|---|---|---|
| Medical1 | Max. Contrast | 7.8448 | 0.5823 | 28921 | 4292.0292 | 4.5682 | 19256.7633 | 0.7469 |
| Knee | 7.8279 | 0.5820 | 29564 | 4414.5851 | 4.5558 | 19718.7333 | 0.7608 | |
| Max. Detail | 7.7870 | 0.5767 | 29861 | 4481.5297 | 4.4908 | 19935.0599 | 0.7732 | |
| Medical2 | Max. Contrast | 7.1626 | 0.7809 | 20764 | 3818.1387 | 5.5935 | 13726.0797 | 0.6879 |
| Knee | 7.1339 | 0.7809 | 21025 | 3876.9084 | 5.5710 | 13911.8213 | 0.6584 | |
| Max. Detail | 7.1054 | 0.7784 | 21162 | 3905.2624 | 5.5309 | 14008.8062 | 0.6366 | |
| Medical3 | Max. Contrast | 7.4796 | 0.6403 | 501 | 68.4979 | 4.7890 | 227.2635 | 0.7746 |
| Knee | 7.1434 | 0.5984 | 875 | 118.2316 | 4.2747 | 427.0364 | 0.8652 | |
| Max. Detail | 6.4043 | 0.4811 | 1058 | 145.9017 | 3.0808 | 529.6895 | 0.8658 | |
| Medical4 | Max. Contrast | 7.8136 | 0.5744 | 15156 | 1923.4562 | 4.4882 | 9576.7837 | 0.7955 |
| Knee | 7.7803 | 0.5708 | 16008 | 2042.4497 | 4.4411 | 10157.2992 | 0.8073 | |
| Max. Detail | 7.6851 | 0.5563 | 16190 | 2081.7596 | 4.2751 | 10286.2666 | 0.8092 | |
| Medical5 | Max. Contrast | 4.1156 | 0.4317 | 1017 | 117.5567 | 1.7767 | 495.9842 | 0.5690 |
| Knee | 4.1156 | 0.3829 | 1486 | 182.3434 | 1.5757 | 763.2875 | 0.6173 | |
| Max. Detail | 4.1156 | 0.3346 | 1729 | 217.4919 | 1.3772 | 905.4307 | 0.6017 | |
| Medical6 | Max. Contrast | 7.6461 | 0.4482 | 16721 | 2343.1399 | 3.4268 | 10709.4627 | 0.7703 |
| Knee | 7.6442 | 0.4482 | 16732 | 2347.1401 | 3.4259 | 10717.7392 | 0.7703 | |
| Max. Detail | 7.6363 | 0.4482 | 16742 | 2346.8756 | 3.4224 | 10724.0633 | 0.7702 | |
| Medical7 | Max. Contrast | 7.7687 | 0.6045 | 6378 | 970.3478 | 4.6965 | 3831.1820 | 0.7633 |
| Knee | 7.7658 | 0.6047 | 6389 | 972.1292 | 4.6958 | 3838.3433 | 0.7612 | |
| Max. Detail | 7.7591 | 0.6047 | 6389 | 972.2033 | 4.6916 | 3838.3663 | 0.7607 | |
| Medical8 | Max. Contrast | 6.7448 | 0.4860 | 354 | 45.3329 | 3.2779 | 150.7481 | 0.7133 |
| Knee | 6.1648 | 0.4642 | 496 | 64.1421 | 2.8618 | 222.8542 | 0.8958 | |
| Max. Detail | 5.3591 | 0.4163 | 585 | 74.7138 | 2.2309 | 268.6718 | 0.9375 | |
| Medical9 | Max. Contrast | 5.6986 | 0.4041 | 29117 | 7292.7190 | 2.3027 | 20000.4591 | 0.8336 |
| Knee | 5.5245 | 0.4116 | 29587 | 7479.4364 | 2.2737 | 20352.2764 | 0.8403 | |
| Max. Detail | 5.3411 | 0.4130 | 29762 | 7561.0648 | 2.2061 | 20485.1491 | 0.8429 | |
| Medical10 | Max. Contrast | 7.7175 | 0.5087 | 16293 | 2996.5663 | 3.9260 | 10606.1673 | 0.8023 |
| Knee | 7.5585 | 0.5002 | 16562 | 3094.9172 | 3.7810 | 10803.7683 | 0.8674 | |
| Max. Detail | 7.3872 | 0.4864 | 16812 | 3100.2399 | 3.5931 | 10968.0618 | 0.8806 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





