1. Introduction
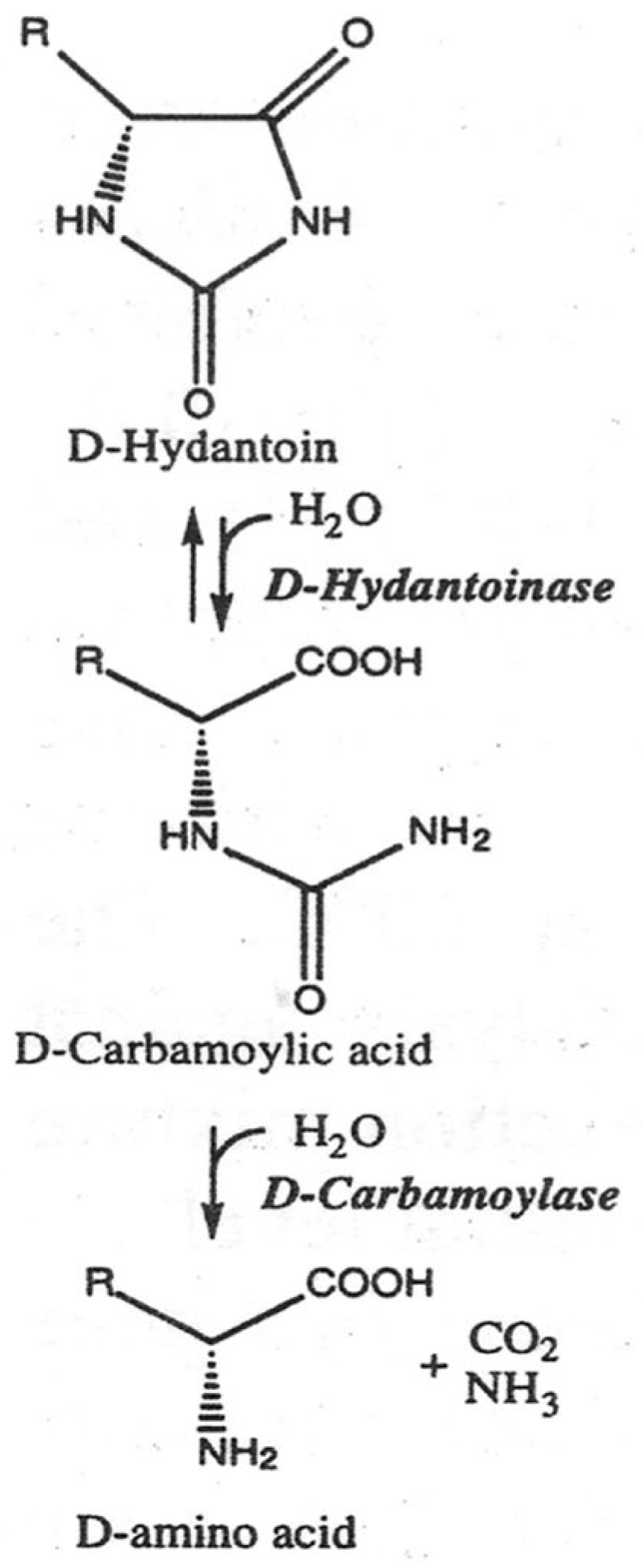
The production of optically pure D-amino acids and their derivatives by enantioselective ring opening of hydantoins and subsequent addition to a decarbamoylated substrate is known as the “hydantoinase process” (
Figure 1) [
1,
2]. This advantageous biotechnological process is based on the use of the enzymes D-hydantoinase and D-carbamoylase, where D-cabamoylase is the rate-limiting enzyme in a two-step reaction system [
3,
4]. D-hydantoinase (EC 3.5.2.2) catalyzes the stereospecific cleavage of chemically synthesized 5-substituted hydantoin to form the substrate for D-carbamoyl amino acid. D-carbamoylase (N-carbamoyl-D-amino acid amidohydrolase) (EC 3.5.1.77) hydrolyzes the resulting D-carbamoyl amino acid to form the desired D-amino acid.
The first mentions of eukaryotic organisms exhibiting the ability to break down hydantoin date back to 1926 [
6] and this information was then used to study the enzymes involved in the catalytic process [
7,
8]. The first mention of bacterial D-carbamoylase from
Agrobacterium was noted in the synthesis of D-amino acids in 1979 [
9]. Then D-carbamoylases were described in other microorganisms, including
Arthrobacter sp. [
10],
Pseudomonas sp. [
11],
Blastobacter sp. [
12] and
Comamonas sp. [
13]. Subsequent studies allowed the cloning and expression of the foreign bacterial D-carbamoylase gene and the characterization of the overexpressed protein in
E. coli cells [
10,
12,
14,
15]. The possibility of enantioselective racemization of hydantoin derivatives using two enzymes naturally aroused applied interest in the biotechnological production of optically pure D-amino acids [
16].
Biochemical studies have shown that D-carbamoylase can have different structures: homodimers or homotetramers with subunits of 32-40 kDa [
4] and homotrimers with subunits of 40 kDa [
13]. The optimal pH for D-carbamoylase enzymatic activity ranges from 7.0 to 9.0 [
13,
15]. Enzyme activity is inhibited by a number of metal ions; however, the enzyme is reactivated by reducing agents, and chelating agents do not affect its activity, suggesting that cations do not play a significant role in enzyme activity [
17,
18]. Using random and site-directed mutagenesis of D-carbamoylase from the mesophilic bacterium
Agrobacterium sp. KNK712, thermostable mutants up to 71.4°C were obtained [
19,
20]. These data indicated the possibility of modulating the catalytic functions of bacterial D-carbamoylase to adapt it to the technological process.
Crystal structures of wild-type and mutant D-carbamoylases were generated for the protein from
Nitratireductor indicus [
21],
Agrobacterium sp. KNK712 [
22] and
A. radiobacter [
23], which are available in data banks. The conserved amino acid sequence of Cys-Glu-Lys indicates its critical role in the catalytic activity of the protein in bacteria. It is assumed that the reaction mechanism of the Cys-Glu-Lys triad occurs through the process of acylation-deacylation [
22,
23].
The search for carbamolyase activity in thermostable bacteria allowed us to detect L-carbamoylase in a thermostable bacterium
Bacillus stearothermophilus, but the enzyme did not meet the requirements for the production of D-amino acids [
24]. However, we later also cloned and studied in detail the gene encoding D-hydantoinase from the thermophilic bacterium
Geobacillus stearothermophilus as a candidate enzyme for the first step of the “hydantoinase process” [
25,
26].
In this study, we show that the D-carbamoylase gene of Pseudomonas sp. strain KNK003A synthesizes an enzyme with activity toward bulky N-carbamoylated amino acids in E. coli host cells. Structural analysis of the enzyme’s three-dimensional structure provides important information useful for production of the desired D-amino acid from D-hydantoins.
2. Materials and Methods
2.1. Reagents and Substrates
Amino acid substrates N-carbamoyl-D-alanine, N-carbamoyl-D-leucine, N-carbamoyl-D-valine, N-carbamoyl-D-phenylalanine were synthesized in the Scientific Technological Centre of Organic and Pharmaceutical Chemistry. The description of the N-carbamoyl-D-amino acids we synthesized is presented in Supplementary
Figures S1–S4. Melting points were determined on a hot stage Boetius PHMK 76/0904 (GDR). The IR spectra were recorded on a Thermo Nicolet NEXUS FTIR spectrometer (USA) in thin films (Nujol). 1H and 13C NMR spectra were recorded on a Varian Mercury-300 VX spectrometer (USA) in DMSO-d6/CCl4, 1:3. Chemical shifts are reported in δ values (ppm) relative to tetramethylsilane as internal standard. Coupling constants (
J values) are given in Hertz (Hz). The signals are reported as follows: s (singlet), d (doublet), dd (double doublet), spt (septet), m (multiplet), br (broad). All chemicals used were of analytical or reagent grade. Carbamoyl derivatives were prepared using the D-amino acids from Reanal (Budapest, Hungary). To confirm the structure of the synthesized molecules,
1H and
13C NMR analyzes were carried out (
Figure S1-S4). Other N-carbamoyl derivatives of amino acids were purchased from Sigma-Aldrich (St. Louis, MO, USA). All chemicals were of analytical grade.
