1. Introduction
Many respiratory diseases affect the respiratory system, such as chronic obstructive pulmonary disease (COPD), asthma, pneumonia, bronchiectasis, bronchiolitis, and upper/lower respiratory tract infections. These diseases are at the top of the list when considering global deaths, emphasizing the importance of accurate and early diagnosis of them. Correct diagnosis and early treatment of respiratory diseases can significantly improve the health status of patients, reduce healthcare costs, and improve quality of life. Among the various diagnostic tools available, analysis of respiratory sounds by auscultation is a basic method for identifying respiratory abnormalities. Respiratory sounds such as roughness, coarse crackling, monophonic wheeze, polyphonic wheeze, stridor, bronchus, and squawk provide valuable clues about the respiratory system. Upper Respiratory Tract Infection (URTI), Chronic Obstructive Pulmonary Disease (COPD), Bronchiectasis, Pneumonia, Bronchiolitis, Asthma, and Lower Respiratory Tract Infection are among the most common respiratory diseases that can be detected by auscultation methods. Traditional auscultation relies heavily on the experience and interpretation capability of the physician, which can lead to variabilities in diagnosis. Digital stethoscopes and advanced signal processing algorithms provide a more objective analysis of respiratory sounds. They also paved the way for the use of automatic decision-making algorithms.
Recent advancements in machine and deep learning algorithms encouraged researchers working on respiratory sound analysis to develop automated classification systems. Studies in this field fall under two main groups. The first group includes the classification of respiratory diseases such as asthma, COPD, etc. The second group focuses on classifying respiratory sounds as crackle, wheeze, etc. Under these main topics, many valuable studies exist in the literature. Shuvo et al. used a lightweight convolutional neural network (CNN) model, which demonstrates significant efficacy in classifying respiratory auscultation sounds [
1]. The model employs a hybrid approach utilizing empirical mode decomposition and continuous wavelet transform, achieving an accuracy of 98.92% in 3-class chronic disease classification and 98.70% in six-class pathological classification. Naqvi and Choudhry presented an automated low-cost diagnostic method for Chronic Obstructive Pulmonary Disease (COPD) and pneumonia, utilizing respiratory sound (LS) analysis from the ICBHI open-access LS database [
2]. The method achieved a classification accuracy of 99.7%. García-Ordás proposed a novel approach utilizing a Variational Convolutional Autoencoder (VAE) combined with a Convolutional Neural Network (CNN) to classify respiratory sounds into healthy, chronic disease, and non-chronic disease categories as well as six specific pathologies. They achieved performance improvements over state-of-the-art methods with a reported F-Score of 0.993 in the ternary classification [
3]. Fraiwan et al. investigated the classification of respiratory diseases using respiratory sound signals achieving an accuracy of 98.27% with boosted decision trees, which outperformed traditional classifiers such as support vector machines [
4]. Pham et al. presented a robust deep-learning framework for the analysis of respiratory anomalies and the detection of respiratory diseases using auscultation recordings. They got an 84% ICHBI score that averages specificity and sensitivity metrics which surpasses the previous state-of-the-art result of 72% [
5]. In another study by Pham et al., an inception-based deep learning model was developed to detect respiratory anomalies and respiratory diseases from audio recordings utilizing the ICBHI benchmark dataset. The model achieved competitive scores of 0.53/0.45 ICHBI score for respiratory anomaly detection and 0.87/0.85 for disease prediction, outperforming several state-of-the-art systems [
6]. Kababulut et al. introduced a clinical decision support system for respiratory disease identification using decision tree algorithms and Shapley-based feature selection to improve performance. Their findings highlight that effective feature selection significantly enhances classification performance in respiratory disease detection [
7]. Sfayyih et al. analyzed deep learning applications in respiratory sound analysis, focusing on the effectiveness of CNNs in classifying respiratory sounds. The authors conclude that deep learning techniques show high accuracy in diagnosing respiratory conditions, underscoring AI's potential in medical diagnostics [
8].
