Introduction Background
Cryo-EM is a technique that involves imaging biological samples at cryogenic temperature using electron beams to produce 3D structures of biological macromolecules. It first rapidly freezes biological samples (e.g., proteins and viruses) to preserve their native state and then images them with an electron microscope to capture numerous 2D projections. These projections are computationally combined to reconstruct a detailed 3D structure of the sample [
1,
2]. Cryo-EM has revolutionized structural biology by allowing researchers to visualize intricate large molecular structures, leading to significant advancements in understanding the functions of proteins, viruses, and their dynamic assemblies.
Over the past decade, cryo-EM technology and data analysis have advanced rapidly. These advancements encompass improvements in microscope hardware, sample preparation methods, image processing techniques, and software algorithms. Notably, the enhancement of optical performance in electron microscopes has enabled reaching atomic resolutions of 1.25 Å, surpassing the previous 1.5 Å barrier. Additionally, innovations in image processing powered by artificial intelligence (AI) and machine learning (ML) (particularly deep learning) have been crucial, advancing the state of the art in cryo-EM reconstruction workflows [
3]. Furthermore, the development of user-friendly software packages and AI/ML-based tools has streamlined data processing, leading to high-quality 3D reconstructions and the progress towards the complete automation of cryo-EM data processing pipelines. Moreover, the advancements in cryo-sample preparation methods have addressed challenges such as protein denaturation and aggregation, ensuring higher quality and reproducibility of samples for 3D reconstruction [
1,
4]. These combined advancements have propelled cryo-EM towards routine high-resolution protein structure determination, marking a significant milestone in structural biology.
2. What is Cryo-EM Particle Picking?
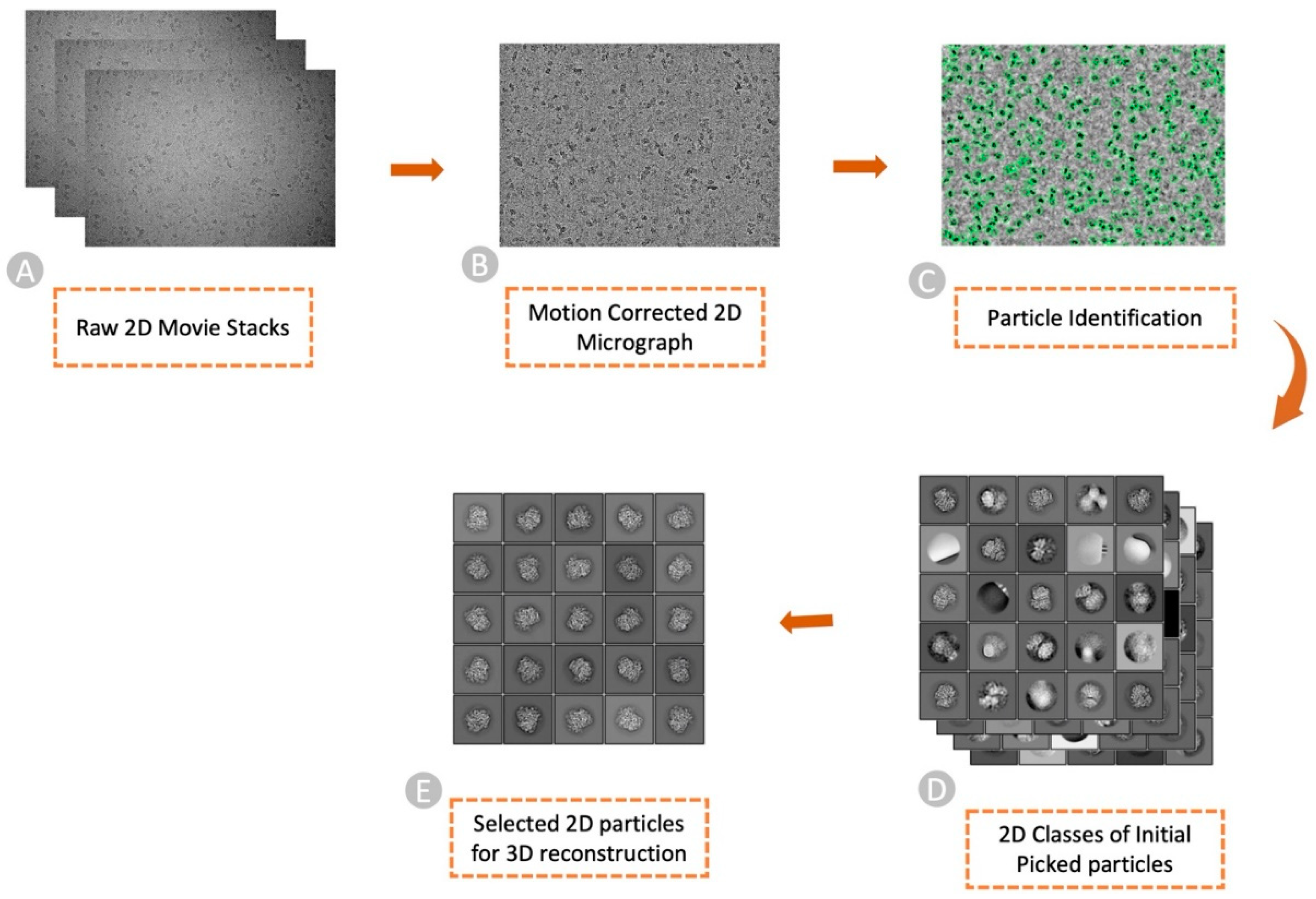
Particle picking is the first critical step in the cryo-EM data analysis, which involves identifying and selecting individual particles of interest (