Gibson Assembly® Cloning Kit was purchased from New England Biolabs (Hitchin, UK); ROTI®PolTaqSMix was purchased from Carl Roth GmbH + Co. KG (Karlsruhe, Germany). GeneJET PCR Purification Kit was purchased from Thermo Scientific™ (Waltham, Massachusetts, USA), protein Ni-NTA Purification System K95001 from Thermo Fisher scientific (Altrincham, UK) and Prestained Protein Ladder 10-180 kDa from Thermo ScientificTM PageRulerTM (Boston, Massachusetts, USA). Protein-Markers 10-310 kDa (ROTI®Mark TRICOLOR) was purchased from Carl Roth (Karlsruhe, Germany). DNA ladder 1 kb was purchased from Solis BioDyne (Tartu, Estonia). Isopropyl β-D-1-thiogalactopyranoside (IPTG) and N-lauroylsarcosine sodium salt were provided by Sigma-Aldrich (St. Louis, MO, USA).
2.2. Cloning of D-Carbamoylase Gene
Pseudomonas sp. strain KNK003A is characterized by stable N-carbamoyl-D-amino acid amidohydrolase at 45°C [
27]. We amplified the gene coding D-carbamoylase of this strain (GenBank: BAD00008) PCR using optimized for DNA cloning N-terminal primer F-5′GGAGATATACAT
ATGACCCGTATTGTTAACGCTGC3′ and C-terminal primer R-5′GTGGTGGTGCTCGAG
CTGCGGCGGCGGCACC3′ carrying His-tag sequence. Region of homology with the D-carbamoylase gene of
Pseudomonas sp. strain KNK003A is underlined in both primers used. DNA amplification was carried out by PCR [
28] with initial denaturation at 95°C for 2 min, 30 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s, and elongation at 72°C for 30 s. The purified PCR product was cloned according to protocol [
29] into the expression vector pET24a(+), cleaved at the
NdeI and
XhoI restriction sites. The constructed plasmid pET24a(+)-
PsDcase was transferred into
E. coli strain BL21 Star by selecting kanamycin-resistant colonies on Luria-Bertani (LB) agar. The presence of the recombinant plasmid was confirmed using PCR analysis of transformed colonies (
Figure S5) [
30]. The recombinant
E. coli BL21 Star/pET24a(+)-
PsDcase cells were grown until OD
600 0.5, and D-carbamoylase synthesis was induced with 0.5-1 mM IPTG at 30°C followed by incubation for a further 6-20 h.
2.3. Purification of Recombinant D-Carbamoylase
Cells were collected by centrifugation at 8000 g for 20 min, suspended in 100 mM phosphate buffer (pH 7.0) containing 1 mM DTT, and sonicated on ice for 5 min with 30 s on/off cycles at 25 kHz (Cole-Parmer® 500-Watt Ultrasonic Processor). The lysate was cleared by centrifugation at 35,000 g for 30 min at 4°C and additionally filtered through a syringe filter with a pore size of 0.45 μm. The resulting supernatant, corresponding to the soluble protein fraction of the crude extract, was analyzed by SDS-PAGE and further used to measure D-carbamoylase activity. Polyhistidine-labeled D-carbamoylase was purified using the Ni-NTA purification system and tested for enzymatic activity.
2.4. D-Carbamoylase Activity Assay
D-carbamolase activity was measured in the soluble fraction of the crude extract of purified protein in 100 mM phosphate buffer (pH 7.0) containing 1 mM DTT at 45°C. Reactions were started by adding 0.1 ml of the appropriate substrate consisting of 100 mM N-carbamoyl-D-amino acid or 200 mM N-carbamoyl-DL-amino acid in 100 mM phosphate buffer (pH 7.0), after preincubating the reaction solutions at 45°C for 10 minutes. After 15-120 min incubation, the reaction was stopped by adding 30% trichloroacetic acid (TCA) to a final concentration of 3% w/v. D-carbamoylase activity was determined using the orthophthalaldehyde (OPA) reagent [
31]. Specifically, 3 mL of freshly prepared OPA reagent (0.1 M borate buffer, pH 9.6, 2.5 mM OPA, and 2.5 mM β-mercaptoethanol) was added to each sample, followed by incubation at 20°C for 30 min. D-amino acid concentrations were determined spectrophotometrically (Thermo Scientific Genesys 50 UV-vis spectrophotometer) based on the absorbance of the OPA reagent at 340 nm. Enzyme activity was expressed in units (U), with 1 U defined as the conversion of 1 μmol of substrate per minute. The specific activity of the enzyme was expressed in U/mg protein. All measurements were performed in at least three separate experiments with duplicates.
2.5. Solving of Inclusion Bodies
Significant amounts of D-carbamoylase were extracted from inclusion bodies (IBs) by modified method as described by Peternel et al. [
32]. The insoluble protein fraction IBs of the crude extract was separated by centrifugation at 10,000×g for 20 min at 4°C. To dissolve the insoluble fraction, 80 μL of 0.2% N-lauroylsarcosine sodium salt solution prepared in 100 mM phosphate buffer (pH 7.0) was added to each 1 mg of wet sediment. The resulting mixture was incubated for 24 hours at 20°C with shaking at 100 rpm and centrifuged for 15 minutes at 4400×g. The resulting supernatant, corresponding to the solubilized target protein, was analyzed by SDS-PAGE and tested for D-carbamoylase activity.
2.6. Protein Assays
D-carbamoylase expression levels were analyzed by SDS-PAGE and the gels were stained with Coomassie Brilliant Blue R-250 to visualize protein bands. Cell-free extract was prepared by sonicating the cells and, after centrifugation, the supernatant was collected as the soluble fraction. The pellet remaining from this sample was resuspended in 0.5 ml of a buffer solution consisting of 50 mM Tris-Cl (pH 7.2), 5% glycerol, 5% SDS and β-mercaptoethanol. The suspended solution was centrifuged again and the supernatant was considered the insoluble fraction. Protein concentration was determined using the described methods of Groves and Davis, and Bradford [
33].
The UniProt database was used to screen purified D-carbamoylase against available D-carbamylases downloaded from GeneBank. The alignment tools T-coffee [
34] and MView [
35] were used to align D-carbamoylase sequences. Homology modeling was performed using the SWISS-MODEL web server [
36].
2.7. Molecular Docking
Ligand structures were generated using ChemBioDraw Ultra 12.0 or downloaded from the PubChem database. Energy minimization of ligand structures was carried out in OpenBabel [
37] using the MMFF94 force field. For molecular docking, we used the Gnina 1.0 program, which uses a blind docking approach [
38]. Discovery Studio 2021 software was used to visualize protein-ligand interactions.
2.8. Molecular Dynamics
Molecular dynamics (MD) simulations were performed using the GROMACS 2023 simulation package [
39]. The ligand molecule topology files were generated by the online server Swissparam [
40]. The water chamber was created at a distance of at least 10 Å from the complex using the Tip3p water model and applying boundary conditions. To neutralize the system, Na
+ and Cl
- ions were added to the system. MD was conducted to verify the quality of the model structure by testing its stability through 100 ps simulations at a constant temperature of 300 K (NVT - constant Number of particles (N), Volume (V), and Temperature (T)). Then NPT (constant Number of particles (N), Pressure (P) and Temperature (T) optimization was carried out for 100 ps. MD was performed for 100 ns using a Charm27 force field. The gmx MMPBSA tool, based on AMBER’s MMPBSA.py [
41], was used to perform the final free energy calculations.
3. Results
3.1. Gene Cloning, D-Carbamoylase Expression and Purification
The carbamoylase gene of
Pseudomonas sp. strain KNK003A was amplified by PCR, and the purified PCR product was cloned into the expression vector pET24a(+) to obtain the recombinant plasmid pET24a(+)-
PsDcase. This plasmid was transferred into
E. coli BL21 Star cells and the presence of the recombinant plasmid was confirmed by direct colony PCR analysis [
30].
