Submitted:
24 August 2024
Posted:
26 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
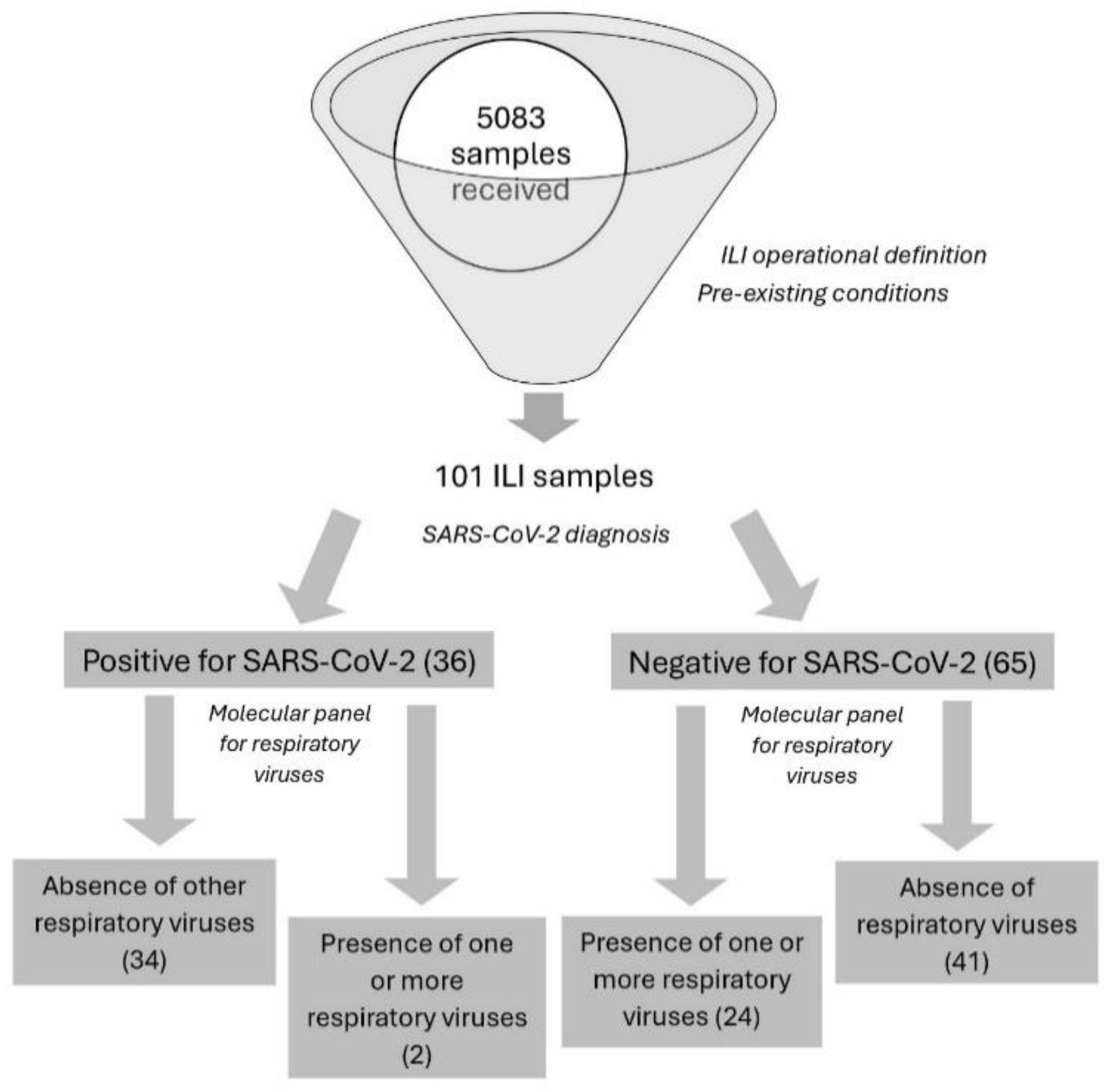
2.1. Sample Reception
2.2. Selection Criteria
2.3. Total RNA/DNA Extraction and RT-qPCR for Respiratory Viruses
3. Data Analysis
4. Results
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Declaration of conflict of interest
References
- Charlton CL, Babady E, Ginocchio CC, Hatchette TF, Jerris RC, Li Y, et al. Practical guidance for clinical microbiology laboratories: Viruses causing acute respiratory tract infections. Clin Microbiol Rev 2019;32:e00042-18. [CrossRef]
- Fernandes-Matano L, Monroy-Muñoz IE, Angeles-Martínez J, Sarquiz-Martinez B, Palomec-Nava ID, Pardavé-Alejandre HD, et al. Prevalence of non-influenza respiratory viruses in acute respiratory infection cases in Mexico. PLOS ONE 2017;12:e0176298. [CrossRef]
- Sánchez DCM, Domínguez PAP, Ira E. Vigilancia Y ANÁLISIS del Riesgo en SALUD PÚBLICA PROTOCOLO DE VIGILANCIA. EN Salud Publ INFECCIÓN Respir AGUDA (IRA) 2017.