Figure 1), such as biological macromolecules (usually proteins, nucleic acids, lipid complexes, and viruses), from electron micrographs to reconstruct their 3D structures. This review is focused on picking protein particles. Accurate particle picking is crucial for obtaining high-resolution structures, aiding in understanding biological processes at the molecular level and advancing drug discovery and disease research [
5,
6,
7,
8]. In the early days, manual particle picking was the main approach used in the cryo-EM data analysis, requiring researchers to visually identify individual particles and manually mark their positions (as exemplified in
Figure 1C) to facilitate further analysis and reconstruction of their 3D structures [
2]. This approach is labor-intensive and challenging as the particles are barely visible to the naked eyes in complex cryo-EM micrographs.
Following the manual particle picking, template-based picking methods [
9,
10] emerged as a more efficient alternative, by using some manually predefined particle templates to automatically recognize and extract more particles from micrographs. This approach reduced the time and subjectivity associated with manual picking, allowing for higher throughput and more consistent results.
While the template-based picking improved efficiency, it faces challenges such as template variability and difficulty in handling heterogeneous samples, leading to inaccurate and incomplete particle selection. These limitations spurred the emergence of AI-based particle picking methods, which leverage machine learning algorithms to automatically identify particles from cryo-EM micrographs, overcoming the shortcomings of manual and template-based approaches. AI-based picking offers enhanced accuracy, scalability, and adaptability to diverse sample types, significantly advancing cryo-EM data processing and comprehensive structural analysis of biological macromolecules.
3. The Challenges in Particle Picking and the Resources to Tackle Them
Particle picking in cryo-EM poses several challenges related to the complex nature of cryo-EM micrographs. These challenges include the low signal-to-noise ratio (SNR) of the original micrographs, resulting in highly noisy images due to radiation damage and various noise sources such as low contrast, particle overlap, ice contamination, and amorphous carbon [
2,
11]. Additionally, the intricate shapes of proteins, variations in particle sizes and orientations, and the presence of artifacts further complicate the particle picking process [
11].