To overexpress D-carbamoylase, individual recombinant colonies of
E. coli BL21 Star/pET24a(+)-
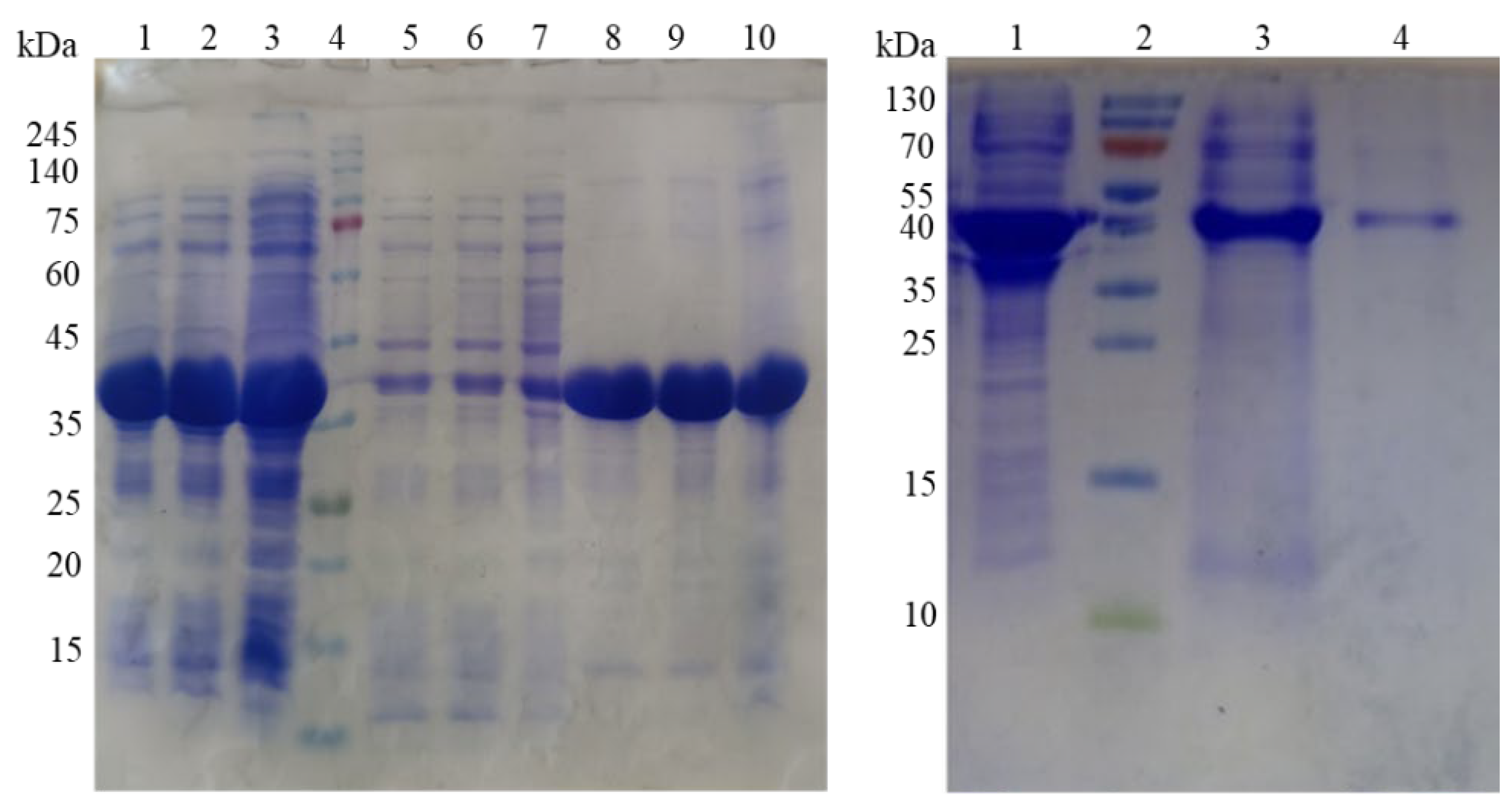
PsDcase cells were induced with 0.75 mM IPTG at 30°C, followed by incubation for 6, 8, and 20 h. The centrifuged cells were disrupted by sonication, the crude extract was separated by centrifugation, and the supernatant and precipitated fractions were subjected to SDS-PAGE analysis, which showed a high expression of the target protein (
Figure 2A). However, only a small portion of the D-carbamoylase produced was present in the soluble fraction, while a significant portion remained trapped in the IBs.
Protein aggregation occurs regardless of the bacterial species [
42,
43]. In bacteria in which recombinant proteins are not secreted into the medium, aggregation of recombinant proteins leads to the formation of IBs [
44]. Over the past decades, several studies have reported that it is often difficult to achieve overexpression of soluble carbamoylase in heterologous bacterial expression systems, especially in
E. coli [
45,
46,
47]. Poorly soluble functional proteins can be extracted from insoluble protein aggregates using mild detergents [
48]. N-lauroylsarcosine sodium is one of the most commonly used agents for the extraction of nondenaturing protein from IBs. We extracted a significant amount of the target protein from IBs using 0.2% N-lauroylsarcosine sodium solution, but the solubilized protein D-carbamoylase had low biological activity.
Another possibility to solve the problem of insoluble protein is to reduce the expression level of the target protein to prevent the formation of insoluble aggregates. The use of low temperatures to prevent the formation of IBs was first demonstrated for human proteins, interferon α2 and human interferon γ [
49]. We induced the production of our D-carbamoylase at a lower temperature, this did not produce the expected result at 25 °C, D-cabamoylase still remained aggregated. The absence of the effect of solubilization of recombinant proteins due to a decrease in cell growth temperature was also observed during the purification of other proteins [
46,
50].
Finally, the inability to obtain soluble and active enzyme using two different approaches was partially overcome by purifying D-carbamoylase using the Ni-NTA protocol PageRuler
TM (Thermo Scientific
TM), which resulted in a fairly pure and active enzyme despite its loss during purification (
Figure 2B).
3.2. Virtual Screening of Possible Carbamoylases
We identified 32 proteins corresponding to the term “D-carbamoylase” in the UniProt database. The study of the properties of proteins suggested that D-carbamoylase of
Ensifer adhaerens S-5 and
Pseudomonas sp. strain KNK003A may be suitable enzymes in terms of compatibility with D-hydantoinase of the thermophilic bacterium
Geobacillus stearothermophilus, which we studied [
26]. In particular, the enzyme’s ability to open bulky N-carbamoylamino acids such as N-carbamoyl-D-phenylalanine, N-carbamoyl-p-hydroxy-D-phenylglycine, and N-carbamoyl-D-phenylglycine was considered an advantage.
Multiple sequence analysis of the mentioned enzymes with
Agrobacterium sp. KNK712 (GenBank: P60327) and
Arthrobacter crystallopoietes DSM 20117 (GenBank: AAO24770.1) carbamoylases revealed highly conserved regions (
Figure S6). High similarity was observed, between aligned four sequences, especially in the regions responsible for the catalytic activity of the enzyme. It is important to note that the catalytic triad Cys-Glu-Lys [
22,
23] is presented as Cys172, Glu47 and Lys127 in
Pseudomonas sp. strain KNK003A D-carbamoylase.
3.3. Molecular Docking of D-Carbamoylase
To understand interactions between ligands and enzyme of interest, we generated a model of
Pseudomonas sp. strain KNK003A D-carbamoylase monomer. Mutant D-carbamoylase from
N. indicus C115 (PDB code: 6le2, resolution 2.14 Å) was used as a template for the homology model of
Pseudomonas sp. strain KNK003A [
27] D-carbamoylase model due to its highest sequence similarity (0.48), Seq Id (60.33) and coverage (0.98) (see
Figure S6).