It is widely known that feature extraction has a substantial impact on the efficiency of clinical decision systems. The literature presents numerous diverse feature extraction methods. Classical methods such as Fourier Transform [
9], Empirical Mode Decomposition [
1], Wavelet Transform [
1,
6], and Mel-Frequency Cepstral Coefficients (MFCC) [
10,
11,
12,
13,
14,
15] are among the most commonly used methods for feature extraction in respiratory sound classification field. The proper selection
of the most descriptive feature subsets from all extracted features has also been very important for the success of the classification system. Through feature selection, the computational burden on the classifier is reduced by employing a smaller feature size, and also classification performance is increased. Therefore, finding effective feature selection (FS) methods has been an extensively studied topic. Abdel-Basset et al. discussed the critical role of feature selection in enhancing the performance of machine learning algorithms, particularly in the context of high-dimensional datasets. They categorized feature selection methods into wrapper, filter, and embedded approaches, highlighting that wrapper methods, while computationally intensive, often yield superior subsets of features tailored to specific classifiers [
16]. Kang et al. provide a comprehensive overview of FS techniques, highlighting their significance in managing the challenges posed by high-dimensional datasets [
17]. Iqbal et al. presented a comprehensive approach to feature extraction and selection from physiological signals [
18].
In recent years, the application of nature-inspired metaheuristic algorithms for feature selection has gained significant attention within the machine learning community. Metaheuristic methods range from well-established techniques like genetic algorithm (GA) and particle swarm optimization (PSO) to newer and more creative approaches such as grey wolf optimization (GWO), teaching learning-based optimization (TLO), whale optimization algorithm (WOA), and Equilibrium optimizer (EO). FS with metaheuristic methods was found to be superior to classical techniques in many studies. The comprehensive review conducted by Nssibi et al. evaluated various metaheuristic techniques, highlighting their effectiveness in navigating the complex search space associated with feature selection tasks [
19]. Sathiyabhama et al. introduced a novel computer-aided diagnosis (CAD) system that employs a GWO and Rough Set-based approach to identify abnormalities in mammogram images effectively [
20]. Kang et al. introduced the Two-Stage Teaching-Learning-Based Optimization (TS-TLBO) algorithm, which demonstrates significant improvements in classification accuracy [
17]. Nadimi-Shahraki et al. presented an enhanced version of the Whale Optimization Algorithm (E-WOA) specifically tailored for medical feature selection with a focus on the COVID-19 case study. The experimental results demonstrated that E-WOA significantly outperforms traditional WOA variants and other well-known optimization algorithms [
21]. Chen et al. introduced a novel approach that combines Particle Swarm Optimization (PSO) with the 1-Nearest Neighbor (1-NN) classifier, demonstrating its effectiveness on various life science datasets [
22]. Elgamal et al. introduced an enhanced version of the Harris Hawks Optimization (HHO) algorithm, termed Chaotic Harris Hawks Optimization (CHHO), which integrates chaotic maps and Simulated Annealing (SA) to address the limitations of the standard HHO [
23]. Rajammal et al. presented a Binary Improved Grey Wolf Optimizer (BIGWO) that integrates a mutation operation and an Adaptive k-nearest Neighbour (AkNN) algorithm to enhance feature selection efficacy [
24]. Prabhakar and Won propose several innovative techniques including metaheuristics feature selection methods for classification in telemedicine applications. The study highlights the effectiveness of these methods in analyzing respiratory sounds and showcases the potential for enhanced diagnostic capabilities in healthcare settings [
25]. Abedi et al. developed an innovative algorithm that utilizes GA and support vector machine (SVM) classification to analyze thoracic respiratory effort and oximetric signal features [
26]. Álvarez et al. conducted a comprehensive study for detecting OSA patients. The authors employed GA for feature selection achieving very high diagnostic accuracy [
27]. All of these studies show that metaheuristic feature selection methods have been successful in addressing the difficulties presented especially by high-dimensional data. Classifier models using features selected by metaheuristic methods enhance prediction accuracy, decrease computing costs, and clarify the process by eliminating unimportant features.
