Submitted:
22 August 2024
Posted:
23 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Data Sets, Gene Identification and Enrichment
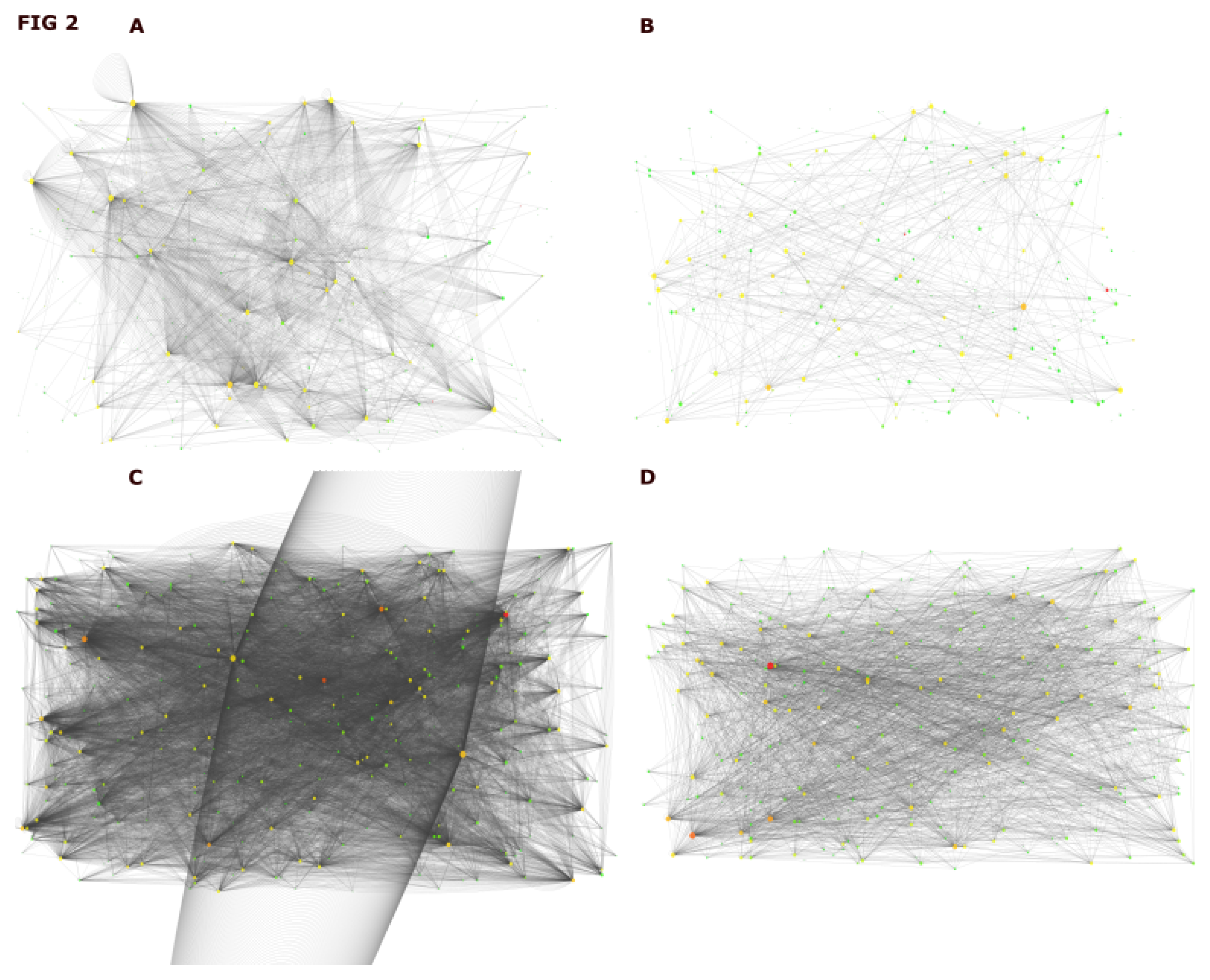
2.2. Assembly of Interactome for DIAMOnD Genes Identification
2.3. Identification of DIAMOnD Genes
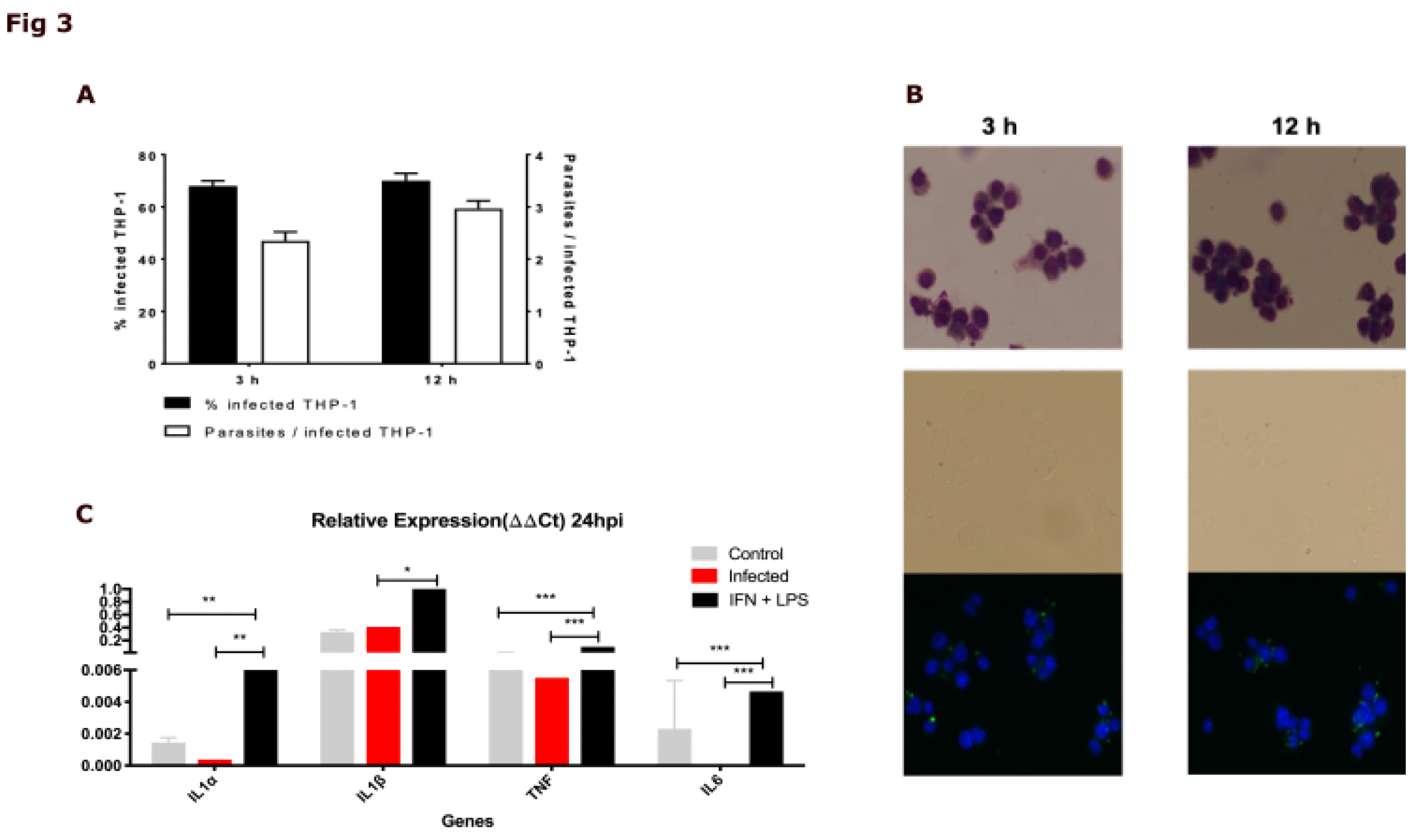
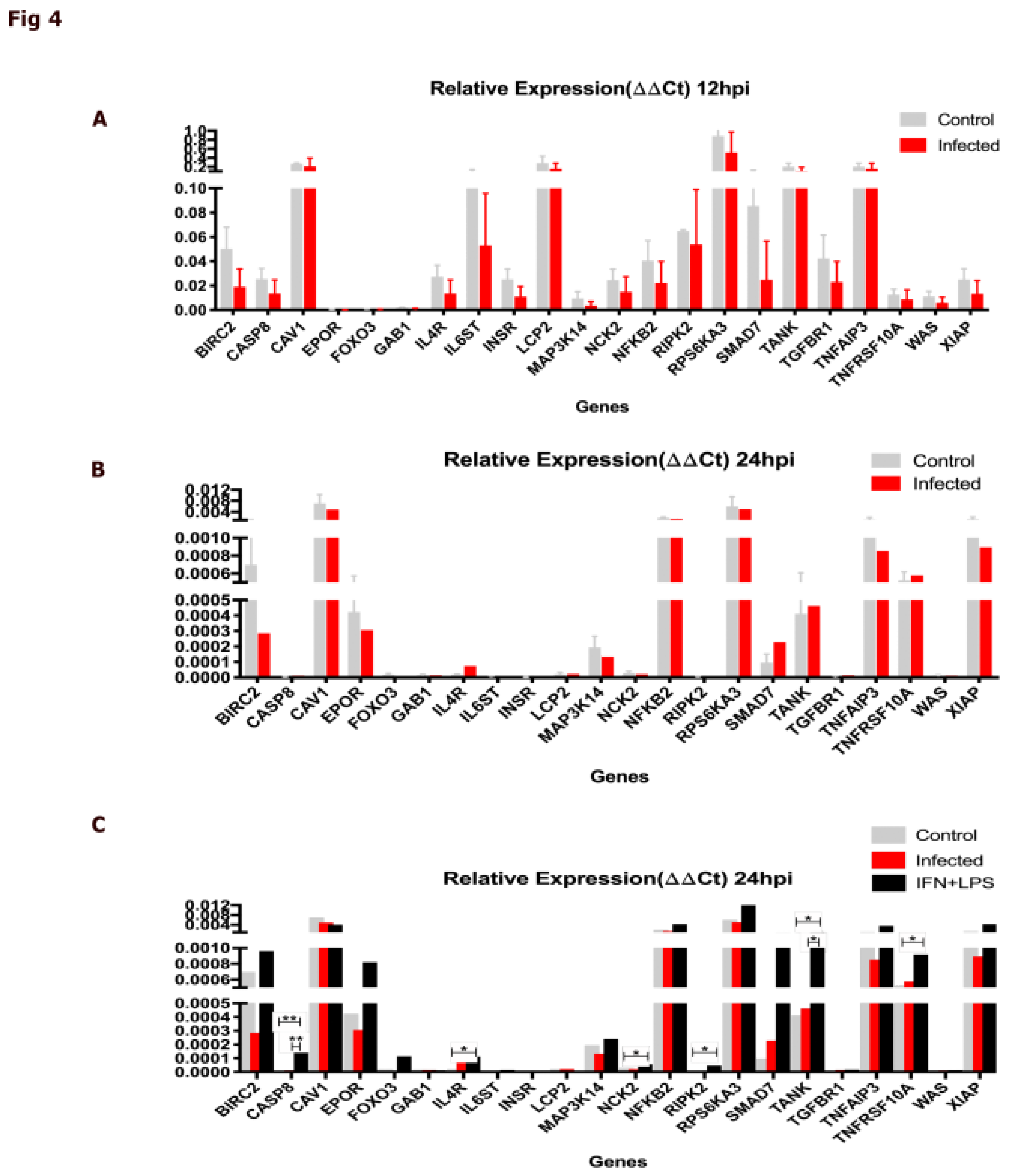
2.4. The DIAMOnD Genes Are Expressed by Human Macrophages Infected with Leishmania major
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Data source and Filtering
5.2. Enrichment Analysis
5.3. Network Analysis
5.4. Biological Validation
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Arenas, R.; Torres-Guerrero, E.; Quintanilla-Cedillo, M.R.; Ruiz-Esmenjaud, J. Leishmaniasis: A review. F1000Research 2017, 6, 1–15. [Google Scholar] [CrossRef]
- Oryan, A.; Akbari, M. Worldwide risk factors in leishmaniasis. Asian Pacific Journal of Tropical Medicine 2016, 9, 925–932. [Google Scholar] [CrossRef]
- Bogdan, C.; Röllinghoff, M. The immune response to Leishmania: Mechanisms of parasite control and evasion. International Journal for Parasitology 1998, 28, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Gregory, D.J.; Forget, G. Subversion mechanisms by which Leishmania parasites can escape the host immune response: A signaling point of view. Clinical Microbiology Reviews 2005, 18, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Arango Duque, G.; Descoteaux, A. Leishmania survival in the macrophage: Where the ends justify the means. Current Opinion in Microbiology 2015, 26, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Alves-Ferreira, E.V.; Toledo, J.S.; De Oliveira, A.H.; Ferreira, T.R.; Ruy, P.C.; Pinzan, C.F.; Santos, R.F.; Boaventura, V.; Rojo, D.; López-Gonzálvez,; et al. Differential Gene Expression and Infection Profiles of Cutaneous and Mucosal Leishmania braziliensis Isolates from the Same Patient. PLoS Neglected Tropical Diseases 2015, 9, 1–19. [Google Scholar] [CrossRef]