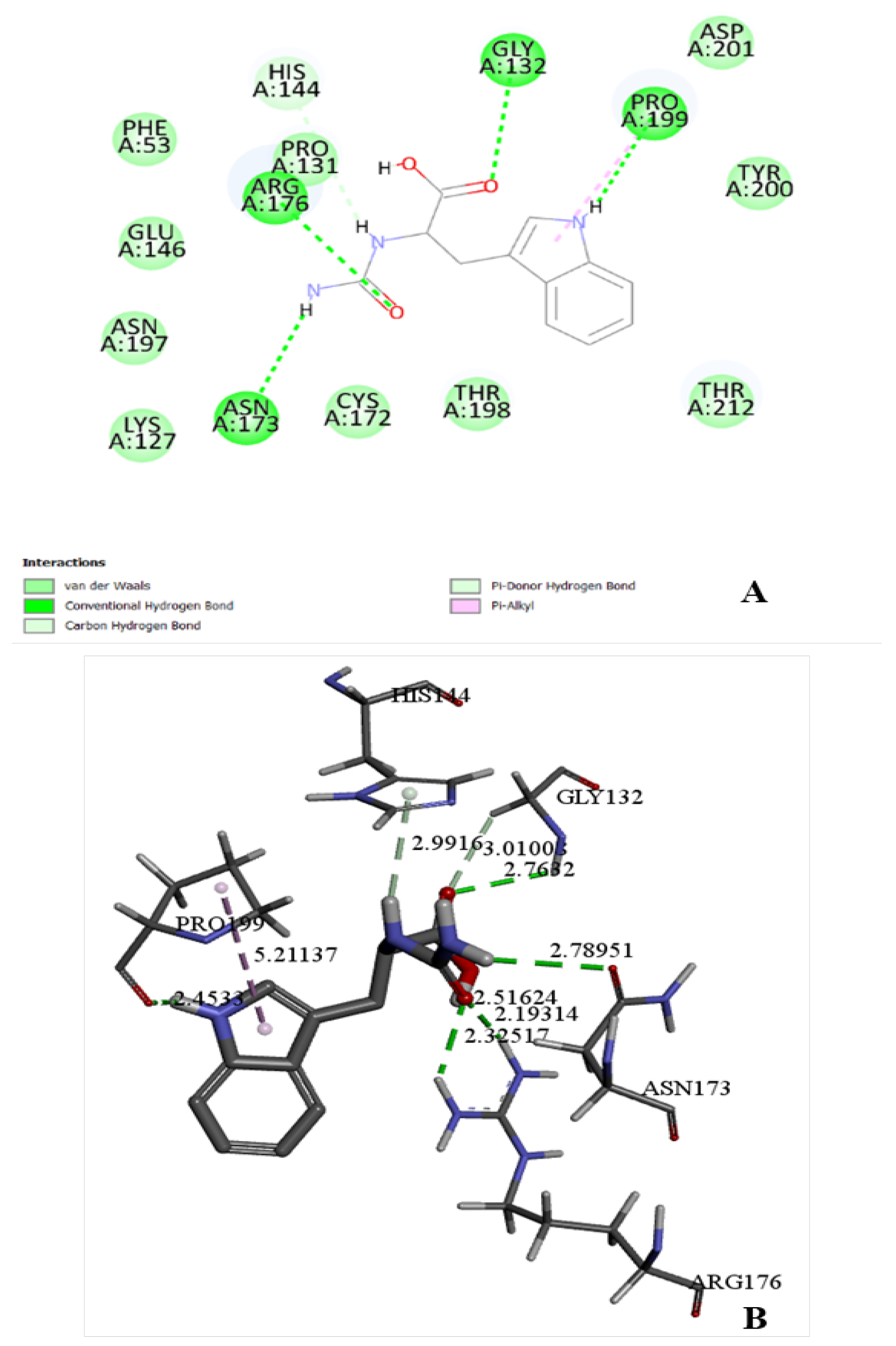
Molecular docking of D-carbamoylase was used to detect the binding pocket of the enzyme and the correct orientation of the ligand in the pocket. The
Pseudomonas sp. strain KNK003A enzyme model suggested the possibility of exhibiting activity towards N-carbamoyl-D-tryptophan (
Figure 3A). To select the correct position of N-carbamoyl-D-tryptophan in the binding pocket, we considered the protein conformation with the lowest binding energy and ligand orientation. From the various positions proposed by the docking algorithm, a position was selected that allows the formation of hydrogen bonds of the amino acids Arg176 and Asn173 with the exposed carbamoyl group of the ligand, necessary for catalysis. The chosen position allows the target group of the ligand to interact closely with the catalytically important amino acids Lys127 and Cys172.
N-carbamoyl-para-hydroxy-D-phenylglycine, N-carbamoyl-D-valine, N-carbamoyl-D-methionine, N-carbamoyl-D-phenylglycine, N-carbamoyl-D-phenylalanine, N-carbamoyl-D-leucine, N-carbamoyl-D-alanine, N-carbamoyl-beta-alanine, and N-carbamoyl-D-glycine are among the ligands that have affinity for D-carbamoylase and can potentially be degraded by the enzyme. The docking image in
Figure 3B shows that D-carbamoylase has an affinity for bulky substrates, indicating that there is enough space in the binding pocket to accommodate relatively large substrates. In addition, given the hydrophobic structure of N-carbamoyl-D-tryptophan and other ligands, the hydrophobic nature of the pocket can be assumed. The binding of N-carbamoyl-D-tryptophan to D-carbamoylase was previously described for D-hydantoinase [
26], which is important for harmonizing the catalytic potential of both enzymes in the “hydantoinase process”. Thus, molecular docking showed that the modeled D-carbamoylase has significant affinity for a wide range of substrates. Further modeling studies were continued using N-carbamoyl-D-tryptophan as a substrate.
3.4. Molecular Dynamics of D-Carbamoylase Binding to Carbamoylated Amino Acids
To confirm the role of putative amino acids in substrate binding and D-carbamolase catalysis, 100 ns molecular dynamics simulations of the interaction of the
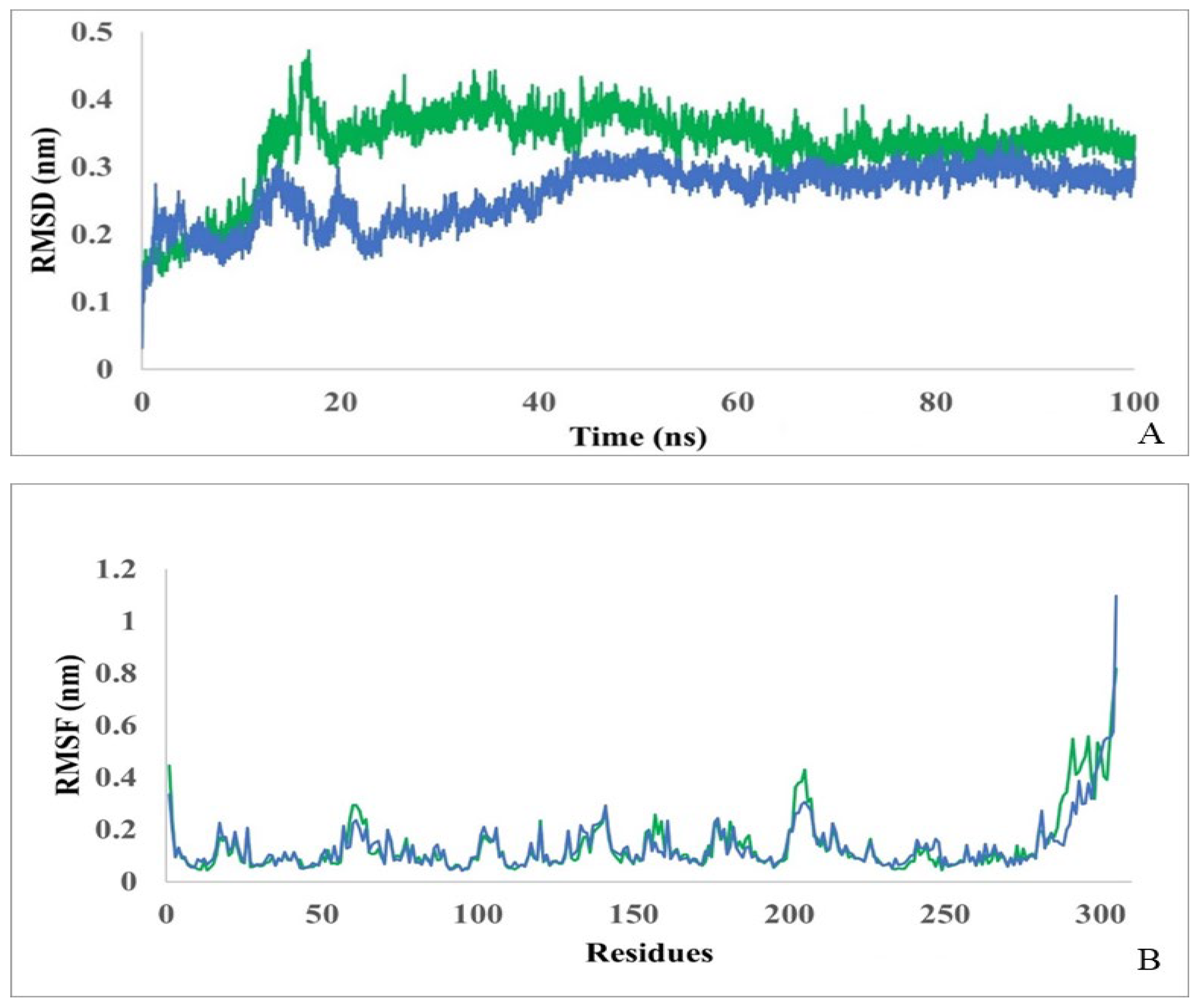
Pseudomonas sp. strain KNK003A enzyme alone and in complex with N-carbamoyl-D-tryptophan were performed. To assess the deviation from the primary structure and stability of the protein and complex, the root mean square deviation (RMSD) was monitored. As shown in
Figure 4A, stabilization of a complex protein-ligand is achieved after ~50 ns. The average RMSD for the protein alone was calculated to be 0.26 nm and for the complex to be 0.33 nm. From the RMSD analysis, it can be concluded that the molecular dynamics trajectories are stable throughout the simulation, are within the acceptable range, and the data can be used for further analysis.