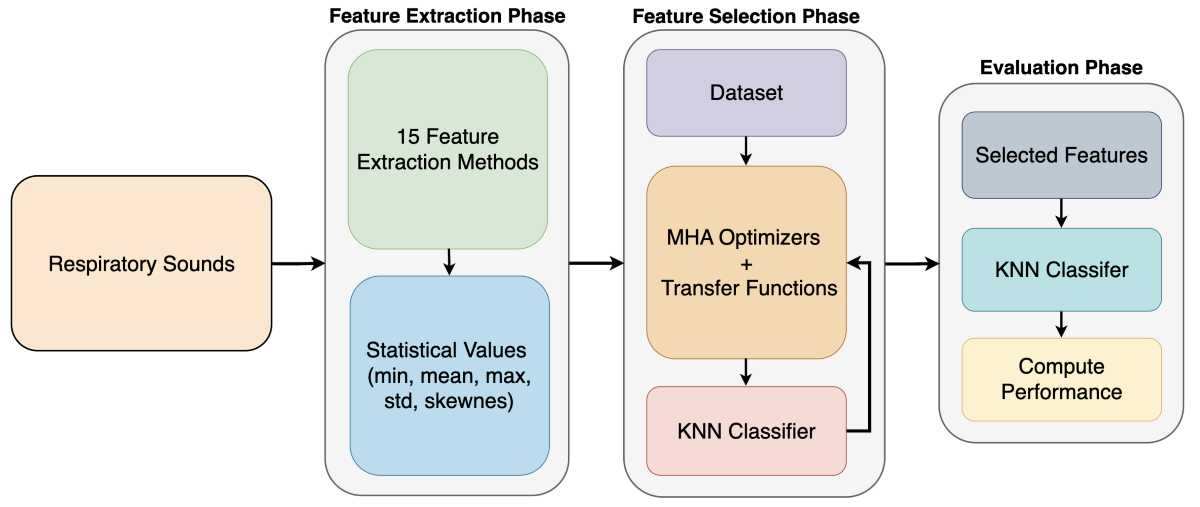
Though successfully used for feature selection in many studies, there is not much research on the utilization of metaheuristic feature selection methods in respiratory disease classification. This study aims to conduct a detailed and comparative analysis of metaheuristic optimization methods in respiratory disease classification. For this purpose, various features were extracted from audio recordings obtained from the publicly available ICBHI 2017 Respiratory Sound Database [
28] using 15 frequently used feature extraction techniques. Then, by employing diverse statistical metrics on the collected numerical data, a new feature set was acquired. Next, to determine the best features that enhance classification performance, six well-known metaheuristics methods were employed with eight transfer functions. Finally, the performances of each method were measured and compared with each other using a simple and identical k-NN classifier. In this study, two different classification problems were examined. The first one is a binary classification task (respiratory disease vs healthy) while the second one is a multi-class task (healthy, chronic respiratory disease, nonchronic respiratory disease). Since the database