- Rodriguez, N.E.; Chang, H.K.; Wilson, M.E. Novel Program of Macrophage Gene Expression Induced by Phagocytosis of Leishmania chagasi. Infection and Immunity 2004, 72, 2111–2122. [Google Scholar] [CrossRef] [PubMed]
- Ovalle-Bracho, C.; Franco-Muñoz, C.; Londoño-Barbosa, D.; Restrepo-Montoya, D.; Clavijo-Ramírez, C. Changes in macrophage gene expression associated with Leishmania (Viannia) braziliensis infection. PLoS ONE 2015, 10, 1–13. [Google Scholar] [CrossRef]
- Ontoria, E.; Hernández-Santana, Y.E.; González-García, A.C.; López, M.C.; Valladares, B.; Carmelo, E. Transcriptional profiling of immune-related genes in Leishmania infantum-infected mice: Identification of potential biomarkers of infection and progression of disease. Frontiers in Cellular and Infection Microbiology 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Rabhi, I.; Rabhi, S.; Ben-Othman, R.; Aniba, M.R.; Trentin, B.; Piquemal, D.; Regnault, B.; Guizani-Tabbane, L.; Attia, H.; Ben Miled, S.; et al. Comparative analysis of resistant and susceptible macrophage gene expression response to Leishmania major parasite. BMC Genomics 2013, 14, 1–11. [Google Scholar] [CrossRef]
- Petti, M.; Punzi, C.; Alfano, C.; Farina, L.; Astolfi, L.; Paci, P.; Guzzi, P.H.; Castiglione, F.; Tieri, P. Network Inference and Reconstruction in Bioinformatics. Reference Module in Life Sciences 2024, 1–14. [Google Scholar] [CrossRef]
- Phan, T.N.; Park, K.H.P.; Shum, D.; No, J.H. Identification of Leishmania donovani PEX5-PTS1 Interaction Inhibitors through Fluorescence Polarization-Based High-Throughput Screening. Molecules 2024, 29, 1–13. [Google Scholar] [CrossRef]