- Stockwell MS, Reed C, Vargas CY, Wang L, Alba LR, Jia H, et al. Five-year community surveillance study for acute respiratory infections using text messaging: Findings from the MoSAIC study. Clin Infect Dis 2022;75:987–95. [CrossRef]
- Bello-Lemus Y, Anaya-Romero M, Gómez-Montoya J, Árquez M, González-Torres HJ, Navarro-Quiroz E, et al. Comparative analysis of in-house RT-qPCR detection of SARS-CoV-2 for resource-constrained settings. Diagnostics 2022;12:2883. [CrossRef]
- Guia para la Vigilancia por Laboratorio de Virus Respiratorios.pdf n.d.
- Respiratory viruses identified from hospitalized patients in an institution of higher complexity. Interdisciplinary Journal of Epidemiology and Public Health n.d. Available online: https://revistas.unilibre.edu.co/index.php/iJEPH/article/view/5056 (accessed on 31 January 2023).
- Suryadevara M, Domachowske JB. Epidemiology and seasonality of childhood respiratory syncytial virus infections in the tropics. Viruses 2021;13:696. [CrossRef]
- Darniot M, Pitoiset C, Millière L, Aho-Glélé LS, Florentin E, Bour J-B, et al. Different meteorological parameters influence metapneumovirus and respiratory syncytial virus activity. J Clin Virol 2018;104:77–82. [CrossRef]
- Instituto Nacional de Salud. Infeccion respiratoria aguda PE IX 2021. Available online: https://www.minsalud.gov.co/salud/Paginas/Infecciones-Respiratorias-Agudas-(IRA).aspx (accessed on 31 January 2023).
- Instituto Nacional de Salud. Infeccion respiratoria aguda PE IX 2021. Available online: https://www.minsalud.gov.co/salud/Paginas/Infecciones-Respiratorias-Agudas-(IRA).aspx (accessed on 31 January 2023).
- Instituto Nacional de Salud. Infeccion respiratoria aguda PE IX 2021. Available online: https://www.minsalud.gov.co/salud/Paginas/Infecciones-Respiratorias-Agudas-(IRA).aspx (accessed on 31 January 2023).
- Sánchez DC, Fuentes SMA Informe DE evento infeccion respiratoria AGUDA, Colombia 2020 2019.
- PortalSivigila. Indicadores COVID-19. Available online: http://portalsivigila.ins.gov.co/ (accessed on 31 January 2023).
- Londono-Avendano MA, Peláez-Moreno M, López Medina E, Moreno Turriago MS, Parra Patiño B. Transmission of respiratory syncytial virus genotypes in Cali, Colombia. Influenza Other Respir Viruses 2021;15:521–8. [CrossRef]
- Cruz JS, de Souza Luna LK, Alves VRG, Conte DD, Bellei NC. Viral load of respiratory syncytial virus among children from primary care and hospital settings admitted to a university hospital in Brazil (2009–2013). J Med Virol 2021;93:3397–400. [CrossRef]
- Jain S, Williams DJ, Arnold SR, Ampofo K, Bramley AM, Reed C, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med 2015;372:835–45. [CrossRef]
- Benavides Osorio J Etiología viral y factores de riesgo asociados a las infecciones respiratorias agudas en niños menores de 5 años ingresados al hospital Universidad del Norte entre 2016 Y 2017. Master’s thesis. Universidad del Norte.