One key step to tackle the challenges is the creation of high-quality datasets of cryo-EM micrographs to train and test particle picking methods. EMPIAR [
12] is a large, important database, storing high-quality raw electron microscopy data (e.g., cryo-EM micrographs) from various EM techniques. CryoPPP [
2] is an expert-curated dataset comprised of cryo-EM micrographs with labeled protein particle coordinates to facilitate the training of AI and ML models for automated particle picking. The dataset includes raw micrographs, motion correction files adjusted for imaging artifacts, particle stacks of manually identified protein particles, and ground truth labels (coordinates) detailing true positives and common false positives like ice contamination and carbon edges.
In addition to data resources, recent advancements in computer vision techniques can be used to preprocess cryo-EM micrographs to enhance their quality for particle picking. Key tools include OpenCV [
13], offering functions for image noise reduction, edge detection, and contrast enhancement crucial for protein particle visibility. Similarly, for machine learning developers, Scikit-Image provides various denoising and segmentation tools like Non-Local Means and watershed segmentation, which can be used in pre-processing cryo-EM micrographs.
4. Emergence of AI in Particle Picking
Over the last two decades, AI methods, particularly machine learning and deep learning techniques, have been developed to automate and improve particle picking efficiency and accuracy. In this section, we delve into the AI-based methods, including both classical methods (summarized in
Table 1) and advanced deep learning methods (summarized in
Table 2) that have gradually advanced particle picking in cryo-EM. We examine their principles, performance, and potential to overcome existing hurdles in the field.
5. Classical Particle Picking Methods
Earlier in 2004, Mallick et al. presented a pioneering machine learning approach for automated particle picking in cryo-EM micrographs, employing the Adaboost learning algorithm [
14]. Their discriminative algorithm learned essential features of particle appearance from training examples, enabling generic detection unrestricted by particle shape or size. It trained a cascaded classifier using Adaboost, which was augmented by rotating test micrographs over various orientations to enhance detection versatility. However, the method encountered difficulties in assessing particle detection performance due to variability in particle labeling by different microscopists and the complexity of protein shapes in micrographs. The dataset used by the method only included ground truth labels only for particles of rectangular shape, leading to suboptimal performance of recognizing particles of circular shapes and potential biases in evaluation. Additionally, the method's reliance on a specific feature type (rectangular features derived from templates) and a single training methodology, Adaboost, was noted as a limitation, suggesting that exploring more feature types and training approaches might offer greater accuracy.
gEMpicker [
15] is a particle picking method leveraging parallel programming techniques distributed across GPUs and CPU cores in a computer cluster to efficiently gather and combine cluster calculation results, thereby enhancing overall picking throughput for generation of higher resolution 3D cryo-EM density maps from picked particles. gEMpicker's performance was evaluated with various Fast Fourier Transform (FFT) libraries. Given that most computational costs stemmed from FFT-based normalized cross-correlation (NCC) calculations, the choice of FFT library significantly influenced the performance. This study found that FFT size and zero-padding were important for efficiency optimization.
Langlois et al.'s method, AutoPicker [
16], represented an advancement in selecting particle images in low-contrast conditions, outperforming manual selection on close-to-focus micrographs and leading to improved or comparable 3D reconstruction resolution. Leveraging unsupervised learning, AutoPicker minimized manual intervention, requiring only the approximate size of the macromolecule to be picked as input. Employing template matching and unsupervised learning, AutoPicker identified potential particles while excluding contaminants and noise windows, thereby enhancing particle selection accuracy. Its View Classifier (ViCer) further refined candidate particles, particularly in cases with contaminants, contributing to improved accuracy in noisy micrographs. The integration of real-time feedback mechanisms and adaptive algorithms was proposed to enhance the system's performance over time for effective handling of diverse macromolecules.
APPLE Picker [
17], developed in 2018, tackled cryo-EM particle picking with a template-free approach, reducing user effort and bias. By leveraging cross-correlation with a reference set derived from cryo-EM micrographs themselves, it efficiently identified particles and minimized false positives. However, this dependence on cross-correlation could be susceptible to noise mimicking particles, and the method's effectiveness can vary substantially with respect to particle complexity.