- Phan, T.N.; Park, K.P.; Benítez, D.; Comini, M.A.; Shum, D.; No, J.H. Discovery of novel Leishmania major trypanothione synthetase inhibitors by high-throughput screening. Biochemical and Biophysical Research Communications 2022, 637, 308–313. [Google Scholar] [CrossRef]
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S.; et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014, 506, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Biswas, D.; Datta, A.; Roy, P.K. Combating leishmaniasis through awareness campaigning: A mathematical study on media efficiency. International Journal of Mathematical, Engineering and Management Sciences 2016, 1, 139–149. [Google Scholar] [CrossRef]
- De Menezes, J.P.B.; Khouri, R.; Oliveira, C.V.S.; De Oliveira Almeida Petersen, A.L.; De Almeida, T.F.; Mendes, F.R.; Do Amor Divino Rebouças, A.; Lorentz, A.L.; Luz, N.F.; Lima, J.B.; et al. Proteomic analysis reveals a predominant NFe2L2 (Nrf2) signature in canonical pathway and upstream regulator analysis of Leishmania-infected macrophages. Frontiers in Immunology 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 2003, 13, 2498–504. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Toro, J.; Prieto, C.; De Las Rivas, J. APID2NET: Unified interactome graphic analyzer. Bioinformatics 2007, 23, 2495–2497. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein.
- Science2021, 30, 187–200. [CrossRef]
- Alonso-López, D.; Gutiérrez, M.A.; Lopes, K.P.; Prieto, C.; Santamaría, R.; De Las Rivas, J. APID interactomes: Providing proteome-based interactomes with controlled quality for multiple species and derived networks. Nucleic Acids Research 2016, 44, W529–W535. [Google Scholar] [CrossRef] [PubMed]