- Co-infección viral respiratoria en niños hospitalizados por infección respiratoria aguda y su impacto en la gravedad clínica. Rev Chil Infect 2012; 29 (2): 169-174. [CrossRef]
- García Corzo JR, Niederbacher Velasquez J, González Rugéles CI, Rodríguez Villamizar LA, Machuca Pérez M, Torres Prieto A, et al. Etiología viral de infección respiratoria aguda en niños menores de 5 años en las provincias comunera y García Rovira de santander. Revista de la Universidad Industrial de Santander Salud 2016;48:240–5. [CrossRef]
- Le-Corre N, Pérez R, Vizcaya C, Martínez-Valdebenito C, López T, Monge M, et al. Relevance of codetection of respiratory viruses in the severity of acute respiratory infection in hospitalized children. Andes Pediatr 2021;92:349–58. [CrossRef]
- Chauhan JC, Slamon NB. The impact of multiple viral respiratory infections on outcomes for critically ill children. Pediatr Crit Care Med 2017;18:e333–8. [CrossRef]
- Rehder KJ, Wilson EA, Zimmerman KO, Cunningham CK, Turner DA. Detection of multiple respiratory viruses associated with mortality and severity of illness in children. Pediatr Crit Care Med 2015;16:e201–6. [CrossRef]
- Asner SA, Science ME, Tran D, Smieja M, Merglen A, Mertz D. Clinical disease severity of respiratory viral co-infection versus single viral infection: A systematic review and meta-analysis. PLOS ONE 2014;9:e99392. [CrossRef]
- Scotta MC, Chakr VC, de Moura A, Becker RG, de Souza APD, Jones MH, et al. Respiratory viral coinfection and disease severity in children: A systematic review and meta-analysis. J Clin Virol 2016;80:45–56. [CrossRef]
- Meskill SD, O’Bryant SC. Respiratory virus co-infection in acute respiratory infections in children. Curr Infect Dis Rep 2020;22:3. [CrossRef]
- PAHO. Influenza regional Update EW 38, 2021 /actualización regional de Influenza SE 38 de 2021. Available online: https://www.paho.org/es/documentos/actualizacion-regional-influenza-semana-epidemiologica-38-octubre-6-2021 (accessed on 31 January 2023).
| Allplex™ Respiratory Panel 1 | Allplex™ Respiratory Panel 2 | Allplex™ Respiratory Panel 3 |
|---|---|---|
| Influenza A (Flu A) | Human adenovirus (AdV) | Bocavirus 1/2/3/4 (BoV) |
| Influenza B (Flu B) | Human metapneumovirus (hMPV) | Human rhinovirus A/B/C (HRV) |
| Respiratory Syncytial Virus A (RSV A) | Human enterovirus (hEV) | Coronavirus 229E |
| Respiratory Syncytial Virus B (RSV B) | Parainfluenza 1 (PIV-1) | Coronavirus NL63 |
| Influenza A-H1N1 (Flu A-H1pdm09) | Parainfluenza 2 (PIV-2) | Coronavirus OC43 |
| Parainfluenza 3 (PIV-3) | ||
| Parainfluenza 4 (PIV-4) |
| Variables | POSITIVE SARS-CoV-2 DIAGNOSIS | |||||||
|---|---|---|---|---|---|---|---|---|
| No hospitalization | Hospitalization | Intensive care unit admission | Deaths | |||||
| n | % | n | % | n | % | n | % | |
| Sex | ||||||||
| Male | 6 | 5.94% | 1 | 0.99% | 1 | 0.99% | - | - |
| Female | 13 | 12.87% | 11 | 10.89% | 2 | 1.98% | 2 | 1.98% |
| Age (years) | ||||||||
| 0 - 5 | 1 | 0.99% | - | - | - | - | - | - |
| 6-11 | - | - | - | - | - | - | - | - |
| 12-18 | 1 | 0.99% | - | - | - | - | - | - |