Table 1.
A list of classical AI methods for particle picking, their techniques and train/test data.
Table 1.
A list of classical AI methods for particle picking, their techniques and train/test data.
| SN |
Method |
Techniques |
Train/Test Data |
Year |
| 1 |
Mallick et al.’s method [14] |
Adaboost Learning Algorithm |
Keyhole Limpet Hemocyanin (KLH) [18] |
2004 |
| 2 |
gEMpicker [15] |
Roseman’s NCC Matching Algorithm |
Keyhole Limpet Hemocyanin (KLH) |
2013 |
| 3 |
Langlois et al.’s method [16] |
Principal Component Analysis (PCA) and Otsu’s Algorithm |
VA-ATPase from T. Thermophilus HB8 And 70S Ribosome from E. Coli, |
2014 |
| 4 |
APPLE picker [17] |
Support Vector Machine |
Β-Galactosidase, T20S Proteasome, 70S Ribosome, and Keyhole Limpet Hemocyanin (KLH) |
2018 |
| 5 |
SuperCryoEMPicker [19] |
Super-Clustering Approach |
80S Ribosome and Beta-Galactosidase Datasets |
2019 |
| 6 |
AutoCryoPicker [20] |
Unsupervised Clustering |
Apoferritin Dataset [21] and Keyhole Limpet Hemocyanin (KLH) Dataset |
2019 |
| 7 |
Li et al [22] |
Segmentation-Aware Synergy Framework |
EMPIAR-10028, EMPIAR-10097, and EMPIAR-10333 |
2022 |
Al-Azzawi et al. proposed two methods in 2019: AutoCryoPicker [
20] and SuperCryoEMPicker [
19]. AutoCryoPicker carried out micrograph cleaning and particle shape detection using a customized Circular Hough Transform algorithm. This technique efficiently identified particle shapes and centers, facilitating accurate particle selection through the creation of bounding boxes. Notably, the method incorporated a series of image preprocessing steps, and introduced an intensity distribution model for particle clustering, demonstrating superior performance over traditional clustering algorithms like K-means and Fuzzy C-Means (FCM) [
20]. SuperCryoEMPicker introduced a super-clustering technique designed to handle complex shapes, irregularities, and low SNR in particle images. The approach consisted of three primary stages: pre-processing, particle clustering, and particle selection. It surpassed traditional clustering methods like k-means, fuzzy c-means (FCM), and intensity-based cluster (IBC) by incorporating super-pixel algorithms. The pre-processing stage involved various image enhancement procedures, such as global intensity adjustment, contrast enhancement, noise suppression, and edge enhancement, aimed at improving cryo-EM image quality and enabling more precise particle detection [
19]. Finally, in the particle picking stage, a final set of particles was selected and picked from the clustered candidates after post-processing steps like binary mask cleaning and particle property measurement.
In 2022, Li et al. introduced a segmentation-aware synergy network, which incorporated an instance-level attention regression module during denoising to address vulnerable feature representation challenges and bridge the gap between denoising and recognition tasks [
22]. By treating particle selection as a semantic segmentation problem, the method enhanced the semantic-level label generation pipeline, overcoming some limitations of traditional mask obtainment techniques, particularly for datasets with missing angle information. The proposed multitask ensemble learning framework enhanced the synergy between denoising and downstream recognition, resulting in reliable location estimations for single particle analysis. The method was trained with pixel-level denoising loss, instance-level denoising loss, and particle segmentation loss. The segmentation loss was implemented as the binary cross-entropy (BCE) loss function for supervised training.