- Ghiassian, S.D.; Menche, J.; Barabási, A.L. A DIseAse MOdule Detection (DIAMOnD) Algorithm Derived from a Systematic Analysis of Connectivity Patterns of Disease Proteins in the Human Interactome. PLoS Computational Biology 2015, 11, 1–22. [Google Scholar] [CrossRef]
- Thuer, E.; Gabaldon, T. Comparative transcriptomics of THP-1 monocytes in response to different pathogens. bioRxiv 2017, 155853. [Google Scholar]
- Gebre-Hiwot, A.; Tadesse, G.; Croft, S.L.; Frommel, D. An in vitro model for screening antileishmanial drugs: the human leukaemia monocyte cell line, THP-1. Acta Tropica 1992, 51, 237–245. [Google Scholar] [CrossRef]
- Rai, R.; Dyer, P.; Richardson, S.; Harbige, L.; Getti, G. Apoptotic induction induces Leishmania aethiopica and L. Mexicana spreading in terminally differentiated THP-1 cells. Parasitology 2017, 144, 1912–1921. [Google Scholar] [CrossRef]
- O’Keeffe, A.; Hyndman, L.; McGinty, S.; Riezk, A.; Murdan, S.; Croft, S.L. Development of an in vitro media perfusion model of Leishmania major macrophage infection. PLoS ONE 2019, 14, 1–20, Soong, L. Subversion and utilization of host innate defense by Leishmania amazonensis. Frontiers in Immunology 2012, 3, 1-7. 10.3389/fimmu.2012.00058. [Google Scholar] [CrossRef]
- Wanasen, N.; Soong, L. L-arginine metabolism and its impact on host immunity against Leishmania infection. Immunologic Research 2008, 41, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Atayde, V.D.; Hassani, K.; da Silva Lira Filho, A.; Borges, A.R.; Adhikari, A.; Martel, C.; Olivier, M. Leishmania exosomes and other virulence factors: Impact on innate immune response and macrophage functions. Cellular Immunology 2016, 309, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Tieri, P.; Zhou, X.; Zhu, L.; Nardini, C. Multi-omic landscape of rheumatoid arthritis: re-evaluation of drug adverse effects. Frontiers in Cell and Developmental Biology 2014, 2, 59. [Google Scholar] [CrossRef]