used is highly imbalanced, F1 and MCC metrics were used as the main performance metrics instead of accuracy. The findings demonstrate that metaheuristic algorithms using correct transfer functions could effectively reduce data dimensionality while enhancing classification accuracy.
This paper is organized as follows.
Section 2 provides information about the materials and methods used, including the feature extraction methods from audio recordings, implementation of metaheuristic feature selection methods and transfer functions, classification stage, and evaluation of the results.
Section 3 conducts a detailed comparative analysis based on the results obtained. Finally,
Section 4 presents the discussion and conclusions of the study.
4. Discussion
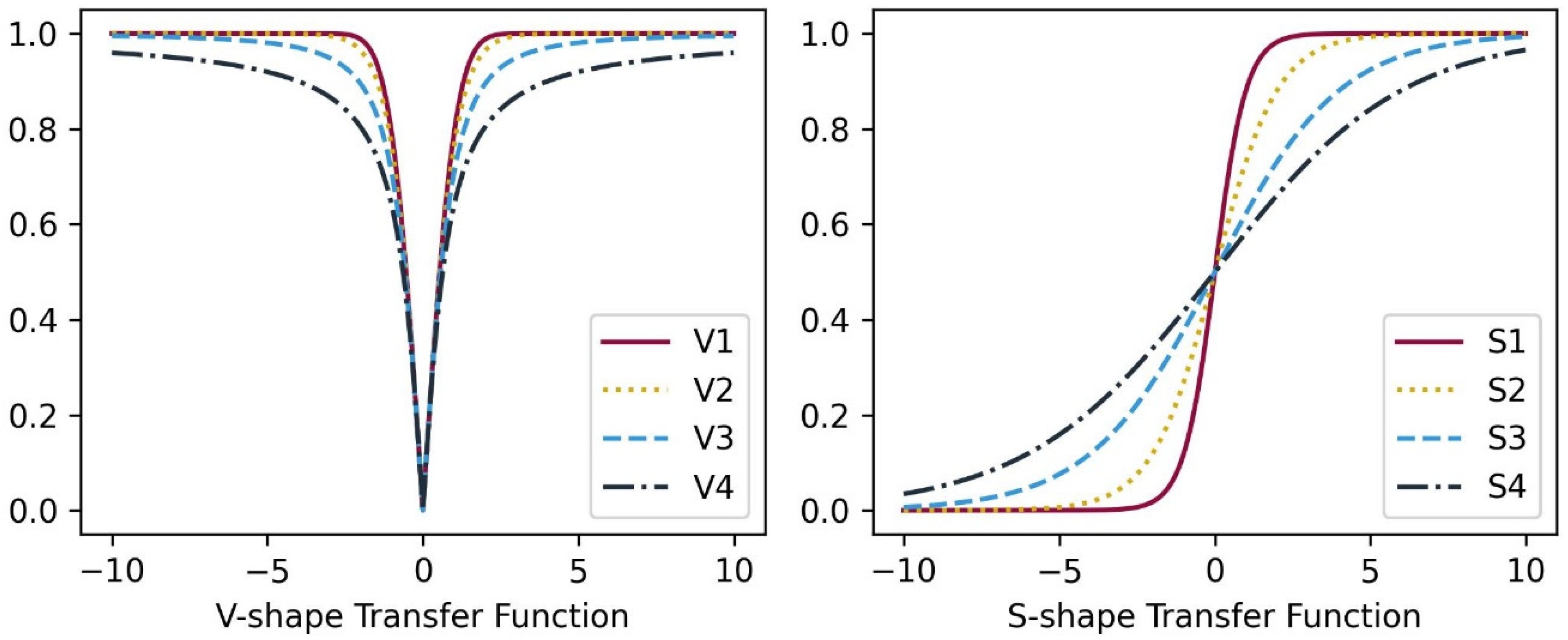
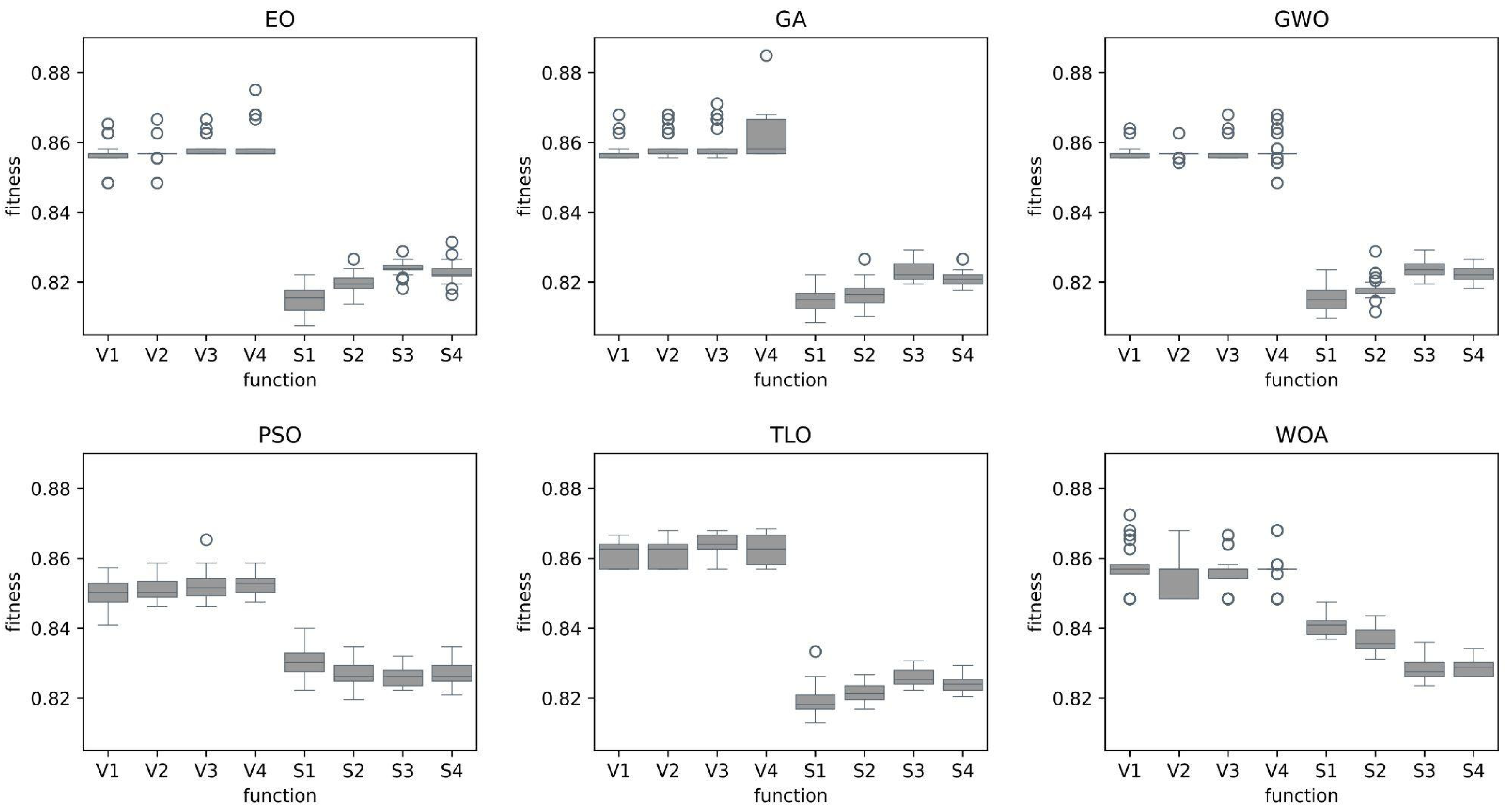
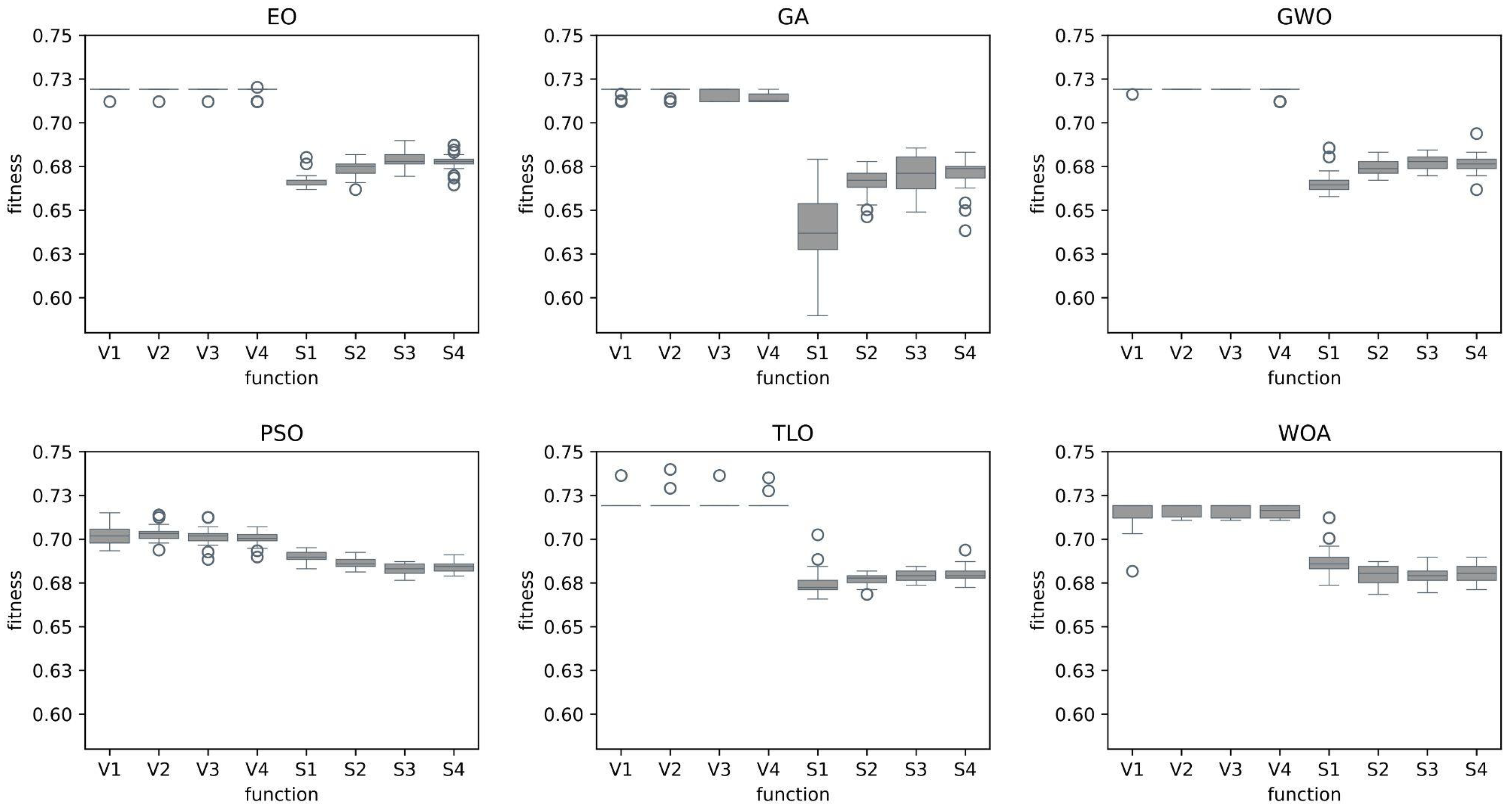
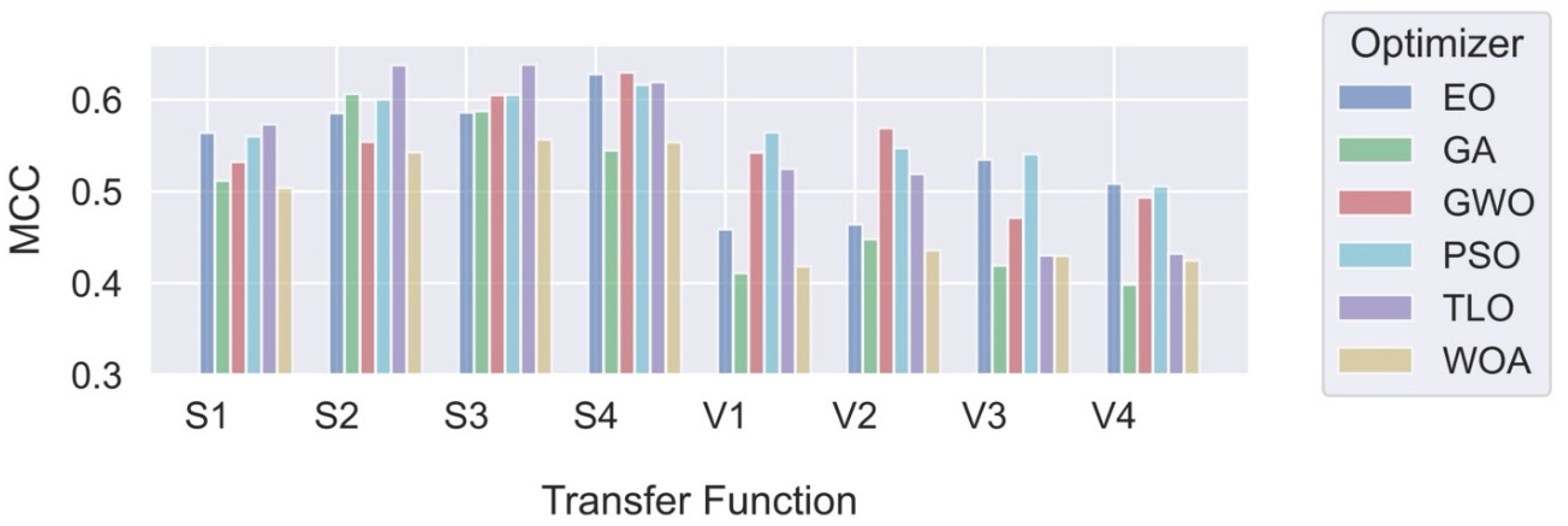
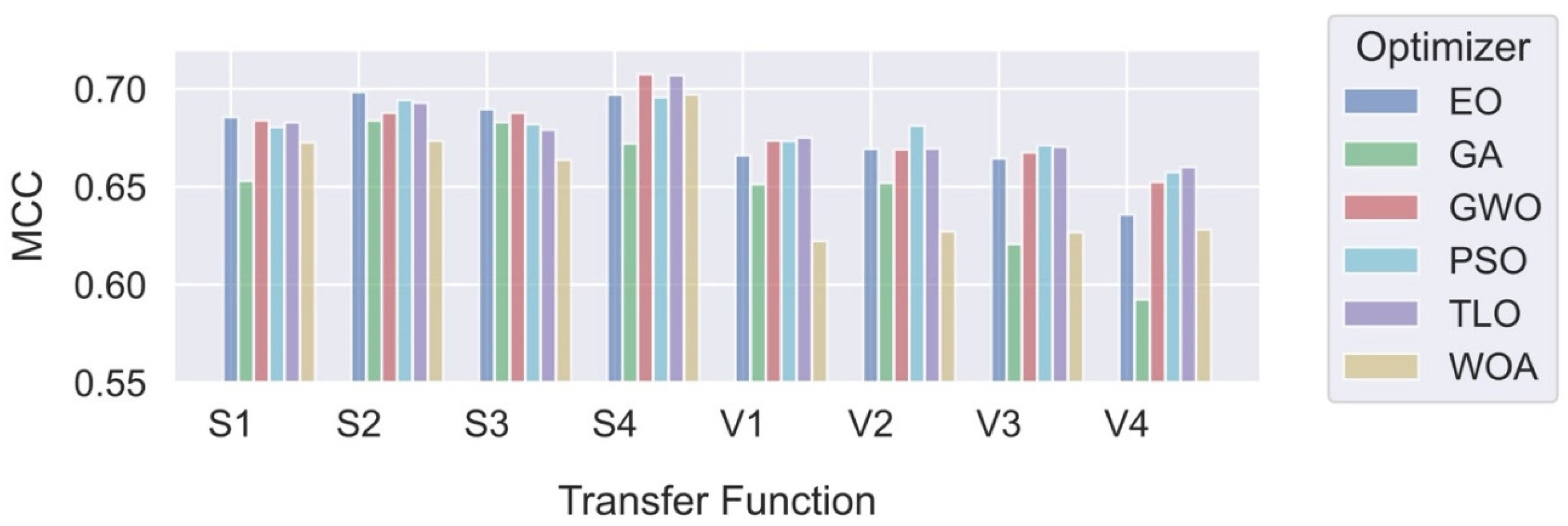
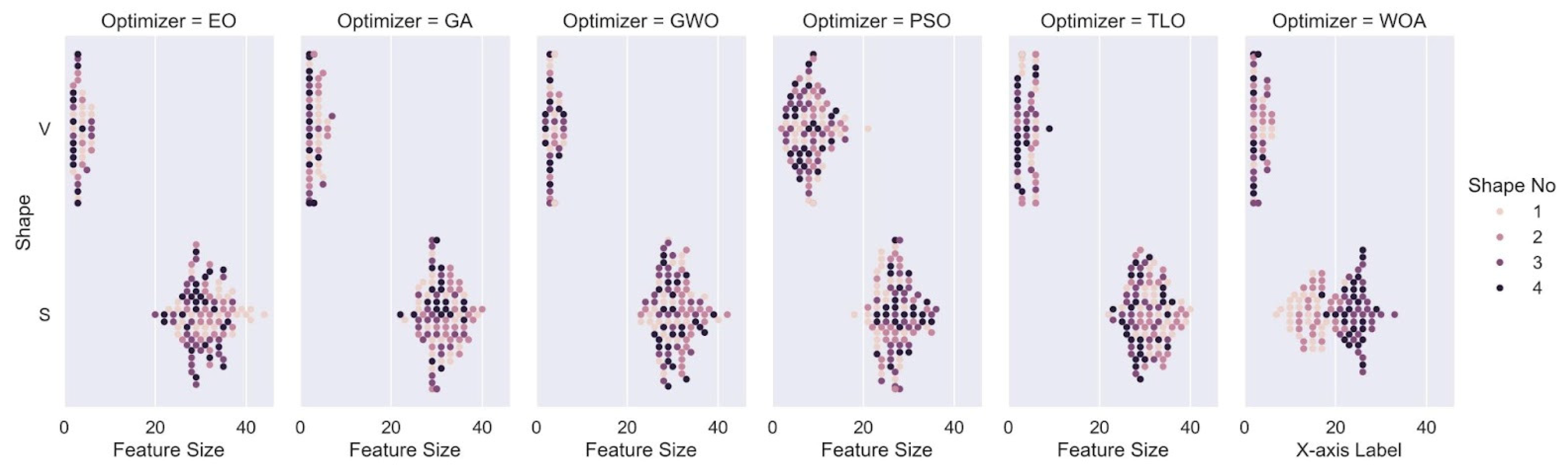
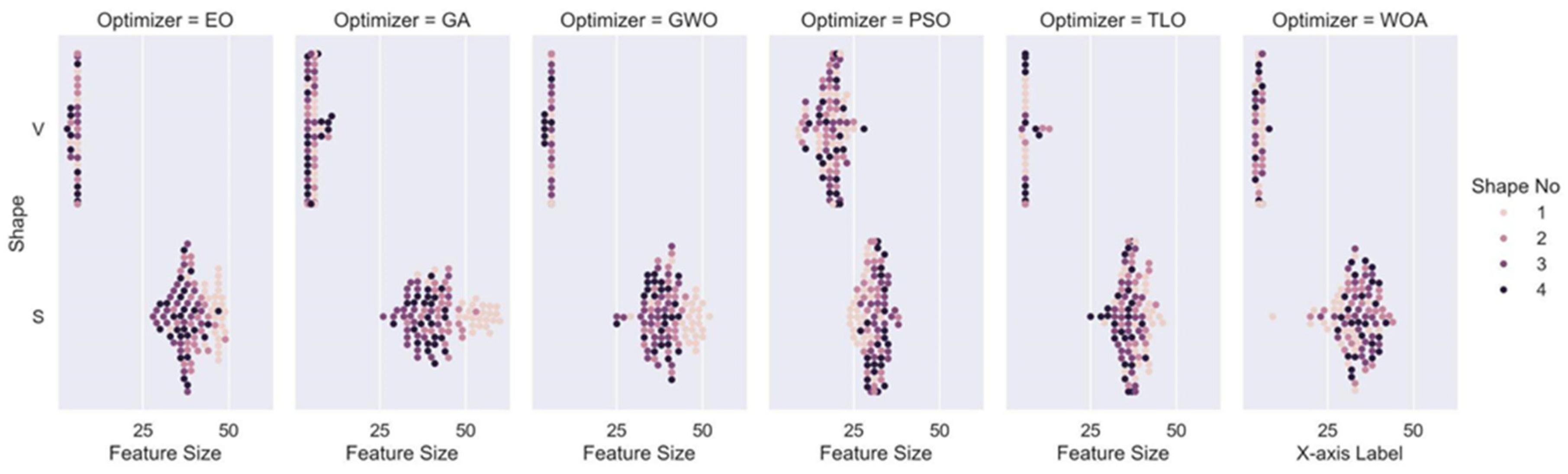
In this study, we explored and compared the performances of nature-inspired metaheuristic algorithms for feature selection in respiratory disease classification. The primary objective is to enhance the classification accuracy