To measure the fluctuations of each amino acid, root mean square fluctuations (RMSF) were also assessed (
Figure 4B). The analysis showed that D-carbamoylase amino acid fluctuations for both models remained within the acceptable range throughout the 100 ns simulation. The average RMSF value for the protein is 0.136±0.02 nm and for complex it is 0.139±0.03 nm. The initial N-terminal and final C-terminal residues in the curves showed higher flexibility compared to the rest of the protein sequence. For amino acids representing the binding cavity, RMSF values were lower than or close to the average RMSF, indicating the rigidity of the active site. Thus, D-carbamoylase RMSF exhibits almost the same flexibility when binding to N-carbamoyl-D-tryptophan, indicating stability of catalytic activity.
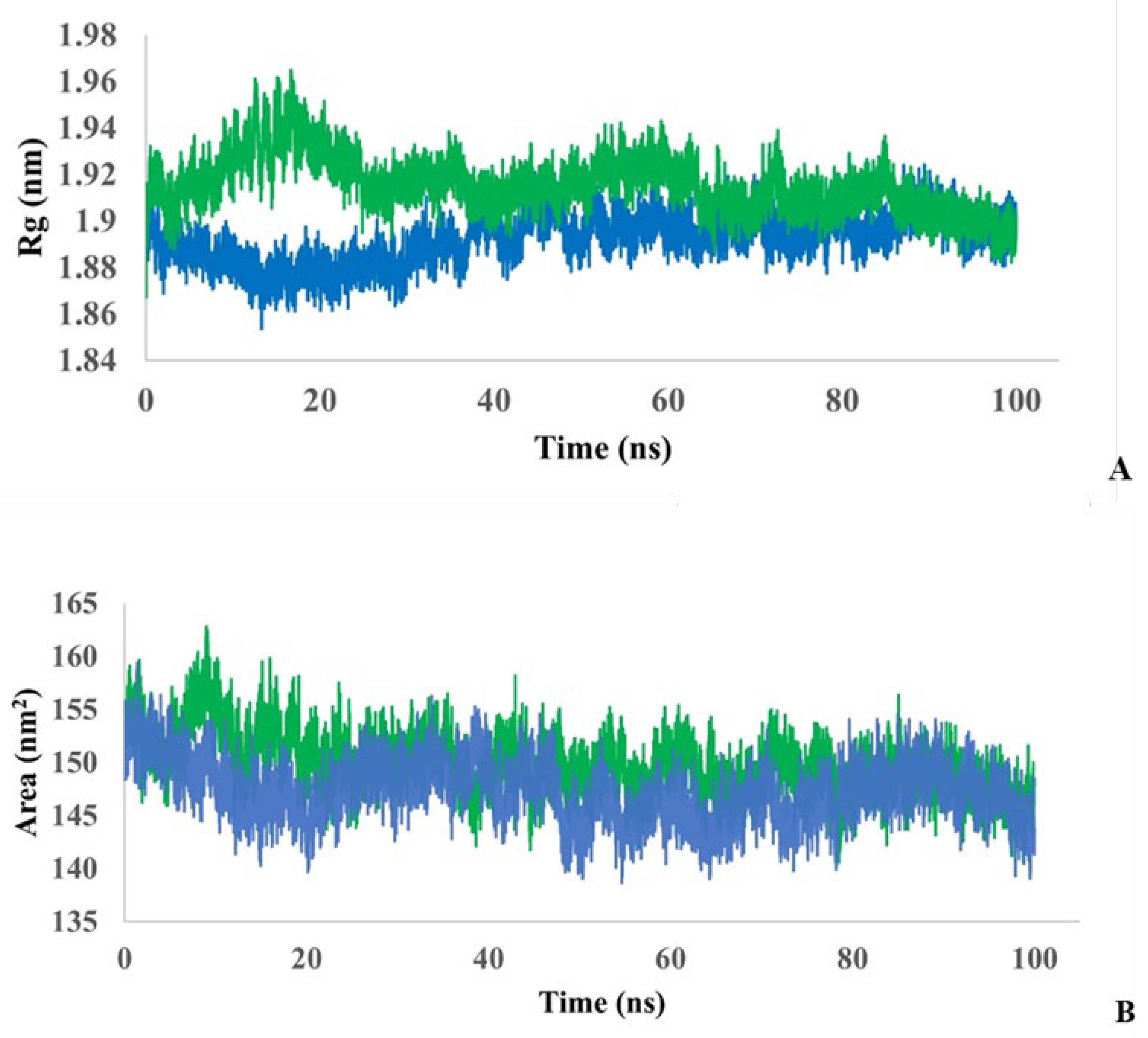
To understand whether the simulated protein stably folds or unfolds over the course of a 100 ns simulation, the radius of gyration (Rg) in the protein and the protein–ligand complex was monitored (
Figure 5A). During the first ~35 ns, the complex is more unfolded compared to the single protein, but then the degree of folding appears to stabilize. The average Rg value for protein is estimated to be 1.89±0.05 nm, for the complex - 1.91±0.05 nm. Thus, both models showed relatively similar folding behavior, but with slightly more flexibility for the protein–ligand complex.