- Sousa, R.; Andrade, V.M.; Bair, T.; Ettinger, N.A.; Guimarães, L.; Andrade, L.; Guimarães, L.H.; Machado, P.R.; Carvalho, E.M.; Wilson, M.E.; et al. Early suppression of macrophage gene expression by Leishmania braziliensis. Frontiers in Microbiology 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Chaussabel, D.; Semnani, R.T.; McDowell, M.A.; Sacks, D.; Sher, A.; Nutman, T.B. Unique gene expression profiles of human macrophages and dendritic cells to phylogenetically distinct parasites. Blood 2003, 102, 672–681. [Google Scholar] [CrossRef]
- Ben Khalaf, N.; De Muylder, G.; Louzir, H.; McKerrow, J.; Chenik, M. Leishmania major protein disulfide isomerase as a drug target: Enzymatic and functional characterization. Parasitology Research 2012, 110, 1911–1917. [Google Scholar] [CrossRef]
- Khalaf, N.B.; De Muylder, G.; Ratnam, J.; Kean-Hooi Ang, K.; Arkin, M.; McKerrow, J.; Chenik, M. A high-throughput turbidometric assay for screening inhibitors of leishmania major protein disulfide isomerase. Journal of Biomolecular Screening 2011, 16, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Pandey, R.K.; Siqueira-Neto, J.L.; Kwon, Y.J.; Freitas-Junior, L.H.; Shaha, C.; Madhubala, R. Proteomic-based approach to gain insight into reprogramming of THP-1 cells exposed to Leishmania donovani over an early temporal window. Infection and Immunity 2015, 83, 1853–1868. [Google Scholar] [CrossRef] [PubMed]