| 19 - 27 | 8 | 7.92% | - | - | - | - | - | - |
| 28 - 59 | 7 | 6.93% | 3 | 2.97% | 2 | 1.98% | 2 | 1.98% |
| 60 - 80 | 2 | 1.98% | 8 | 7.92% | 1 | 0.99% | - | - |
| >80 | - | - | 1 | 0.99% | - | - | - | - |
| Comorbidities | ||||||||
| Autoimmune disease | - | - | - | - | - | - | - | - |
| Diabetes | - | - | - | - | - | - | - | - |
| Immunosuppressed individuals | 1 | 0.99% | - | - | - | - | - | - |
| Digestive system diseases | 1 | 0.99% | - | - | - | - | - | - |
| Cardiovascular system diseases | - | - | 2 | 1.98% | - | - | - | - |
| Nervous system diseases | - | - | - | - | - | - | - | - |
| Respiratory system diseases | 1 | 0.99% | 1 | 0.99% | 1 | 0.99% | 1 | 0.99% |
| Urinary system diseases | - | - | - | - | - | - | - | - |
| Various comorbidities | 1 | 0.99% | - | - | 1 | 0.99% | 1 | 0.99% |
| Human Immunodeficiency virus | - | - | - | - | - | - | - | - |
| None | 15 | 14.85% | 9 | 8.91% | 1 | 0.99% | - | - |
| Molecular diagnosis (respiratory panels) | ||||||||
| Negative | 17 | 16.83% | 12 | 11.88% | 3 | 2.97% | 2 | 1.98% |
| Positive | 2 | 1.98% | - | - | - | - | - | - |
| Variables | No hospitalization | Hospitalization | Intensive care unit admission | |||
|---|---|---|---|---|---|---|
| n | % | N | % | n | % | |
| Sex | ||||||
| Male | 13 | 20.0% | 12 | 18.5% | 12 | 18.5% |
| Female | 8 | 12.3% | 11 | 16.9% | 9 | 13.8% |
| Age groups (years) | ||||||
| 0 - 5 | 5 | 7.7% | 11 | 16.9% | 10 | 15.4% |
| 6 - 11 | 6 | 9.2% | 1 | 1.5% | 1 | 1.5% |
| 12 - 18 | 3 | 4.6% | 1 | 1.5% | 1 | 1.5% |
| 19 - 27 | 1 | 1.5% | 1 | 1.5% | 2 | 3.1% |
| 28 - 59 | 3 | 4.6% | 4 | 6.2% | 5 | 7.7% |
| 60 - 80 | 2 | 3.1% | 3 | 4.6% | 2 | 3.1% |
| >80 | 1 | 1.5% | 2 | 3.1% | - | - |
| Comorbidities | ||||||
| Autoimmune disease | - | - | - | - | 1 | 1.5% |
| Diabetes | 2 | 3.1% | - | - | - | - |
| Immunosuppressed individuals | 1 | 1.5% | 1 | 1.5% | 1 | 1.5% |
| Cardiovascular system diseases | 2 | 3.1% | 1 | 1.5% | ||
| Nervous system diseases | - | - | 1 | 1.5% | 1 | 1.5% |
| Respiratory system diseases | 1 | 1.5% | 3 | 4.6% | 3 | 4.6% |
| Urinary system diseases | - | - | - | - | 1 | 1.5% |
| Human Immunodeficiency virus | - | - | 1 | 1.5% | - | - |
| Various comorbidities | - | - | 3 | 4.6% | 1 | 1.5% |
| Autoimmune disease | - | - | - | - | 1 | 1.5% |
| None | 15 | 23.1% | 14 | 21.5% | 12 | 18.5% |
| Molecular diagnosis (respiratory panels) | ||||||
| Negative | 19 | 29.2% | 9 | 13.8% | 13 | 20.0% |
| Positive | 2 | 3.1% | 14 | 21.5% | 8 | 12.3% |
| Etiological agent | n | % |
|---|---|---|
| RSV A | 4 | 15.38% |
| RSV B | 4 | 15.38% |
| PIV 2 | 1 | 3.85% |
| PIV 3 | 2 | 7.69% |
| HRV | 4 | 15.38% |
| > 1 respiratory virus | 11 | 42.31% |
| AdV, HEV, HRV | 1 | 3.85% |
| RSV A, AdV, HEV | 1 | 3.85% |
| RSV B, HRV | 1 | 3.85% |
| PIV 2, HRV | 1 | 3.85% |
| PIV 4, MPV | 4 | 15.38% |
| PIV 4, MPV, HRV | 1 | 3.85% |
| HRV, Coronavirus subtype 229E, Coronavirus subtype NL63 | 1 | 3.85% |
| HRV, Coronavirus subtype OC43 | 1 | 3.85% |
| Total | 26 | |