To analyze the interaction of the complex and the protein with the solvent, solvent accessible surface area (SASA) was calculated during 100 ns simulations (
Figure 5B). The average SASA value for a single protein was estimated to be 147.3±1.98 nm
2, and for the protein-ligand complex was estimated to be 149.7±2.04 nm
2. Thus, the SASA values are close for both models.
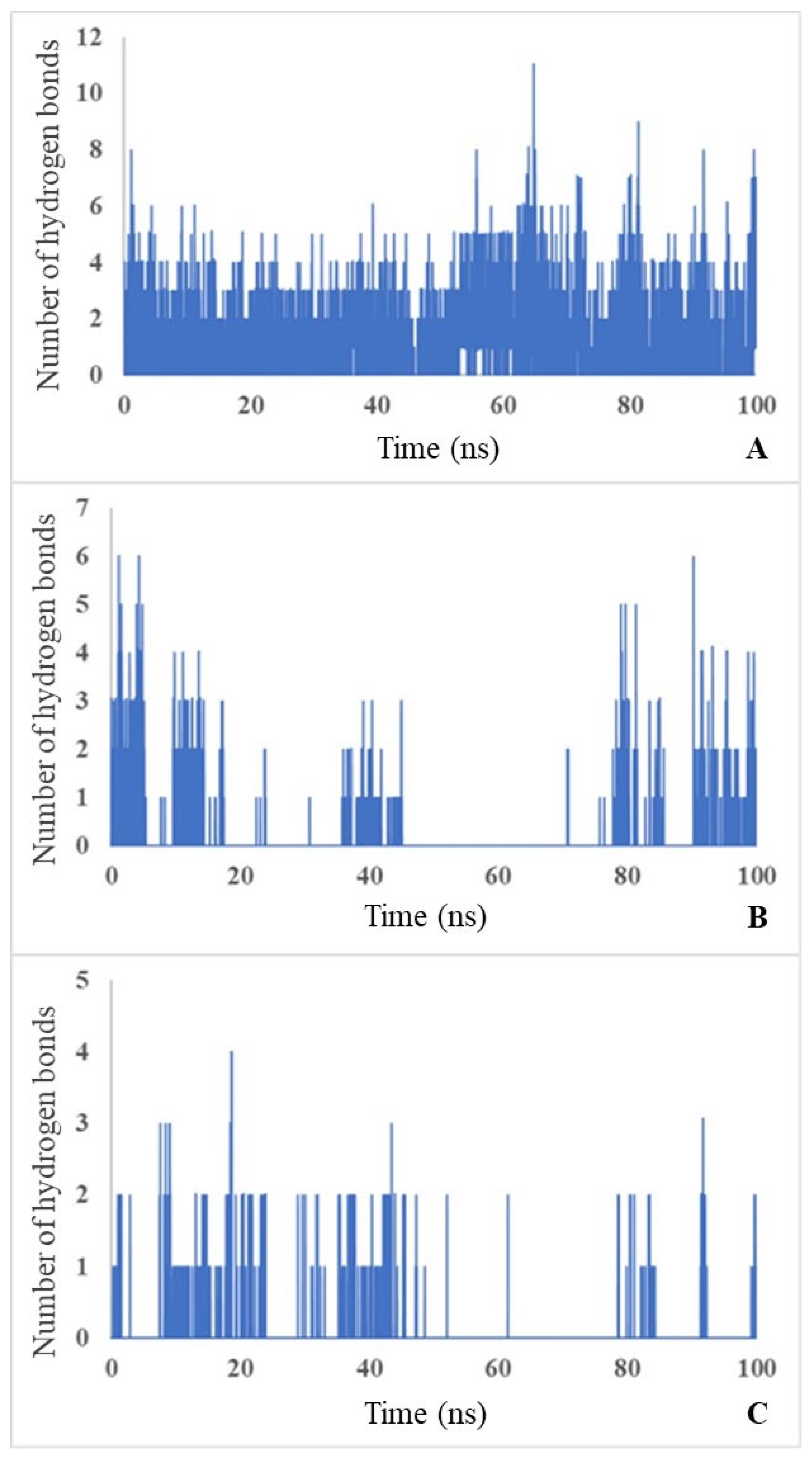
MD simulations showed that carbamoyl-D-tryptophan forms hydrogen bonds with the enzyme D-carbamoylase. Based on the curve of the number of hydrogen bonds over 100-ns simulation time, carbamoyl-D-tryptophan forms up to 11 hydrogen bonds with the protein in 1 ns (
Figure 6A; see the maximum number on the y-axis), 6 hydrogen bonds with Arg176 (
Figure 6B) and 4 hydrogen bonds with Asn173 of the protein in 1 ns (
Figure 6C). The formation of these bonds suggests a stable interaction between the ligand and the protein and confirms the results of molecular docking.
The molecular dynamics of hydrogen bond formation during the interaction of D-carbamoylase with N-carbamoyl-D-tryptophan during 100-ns simulations may be used as an indicator of the appropriate state of the binding site during catalysis.
3.5. Calculation of Molecular Mechanics of Poisson-Boltzmann Surface Area (mmpbsa)
The total binding energy of D-carboxylase with the N-carbamoyl-D-tryptophan complex is estimated to be -13.32±1.57 kcal/mol (see
Supplementary Materials). The large contribution of van der Waals interactions to the total binding energy suggests a hydrophobic contact of the ligand with the receptor.
3.6. D-Carbamoylase Substrate Specificity
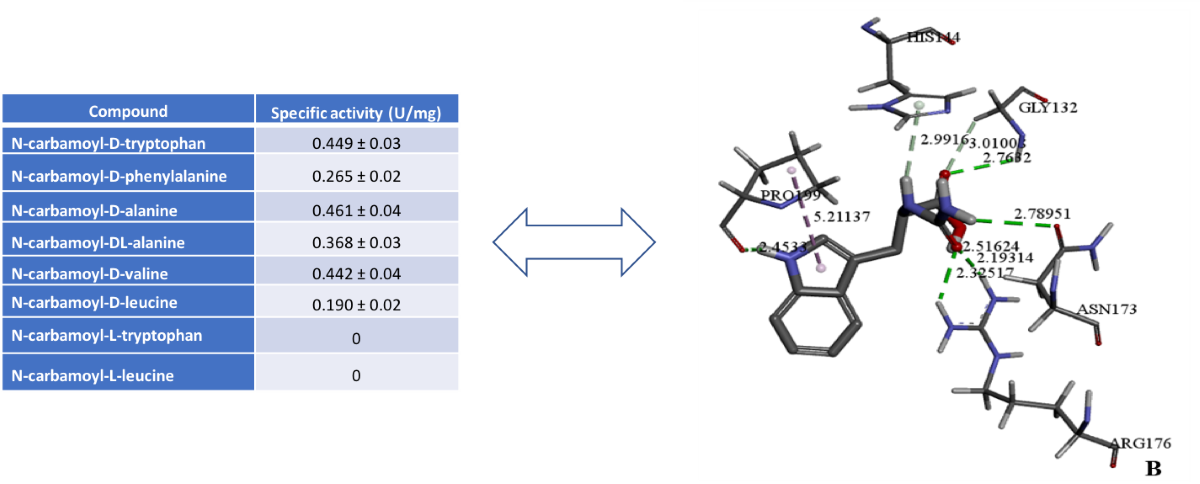
Purified D-carbamoylase was analyzed for enzymatic activity based on the obtained molecular docking data. The highest levels of enzyme activity towards various substrates were detected at 45°C with incubation for 120 min (
Table 1), using IPTG-induced D-carbamoylase incubated at 30°C with shaking for 20 hours.
As expected, D-carbamoylase showed strong specificity for N-carbamoyl-D-amino acids and was not active against the tested N-carbamoyl-L-amino acids. Low specificity was observed for N-carbamoylated DL-amino acids, but is less reflective of the expected two-fold difference between D- and L-amino acids. The enzyme D-carbamoylase showed activity against aliphatic and aromatic N-carbamoylamino acids in vitro, which is in good agreement with in silico data. Additionally, higher activity was observed against bulky substrates such as N-carbamoyl-D-tryptophan, N-carbamoyl-D-phenylalanine, N-carbamoyl-D-valine and N-carbamoyl-D-leucine.
D-carbamoylase belongs to a superfamily of amidohydrolases that catalyze the breakdown of a wide range of N-carbamoyl-D-amino acids. Both in vitro and in silico methods have been used to characterize the structure and behavior of the bacterial enzyme. Molecular docking revealed the affinity of D-carbamoylase for various substrates, and molecular dynamics studies showed stability and constant formation of hydrogen bonds, especially with N-carbamoyl-D-tryptophan, indicating the involvement of the substrate binding site of the enzyme. The total binding energy of the ligand-protein complex is also calculated and the contribution of each energy to this interaction is shown. These data are encouraging indicators for the subsequent production of optically pure D-amino acids.