6. Advanced Deep Learning Methods
In 2016, DeepPicker [
23] introduced an iterative process to train its particle picking model, where top predicted particles from initial training steps were incorporated as new training data. It used a convolutional neural network (CNN) with a pooling layer to manage model complexity and prevent overfitting, thereby enhancing its capability to differentiate correct particles from background noise. Recall, precision, and center deviations were used to evaluate picked particles against ground-truth particles. Additionally, 2D clustering and class averaging were employed to validate the quality and accuracy of the selected particles. However, despite the efforts to mitigate overfitting through the pooling layer and iterative refinement, DeepPicker remained susceptible to overfitting the training data, limiting its generalizability to unseen datasets.
DeepEM [
24] employed a deep CNN for single particle recognition, which was trained with labeled 'good' and 'bad' particles to differentiate desired features from undesired ones, facilitating template-free particle picking and reducing reference particle-dependent bias. The performance evaluation, conducted via precision-recall curves, demonstrated its good accuracy and efficiency in automated particle extraction. However, DeepEM was shown to be effective for feature-rich cryo-EM micrographs but struggled with datasets lacking distinctive particle features.
Xiao et al. presented an approach for automated particle picking in 2017 using Fast R-CNN [
25], a deep learning-based object detection framework [
26]. It employed a 'Cross Molecule Training Strategy' to train the CNN with particles of various protein complexes to capture common latent features in protein particles, facilitating efficient and generalized particle picking without human intervention. The optimization of box-step settings for regions of interest (RoI) balanced GPU memory constraints and accuracy requirements, enhancing the overall performance. Several potential enhancements to this method include exploring advanced image processing techniques for data preprocessing, RoI proposal generation, and false positive reduction to improve efficiency and accuracy. Additionally, investigating alternative neural network architectures or employing ensemble learning methods may further enhance particle picking performance. The continuous refinement of the training strategy, such as incorporating more diverse training datasets or fine-tuning network parameters, could enhance method robustness and adaptability to different protein particle types.
In 2019, five deep learning methods including Pixer, HydraPicker, CrYOLO, Wrap and Topaz were developed, leveraging cutting-edge algorithms in deep learning and computer vision to enhance the accuracy and efficiency of identifying particles.
PIXER [
27] adopted an image segmentation-based approach, utilizing a deep neural network architecture. It first trained a classification network and then leveraged its parameters to expedite the segmentation network's training. During testing, cryo-EM micrographs were inputted into the segmentation network to generate probability density maps. Preliminary particle coordinates were obtained using a grid-based local-maximum method, followed by a classification network to eliminate false positive particles. The classification network was equipped with multiple parallel Atrous convolution channels [
28], enabling the processing of multiscale particles. The segmentation network's performance was evaluated using pixel intersection-over-union criteria on a validation dataset. It used a grid-based local-maximum method to pinpoint particles from the probability density maps, enhancing both accuracy and efficiency in particle selection.
HydraPicker [
29] developed a customized CNN architecture based on ResNet, with larger filters and group normalization layers. Its multi-headed design, comprising shared and dataset-specific heads, facilitated effective handling of unseen particles. Furthermore, the model was trained on different kinds of particles, combining specialized and generic models. Through a fully convolutional design, it divided the model into shared and specialized parts. Exploring limited data scenarios, it introduced a few-shot learning approach, enhancing adaptability to new datasets. While the method presented a couple of advancements in automated particle picking, it carried limitations. Firstly, its focus on specific particle structures during training may limit the model's generalizability to a broader range of datasets and particle types, potentially constraining its applicability in diverse experimental settings. Moreover, the complexity of the proposed methodology, particularly with multiple specialized heads and a shared trunk, may have high computational cost, necessitating careful optimization and resource allocation for practical deployment.
CrYOLO [
30] introduced a ‘You Only Look Once’ (YOLO) framework for automated particle picking, employing a 21-layer CNN for accurate particle detection. By reformulating the classification problem as a regression task, it predicted particle positions in a single pass of the full image, significantly speeding up the process compared to traditional methods. Despite training with a relatively small number of particles per cryo-EM micrograph dataset, CrYOLO achieved high recall and precision rates at a speed of up to five micrographs per second. The method may be further improved via transfer learning by pre-training it on diverse datasets first and then fine-tuning it to improve the generalization capability. Additionally, applying additional data augmentation techniques during training, such as rotation and scaling, might enhance robustness. Furthermore, incorporating domain-specific knowledge or constraints into the model, such as particle size distribution, may improve accuracy and efficiency in particle picking.