while reducing the feature set size, thereby improving the computational efficiency of the model. The experiments were conducted on a dataset consisting of respiratory sound recordings belonging to different diseases. Our approach utilized a variety of metaheuristic algorithms and two different classes of transfer functions to select the most relevant features. We first analyzed the fitness values of eight different transfer functions belonging to two different families for each metaheuristic feature selection algorithm. It is seen that fitness values of V-shaped functions exhibit slightly higher values than S-shaped ones. However, the vertical size of the plots, which represent the dispersion of fitness values in 25 trials (
Figure 6 and
Figure 7), shows that V-shaped functions are generally more stable than S-shaped ones. This leads to the conclusion that fitness values of V-shaped functions do not change much from trial to trial. Then we analyzed the effects of these metaheuristic algorithms and transfer functions on the classification performance. It comes out that MHA methods using S-shaped transfer functions obtain slightly better classification performance than the same methods when they use V-shaped functions. However, using V-shaped transfer functions seems to have two important advantages. First of all, they are much more stable, which means that their fitness values do not change much from trial to trial. Second, the number of features selected by MHA methods when employing V-shaped transfer functions is significantly less than the same methods when they use S-shaped functions.
Finally, we compared the performances of MHA-based feature selection methods with classical methods. Upon analysis of the results, it becomes evident that the proposed MHA-based feature selection methods, with one exception, outperform the classical methods in both binary and multi-class cases. The “Sequential” feature selection method obtained the highest scores in both cases. However, this method uses a very large number of features, increasing the computational burden on the classification system. Optimization methods that achieve the highest MCC scores with S-shaped functions for both cases are GWO and TLO; while the best results with V-valued transfer functions for both cases are obtained with GWO and PSO. Although their performance is slightly less than those using S-shaped functions, the methods used with V-shaped functions achieve this performance by using significantly smaller number of