Our data suggest that the activity of D-carbamoylase can still be increased by site-directed mutagenesis to produce more D-amino acids from the corresponding precursors. Moreover, docking analysis revealed the molecular basis for the increased affinity of D-carbamoylase for bulky N-carbamoyl-D-tryptophan, which may be important for harmonizing its catalytic potential with that of D-hydantoinase in the overall two-enzyme process. Thus, enzymatic and docking analysis give a positive assessment of two bacterial enzymes, D-hydantoinase, described previously [
26], and D-carbamoylase, described in this work, for the implementation of the coupled biotechnological process. D-carbamoylase together with hydantoinase are attractive catalytic enzymes for the biotechnological production of pharmaceuticals.
4. Discussion
Traditional molecular cloning of a gene encoding a desired protein typically requires knowledge of the nucleotide sequence at the beginning and end of the gene. Advances in the chemical synthesis of longer DNA sequences have accelerated the cloning of an entire gene when the desired bacterium is unavailable. Here, we chemically synthesized 1 kb gene encoding D-carbamoylase from Pseudomonas and cloned it into E. coli.
Biochemical analysis established the identity of the D-carbamoylase activity with the originally described enzyme [
27] and also provided additional information on the cleavage spectrum of the new D-carbamoyl derivatives of amino acids. Importantly, the ability of the enzyme to cleave large N-carbamoyl-D-amino acids such as N-carbamoyl-D-tryptophan or N-carbamoyl-D-phenylalanine indicates a real advantage of biotechnological production of D-amino acids over chemical synthesis.
Pseudomonas D-carbamoylase exhibits poor solubility in
E. coli cells, which can be improved to some extent by treatment with sodium N-lauroylsarcosine or by expression of the enzyme in cells grown at 25°C. However, after testing both methods, we preferred the Thermo ScientificTM method using PageRuler
TM to purify the His-tagged protein, which allowed us to obtain sufficiently pure and active D-carbamoylase for further study of the enzymatic activity of the protein. It is important to note that the two-step “hydantoin process” does not include an enzyme purification step.
Detailed virtual analysis of the cloned D-carbamoylase protein provides valuable information about the structural features of the enzyme. Molecular docking revealed the affinity of D-carbamoylase for bulky N-carbamoyl-D-amino acids, which provides sufficient space in the binding pocket to accommodate other bulky substrates. Molecular dynamics of D-carbamoylase binding to N-carbamoyl-D-tryptophan shows rapid, nearly 50 ns, stabilization of the protein-ligand complex. This property is important for the two-step synthesis of large structures of D-tryptophan and other D-amino acids by combining the catalytic potential of D-hydantoinase [
26] and D-carboxylase into a single process.
5. Conclusions
In contrast to chemical synthesis, the two-step biotechnological production of D-amino acids has technological advantages, as the process can be environmentally friendly, recyclable and safe for operating personnel. The enzymes D-hydantoinase and D-carbamoylase, which are involved in various biological processes in bacterial hosts, are promising catalysts for the co-synthesis of D-amino acids and the possible production of pharmaceuticals such as semi-synthetic antibiotics and peptide hormones, as well as pesticides for agronomy. Our in vitro and in vivo data on D-hydantoinase [
25,
26] and D-carbamoylase in the present study predict the possibility of producing pure D-amino acids by the combined action of both enzymes.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1. Figures S1–S4: NMR spectra of the synthesized N-carbamoyl-D-amino acids; Figure S5: Electrophoresis of the PCR-amplified product in colony; Figure S6: Multiple sequence alignment of bacterial D-carbamoylases; Figure S7: Prosa Web z-score for D-carbamoylase model; Calculation of molecular mechanics of Poisson-Boltzmann surface area (mmpbsa).
Author Contributions
Conceptualization, HK, AHo, VS; methodology, HA, AHa, SK; investigation, MP, HK, HA, TS, SA, SK, PH; data curation, MP, HK, HA, AHa; writing - original draft preparation, MP, HK, HA; writing - final draft and editing, V.S. All authors have read and agreed to the published version of the manuscript.
Funding
The research was supported by the Higher Education and Science Committee of Ministry of Education, Science, Culture and Sport (MESCS) of the Republic of Armenia (Research project № 21T-2I289).
Acknowledgments
We express our gratitude to the Committee on Higher Education and Science of the Ministry of Education, Science, Culture and Sports (MESCS).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Syldatk, C.; May, O.; Altenbuchner, J.; Mattes, R.; Siemann, M. Microbial hydantoinases - industrial enzymes from the origin of life? Appl. Microb. Biotechnol. 1999, 51, 293–309. [Google Scholar] [CrossRef] [PubMed]
- Engel, U.; Syldatk, C.; Rudat, J. Stereoselective hydrolysis of aryl-substituted dihydropyrimidines by hydantoinases. Appl. Microb. Biotechnol. 2012, 94, 1221–1231. [Google Scholar] [CrossRef]
- Kim, G. J. and Kim, H. S. Optimization of the enzymatic synthesis of D-p-hydroxyphenylglycine from DL-5-substituted hydantoin using D-hydantoinase and N-carbamoylase. Enzyme Microb. Technol. 1995, 17, 63–67. [Google Scholar] [CrossRef]
- Martínez-Rodríguez, S.; Martínez-Gómez, A.I.; Rodríguez-Vico, F.; Clemente-Jiménez, J.M.; Las Heras-Vázquez, F.J. Carbamoylases: characteristics and applications in biotechnological processes. Appl. Microbiol. Biotechnol. 2010, 85, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Ware, E. The chemistry of the hydantoins. Chem Rev. 1950, 46, 403–470. [Google Scholar] [CrossRef]
- Gaebler, O.H.; Keltch, A.K. On the metabolism of hydantoins and hydantoic acid. J. Biol. Chem. 1926, 70, 763–777. [Google Scholar] [CrossRef]
- Eadie, G.S.; Bernheim, F.; Bernheim, M.L.C. The partial purification and properties of animal and plant hydantoinases. J. Biol. Chem. 1949, 181, 449–458. [Google Scholar] [CrossRef]
- Wallach, D.P.; Grisolia, S. The purification and properties of hydropyrimidine hydrase. J. Biol. Chem. 1957, 226, 277–288. [Google Scholar] [CrossRef]
- Olivieri, R.; Fascetti, E.; Angelini, L.; Degen, L. Enzymatic conversion of N-carbamoyl-D-amino acids to D-amino acids. Enzym. Microb. Tech. 1979, 1, 201–204. [Google Scholar] [CrossRef]
- Möller, A.; Syldatk, *!!! REPLACE !!!*; Schulze, M. Stereo- and substrate-specificity of a D-hydantoinase and D-N-carbamoyl-amino acid amidohydrolase of Arthrobacter crystallopoietes AM2. Enzyme Microb. Technol. 1988, 10, 618–625. [Google Scholar] [CrossRef]
- Yasuhiro, I.; Hirokazu, N.; Yukio, Y.; Kazuyoshi, Y.; Masayuki, T.; Satoni, T. Screening, characterization, and cloning of the gene for N-carbamoyl-D-amino acid amidohydrolase from thermotolerant soil bacteria. Biosci. Biotechnol. Biochem. 1998, 62, 882–886. [Google Scholar]
- Ogawa, J.; Chung, M.C.-M.; Hida, S.; Yamada, H.; Shimizu, S. Thermostable N-carbamoyl-D-amino acid amidohydrolase: screening, purification and characterization. J. Biotechnol. 1994, 38, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, J.; Shimizu, S.; Yamada, H. N-carbamoyl-D-amino acid amidohydrolase from Conamonas sp. E222c purification and characterization. Eur. J. Biochem. 1993, 212, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Grifantini, R.; Pratesi, C.; Galli, G.; Grandi, G. Topological mapping of the cysteine residues of N-carbamyl-D-amino-acid amidohydrolase and their role in enzymatic activity. J. Biol. Chem. 1996, 271, 9326–9331. [Google Scholar] [CrossRef]