Warp [
31] introduced a particle-picking method centered on the BoxNet machine-learning algorithm, a fully convolutional ResNet architecture comprising 72 layers. Trained on both real cryo-EM micrograph data from the EMPIAR repository and synthetic data simulated from Protein Data Bank (PDB) structures, BoxNet adeptly masked out high-contrast artifacts like ethane. The system incorporated a retraining interface, enabling users to fine-tune BoxNet's performance by supplying positive examples of particle positions. Notably, Wrap's BoxNet model had some advantage in dealing with heterogeneous datasets, exhibiting superior performance over some alternative models such as crYOLO or RELION [
32] 3.0's Laplacian of Gaussian approach.
Table 2.
A list of advanced deep learning methods for protein particle picking, their techniques and train/test data.
Table 2.
A list of advanced deep learning methods for protein particle picking, their techniques and train/test data.
| SN |
Approach |
Techniques |
Train/Test Data |
Year |
| 1 |
DeepPicker [23] |
Deep Learning (using Cross-Molecule Training Strategy) |
y-secretase, spliceosome, TRPV1, b-galactosidase, N- ethylmaleimide sensitive factor complex |
2016 |
| 2 |
DeepEM [24] |
Convolutional Neural Network (CNN) |
800 manually selected particle images from the keyhole limpet Hemocyanin (KLH) dataset |
2017 |
| 3 |
Xiao et al.’s method [26] |
Region-based Convolutional Network (R-CNN) |
Gammas, Spliceosome, Trpv1 |
2017 |
| 4 |
Warp [31] |
Convolutional ResNet Architecture |
EMPIAR-10097, EMPIAR-10045, EMPIAR-10078, EMPIAR-10061, EMPIAR-10164, EMPIAR-10153 |
2019 |
| 5 |
SPHIRE-crYOLO [30] |
Deep Learning (Based on YOLO) |
TcdA1 toxin subunit, Drosophila transient receptor channel NOMPC, human peroxiredoxin-3 (Prx3), simulated data of the canonical TRPC4 ion channel, and keyhole limpet hemocyanin (KLH) |
2019 |
| 6 |
HydraPicker [29] |
ResNet Architecture |
Data from Warp [31] |
2019 |
| 7 |
Pixer [27] |
Deep Neural Network |
beta-galactosidase, influenza hemagglutinin trimer, Plasmodium falciparum 80S ribosome, cyclic nucleotide-gated ion channel, and GroEl + TRPV1, KLH, bacteriophage MS2, and rabbit muscle aldolase |
2019 |
| 8 |
Topaz [33] |
Positive U learning CNN |
EMPIAR-10025, EMPIAR-10096, EMPIAR-10028, EMPIAR-10261, EMPIAR-10234, and EMPIAR-10096 |
2019 |
| 9 |
DeepCryoPicker [34] |
Unsupervised Learning |
Apoferritin, KLH, 80S ribosome, β-galactosidase |
2020 |
| 10 |
McSweeney et al.’s method [35] |
Convolutional Neural Networks |
EMPIAR-10204, 10218, 10028, 10335, 10184, and 10059. |
2020 |
| 11 |
DRPnet [36] |
Double Convolutional Neural Network (CNN) Cascade |
TRPV1 dataset (EMPIAR-10005) |
2021 |
| 12 |
TransPicker [37] |
End-to-End Transformer-based Architecture |
EMPIAR-10093, EMPIAR-10017, EMPIAR-10028, EMPIAR-10096, EMPIAR-10406, and EMPIAR-10590 |
2021 |
| 13 |
CASSPER [38] |
Full Resolution Residual Network (FRRN) |
TcdA1 (EMPIAR 10089), HCN1 (EMPIAR 10081), TRPV1 (EMPIAR 10005) and b-galactosidase (EMPIAR 10017) |
2021 |
| 14 |
Urdnet [39] |
U-Net based residual intensive neural network |
71 human 80S ribosomal micrographs, 30 HCN1 micrographs, 24 TcdA1 micrographs, and 16 KLH micrographs |
2022 |
| 15 |
CryoSegNet [40] |
U-Net + SAM Model |
CryoPPP [2] |
2024 |
| 16 |
CryoTransformer [41] |
Transformer + ResNet Architecture |
CryoPPP [2] |
2024 |
Topaz [