| Note: All abbreviations referred in the table stand for the viruses studied as follows, RSV: Respiratory Syncytial Virus; PIV: parainfluenza virus; HRV: human rhinovirus; AdV: Adenovirus; HEV: Human enterovirus; MPV: human metapneumovirus | ||
| Etiological agent | SARS-CoV-2 negative | SARS-CoV-2 positive | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0-5 | 6-11 | 19-27 | 28-59 | 60-80 | >80 | Total | 19-27 | 28-59 | Total | |||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | |||
| RSV A | 3 | 11.54 | - | - | 1 | 3.85 | - | - | - | - | - | - | 4 | - | - | - | - | |
| RSV B | 3 | 11.54 | 1 | 3.85 | - | - | - | - | - | - | - | - | 4 | - | - | - | - | |
| PIV 2 | 1 | 3.85 | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | - | |
| PIV 3 | 1 | 3.85 | - | - | - | - | - | - | - | - | 1 | 3.85 | 2 | - | - | - | - | |
| HRV | 1 | 3.85 | - | - | 1 | 3.85 | - | - | 1 | 3.85 | - | - | 3 | - | - | 1 | 3.85 | 1 |
| >1 respiratory virus | 6 | 23.08 | - | - | - | - | 3 | 11.54 | 1 | 3.85 | - | - | 10 | - | - | - | - | |
| AdV, HEV, HRV | 1 | 3.85 | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | - | |
| RSV A, AdV, HEV | 1 | 3.85 | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | - | |
| RSV B, HRV | - | - | - | - | - | - | 1 | 3.85 | - | - | - | - | 1 | - | - | - | - | |
| PIV 2, HRV | 1 | 3.85 | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | - | |
| PIV 4, MPV | 2 | 7.69 | - | - | - | - | 1 | 3.85 | 1 | 3.85 | - | - | 4 | - | - | - | - | |
| PIV 4, MPV, HRV | 1 | 3.85 | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | - | |
| HRV, Coronavirus subtype 229E, Coronavirus subtype 229E | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 3.85 | - | - | 1 |
| HRV, Coronavirus subtype OC43 | - | - | - | - | - | 1 | 3.85 | - | - | - | - | 1 | - | - | - | - | ||
| 24 | 2 | |||||||||||||||||
| Note: All abbreviations referred in the table stand for the viruses studied as follows, RSV: Respiratory Syncytial Virus; PIV: parainfluenza virus; HRV: human rhinovirus; AdV: Adenovirus; HEV: Human enterovirus; MPV: human metapneumovirus | ||||||||||||||||||
| Comorbidity/ etiological agent | No hospitalization | General hospitalization | Intensive care unit admission | Total | |||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| Immunosuppressed | 1 | ||||||
| RSV A | - | - | - | - | 1 | 3.8 | |
| Nervous system diseases | 1 | ||||||
| PIV 2 | - | - | 1 | 3.8 | - | - | |
| Respiratory system diseases | 4 | ||||||
| PIV 4, MPV | - | - | - | - | 1 | 3.8 | |
| HRV, Coronavirus subtype OC43 | - | - | - | - | 1 | 3.8 | |
| RSV B | - | - | 1 | 3.8 | - | - | |
| HRV | - | - | 1 | 3.8 | - | - | |
| Urinary system diseases | 1 | ||||||
| HRV | - | - | - | - | 1 | 3.8 | |
| Various comorbidities | 2 | ||||||
| PIV 4, MPV | - | - | 1 | 3.8 | - | - | |
| PIV 3 | - | - | 1 | 3.8 | - | - | |
| None | 4 | 15.4 | 9 | 34.6 | 4 | 15.4 | 17 |
| 24 | |||||||
| Note: All abbreviations referred in the table stand for the viruses studied as follows, RSV: Respiratory Syncytial Virus; PIV: parainfluenza virus; HRV: human rhinovirus; AdV: Adenovirus; HEV: Human enterovirus; MPV: human metapneumovirus | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