- Nanba, H.; Ikenaka, Y.; Yamda, Y.; Yajima, K.; Takano, M.; Takahashi, S. Isolation of Agrobacterium sp. strain KNK712 that produces N-carbamyl-D-amino acid amidohydrolase, cloning of the gene for this enzyme, and properties of the enzyme. Biosci. Biotechnol. Biochem. 1998, 62, 875–881. [Google Scholar] [CrossRef]
- Syldatk, C.; Mueller, R.; Siemann, M.; Wagner, F. Microbial and enzymatic production of D-amino acids from DL-5-monosubstituted hydantoins. In Biocatalytic Production of Amino Acids and Derivatives; Rozzell, J.D., Wagner, F., Eds.; Hanser: New York, NY, USA, 1992; pp. 75–127. [Google Scholar]
- Louwrier, A.; Knowles, C.J. The purification and characterization of a novel D(-)-specific carbamoylase enzyme from an Agrobacterium sp. Enzyme Microb. Technol. 1996, 19, 562–571. [Google Scholar] [CrossRef]
- Nozaki, H.; Kira, I.; Watanabe, K.; Yokozeki, K. Purification and properties of D-hydantoin hydrolase and N-car-bamoyl-D-amino acid amidohydrolase from Flavobacterium sp. AJ11199 and Pasteurella sp. AJ11221. J Mol Catal B. 2005, 32, 205–211. [Google Scholar] [CrossRef]
- Ikenaka, Y.; Nanba, H.; Yajima, K.; Yamada, Y.; Takano, M.; Takahashi, S. Increase in thermostability of N-carbamyl-D-amino acid amidohydrolase on amino acid substitutions. Biosci. Biotechnol. Biochem. 1998, 62, 1668–1671. [Google Scholar] [CrossRef]
- Ikenaka, Y.; Nanba, H.; Yajima, K.; Yamada, Y.; Takano, M.; Takahashi, S. Relationship between an increase in thermostability and amino acid substitutions in N-carbamyl-D-amino acid amidohydrolase. Biosci. Biotechnol. Biochem. 1998, 62, 1672–1675. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, G.; Zhou, J.; Ni, J.; Zhang, L.; Hou, X.; Yin, D.; Rao, Y.; Zhao, Y.-L.; Ni, Y. Structure-guided engineering of D-carbamoylase reveals a key loop at substrate entrance tunnel. ACS Catalysis 2020, 10, 12393–12402. [Google Scholar] [CrossRef]
- Nakai, T.; Hasegawa, T.; Yamashita, E.; Yamamoto, M.; Kumasaka, T.; Ueki, T.; Nanba, H.; Ikenaka, Y.; Takahashi, S.; Sato, M.; Tsukihara, T. Crystal structure of N-carbamyl-d-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure 2000, 8, 729–738. [Google Scholar] [CrossRef]
- Wang, W.-C.; Hsu, W.-H.; Chien, F.-T.; Chen, C.-Y. Crystal structure and site-directed mutagenesis studies of N-carbamoyl-d-amino-acid amidohydrolase from Agrobacterium radiobacter reveals a homotetramer and insight into a catalytic cleft. J. Mol. Biol. 2001, 306, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Batisse, N.; Weigel, P.; Lecoq, M.; Sakanyan, V. Two amino acid amidohydrolase genes, encoding L-stereospecific carbamoylase and aminoacylase, are organized into a common operon in Bacillus stearothermophilus. Appl Environ Microbiol. 1997, 63, 763–766. [Google Scholar] [CrossRef]
- Weigel, P.; Mark, F.; Aganyants, H. A. , Sakanyan V. A. Characteristics of thermostable hydantoinases cloned from Geobacilus stearothermophilus. Reports NAS RA 2013, 113, 92–98. [Google Scholar]
- Aganyants, H.; Weigel, P.; Hovhannisyan, Y.; Lecocq, M.; Koloyan, H.; Hambardzumyan, A.; Hovsepyan, A.; Hallet, J.N.; Sakanyan, V. Rational engineering of the substrate specificity of a thermostable D-hydantoinase (dihydropyrimidinase). High-Throughput 2020, 9, 5. [Google Scholar] [CrossRef]
- Ikenaka, Y.; Nanba, H.; Yamada, Y.; Yajima, K.; Takano, M.; Takahashi, S. Screening, characterization, and cloning of the gene for N-carbamyl-D-amino acid amidohydrolase from thermotolerant soil bacteria. Biosci. Biotechnol. Biochem. 1998, 62, 882–886. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; p. 2344. [Google Scholar]
- Gibson, D.G; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, F.; Pereira, H.; Johansson, B. Colony PCR. Methods Mol. Biol. 2017, 1620, 129–139. [Google Scholar]
- Hanczkó, R.; Molnár-Perl, I. Derivatization, stability and chromatographic behavior of ophthaldialdehyde amino acidе and amine derivatives: o-phthaldialdehyde/2-mercaptoethanol reagent. Chromatographia 2003, 57, S103–S113. [Google Scholar] [CrossRef]
- Peternel, Š.; Grdadolnik, J.; Gaberc-Porekar, V.; Komel, R. Engineering inclusion bodies for non-denaturing extraction of functional proteins. Microb. Cell. Fact. 2008, 7, 34. [Google Scholar] [CrossRef]
- Peterson, G.L. Determination of total protein. Meth. Enzymol. 1983, 91, 95–119. [Google Scholar]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for multiple sequence alignments. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.P.; Leroy, C.; Sander, C. MView: A Web Compatible Database search or multiple alignment viewer. Bioinformatics 1998, 14, 380–381. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; Lepore, R.; Schwede, T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: an open chemical toolbox. J. Cheminformatics 2011, 3, 33. [Google Scholar] [CrossRef]
- McNutt, A.T.; Francoeur, P.; Aggarwal, R.; Masuda, T.; Meli, R.; Ragoza, M.; Sunseri, J.; Koes, D.R. GNINA 1.0: Molecular docking with deep Learning. J. Cheminformatics 2021, 13, 43. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software X 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Bugnon, M.; Goullieux, M.; Röhrig, U.F.; Perez, M.A.S.; Daina, A.; Michielin, O.; Zoete, V. SwissParam 2023: A modern Web-based tool for efficient small molecule parameterization. J. Chem. Inf. Model. 2023, 63, 6469–6475. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory. Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Corchero, J.L.; Gasser, B.; Resina, D.; Smith, W.; Parrilli, E.; Vazquez, F.; et al. Unconventional microbial systems for the cost-efficient production of high-quality protein therapeutics. Biotechnol Adv. 2013, 31, 140–153. [Google Scholar] [CrossRef]
- Koszagova, R.; Nahalka, J. Inclusion bodies in biotechnology. J. Microbiol. Biotechnol. Food Sci. 2020, 9, 1191–1196. [Google Scholar] [CrossRef]
- Rinas, U.; Garcia-Fruitos, E.; Corchero, J.L.; Vazquez, E.; Seras-Franzoso, J.; Villaverde, A. Bacterial inclusion bodies: discovering their better half. Trends Biochem. Sci. 2017, 42, 726–737. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Lin, K.Y.; Lu, C.H. Refolding and activation of recombinant N-carbamoyl-D-amino acid amidohydrolase from Escherichia coli inclusion bodies. Process Biochem. 2005, 40, 2135–2141. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Q.; Hu, X.; Yang, J. Using native hydantoinase promoter to induce D-carbamoylase soluble expression in Escherichia coli. Biochem. Eng. J. 2008, 41, 12–16. [Google Scholar] [CrossRef]
- Martinez-Alonso, M.; Garcia-Fruitos, E.; Ferrer-Miralles, N.; Rinas, U.; Villaverde, A. Side effects of chaperone gene co-expression in recombinant protein production. Microb. Cell Fact. 2010, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Singhvi, P.; Saneja, A.; Srichandan, S.; Panda, A.K. Bacterial inclusion bodies: a treasure trove of bioactive proteins. Trends Biotechnol. 2020, 38, 474–486. [Google Scholar] [CrossRef]
- Schein, C.H.; Noteborn, M.H. Formation of soluble recombinant proteins in Escheuichia coli is favored by lower growth temperature. Nature Biotechnol. 1988, 6, 291–294. [Google Scholar] [CrossRef]
- Chao, Y.-P.; Chiang, C.-J.; Lo, T.E.; Fu, H. Overproduction of D-hydantoinase and carbamoylase in a soluble form in Escherichia coli. Appl. Microbiol. Biotechnol. 2000, 54, 348–353. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).