features. Therefore, it seems that if classification performance is the priority, then MHA methods (in particular, GWO and TLO) with S-shaped transfer functions should be the choice. However, if both classification performance and reduced number of features are required, then MHA feature selection methods (in particular, GWO and PSO) using V-shaped transfer functions should be used.
One point needs explanation at this point. The accuracy value is greater than other metrics with classical feature selection methods. This result can lead to the conclusion that metaheuristic methods are worse. However, we want to emphasize that the ACC metric could be misleading in datasets with class imbalance. Therefore, in such datasets, metrics such as MCC or F1 should be used. Our study uses a highly imbalanced dataset. Thus, we used these two metrics for measuring classification performances. This way, we have derived models that can also accurately forecast minority classes. This facilitates the creation of more dependable systems in the field of medical data, particularly in the realm of diagnosing respiratory diseases.
This detailed comparison study shows the superiority of metaheuristic feature selection algorithms in respiratory disease classification compared to classical ones. The use of nature-inspired algorithms provides a robust mechanism for navigating the high-dimensional feature space, ensuring that the selected features contribute maximally to the classification task. Furthermore, the reduction in the number of features not only decreases the computational load but also minimizes the risk of overfitting. This is particularly important in the context of respiratory disease classification, where the diversity and variability of the data can lead to complex decision boundaries. The overall improvement in classification accuracy and computational efficiency underscores the effectiveness of these algorithms in feature selection for respiratory disease classification.
In conclusion, when combined with positive results from the literature for other medical diagnostic applications, it would be correct to say that the integration of nature-inspired metaheuristic algorithms with machine learning models presents a promising avenue for improving the accuracy and efficiency in many classification tasks. Future work could explore the combination of these algorithms with other advanced machine learning techniques, such as ensemble learning, to further enhance the robustness and generalizability of the models.