- Perea-Martínez, A.; García-Hernández, R.; Manzano, J.I.; Gamarro, F. Transcriptomic Analysis in Human Macrophages Infected with Therapeutic Failure Clinical Isolates of Leishmania infantum. ACS Infectious Diseases 2022, 8, 800–810. [Google Scholar] [CrossRef]
- Stamper, B.D.; Davis, M.; Scott-Collins, S.; Tran, J.; Ton, C.; Simidyan, A.; Roberts, S.C. Model-based Evaluation of Gene Expression Changes in Response to Leishmania Infection. Gene Regulation and Systems Biology 2019, 13, 1–8. [Google Scholar] [CrossRef]
- Ty, M.C.; Loke, P.; Alberola, J.; Rodriguez-Cortes, A.; Rodriguez-Cortes, A. Immuno-metabolic profile of human macrophages after Leishmania and Trypanosoma cruzi infection. PLoS ONE 2019, 14, 1–12. [Google Scholar] [CrossRef]
- Ontology, C.T.G.; Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; et al. Gene Ontology: tool for the unification of biology. Nature genetics 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, S.; Yamabe, M.; Yamaguchi, Y.; Kobayashi, Y.; Konno, T.; Tada, K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int J Cancer. 1980; 26:171–176. DOI: 10.1002/ijc.2910260208. International Journal of Cancer 1980, 176, 171–176. [Google Scholar] [CrossRef]
- Roy, G.; Dumas, C.; Sereno, D.; Wu, Y.; Singh, A.K.; Tremblay, M.J.; Ouellette, M.; Olivier, M.; Papadopoulou, B. Episomal and stable expression of the luciferase reporter gene for quantifying Leishmania spp. infections in macrophages and in animal models. Molecular and Biochemical Parasitology 2000, 110, 195–206. [Google Scholar] [CrossRef]
- Seifert, K.; Escobar, P.; Croft, S.L. In vitro activity of anti-leishmanial drugs against Leishmania donovani is host cell dependent. Journal of Antimicrobial Chemotherapy 2010, 65, 508–511. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nature Protocols 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