33] is one of the widely used particle picking methods, which is based on a positive-unlabeled (PU) learning method, minimizing the need for extensive manual labeling by training the classifier with few labeled positive regions and unlabeled regions. It used a Generalized Expectation (GE) criteria to mitigate overfitting, improving prediction accuracy with minimal labeled data. Leveraging CNNs in conjunction with the PU learning, Topaz retrieved more particles at a low false-positive rate. However, manual examination of class averages of true particles or false positives picked by Topaz could be time-consuming and subjective.
In 2020, DeepCryoPicker [
34] was developed to train a deep learning network using 'good' and 'bad' particle examples, similar to DeepEM [
24]. However, the key difference was its use of a sliding window technique to classify sub-images as particles or background. To further improve performance, techniques like Non-Maximum Suppression (NMS) were utilized during testing to reduce false positives. Moreover, the method applied data augmentation and preprocessing steps to enhance the quality of low signal-to-noise ratio micrographs.
In the same year, McSweeney et al. proposed a self-supervised workflow for particle picking, employing an iterative strategy with 2D particle class averaging and a progressively improved CNN [
35]. Automation was achieved through defining a threshold (%/Res: the proportion of particles in a 2D class average to the total particle count), minimizing user input. The iterative process optimized a fine-tuned CNN-based particle picker, enhancing particle picking quality. While offering automated particle picking and integration into single-particle analysis packages, the method required additional 2D and 3D particle classifications to filter out damaged particles. Efficiency limitations were noted for small particles, regardless of the picking program used. Though aiming to eliminate trial-and-error parameters, the dependence on data-dependent parameters such as particle size remained a limitation.
In 2021, TransPicker [
37] introduced a framework leveraging the crDETR module based on a transformer-based detection method for 2D particle picking. Using an improved deformable Transformer [
42], TransPicker adeptly handled particle relocation and global context without using traditional components like anchors or sliding windows [
43], reaching rapid convergence with a combined loss function. The framework addressed limitations in object query numbers through a divide-and-conquer algorithm and incorporated optimizations such as denoising, enhancing (sharpening edges and contours), and adding masks to carbon region to improve accuracy. Moreover, it implemented particle filtering techniques and applied masks on carbon regions and ice contaminants to further reduce false positive ratios.
In the same year, CASSPER [
38] utilized semantic segmentation, a deep learning technique enabling pixel-level classification in cryo-EM micrographs, to streamline particle identification without manual picking. It used the Contrast Limited Adaptive Histogram Equalization (CLAHE) to enhance particle detection in challenging conditions, delivering robust performance. While streamlining particle picking, iterative 2D classification and 3D refinement steps were still necessary for detailed structural analysis, incorporating feedback mechanisms for self-correction and adaptive learning may further enhance CASSPER's robustness and accuracy over time, particularly with complex datasets.
DRPNet [
36], a deep learning-based particle picking network, was tailored to tackle issues like low signal-to-noise ratios and diverse particle characteristics. It employed a fully convolutional regression network (FCRN) to generate continuous distance maps representing particle centers and a classification CNN to refine detections, minimizing false positives. The model treated particle picking as a blob detection problem, considering particles as convex blobs with distinct textures and locating particle centers based on size estimates. Micrograph preprocessing enhanced performance by improving contrast and correcting artifacts. Based on its evaluation, DRPNet improved particle picking in terms of recall, precision, and F-measure. However, it encountered difficulty in the scenarios where particles varied substantially in dimensions and shapes.
UrdNet [
39] developed a U-Net based residual dense neural network that integrated point-level and pixel-level labels to streamline manual particle labeling and enhance the precision of particle picking. This approach combined multiple annotations (point-level and pixel-level annotations) and an improved U-Net architecture to select particles. The network model, comprising 49 convolution layers, four max-pooling layers, four up-sampling layers, and four feature concatenations, facilitated the extraction of global features and deep feature fusion, thereby enabling efficient particle picking. However, it had a limitation in the output definition of the residual block (ambiguity in feature representation), indicating the need for further refinement in the network architecture.
In 2024, CryoTransformer [
41] introduced a self-attention-based transformer model, typically utilized in natural language processing, to cryo-EM particle picking. By combining the transformer model and residual connections within the CNN component, CryoTransformer aimed to capture long-range dependencies/correlation between particles within cryo-EM images, thereby enhancing the accuracy of particle selection. The evaluation of the method on the CryoPPP dataset [
2] involved ablation studies to assess various technical components, including the impact of denoising (pre-processing) micrographs. A set of comprehensive evaluation metrics encompassing machine learning metrics, 2D particle resolution, 3D density map resolution, and particle orientation diversity were applied to assess its performance. The study highlighted the importance of denoising micrographs in particle picking tasks for improving particle picking accuracy. Potential enhancements for CryoTransformer includes leveraging transfer learning techniques, embracing ensemble learning approaches, developing interpretability and visualization methods. Developing a user interface to provide more intuitive controls and real-time feedback during particle picking could streamline the workflow and make the tool more user-friendly for researchers with varying levels of expertise.
Also in 2024, CryoSegNet [
40] was developed to integrate the attention-gated U-Net architecture with a general foundational image segmentation model - the Segment Anything Model (SAM) [
44] - to achieve heightened precision and recall in particle selection. Notably, the method addressed challenges in picking particles for small proteins through fine-tuning with predicted labels from a pre-trained model [
45], improving the accuracy of this difficult task. Moving forward, potential enhancements for CryoSegNet includes fine-tuning the model on the cryo-EM data of diverse proteins, augmenting the training dataset with predicted labels via distillation, and optimizing post-processing steps of selecting particles.
7. A Comparative Study of the AI-based cryo-EM Particle Picking Methods
We conducted a comparative evaluation of some of the aforementioned methods that are updated or widely used in terms of various metrics and visualized the results. The implementation of most of the particle picking methods has become outdated, mainly due to their reliance on idealized cryo-EM micrographs as training data. Additionally, some methods lack publicly available software tools, hindering usability and independent evaluation. Consequently, we focused our evaluation on six recently developed or widely used deep learning methods in the particle picking domain whose source codes are openly accessible: Deep Picker, CrYOLO, Topaz, CASSPER, CryoTransformer, and CryoSegNet.
To ensure a fair comparison of these six methods, we utilized the same set of training and test data from the CryoPPP dataset to train and test them. Specifically, the micrographs of 22 proteins (EMPIAR IDs) from CryoPPP were used for training, while 4 EMPIAR IDs were reserved for testing. Deep Picker was trained using the default parameters in CryoSPARC [
46], CrYOLO was trained using the 'PhosaurusNet' architecture, and Topaz was trained with the 'ResNet16' architecture. CASSPER, CryoTransformer, and CryoSegNet, were trained using their default parameters.
Traditional metrics like F1-score, precision, and recall can only literally measure how accurately protein particles can be picked from a pure machine learning perspective but cannot directly measure how well the picked particles can be used to build the density maps of the proteins, the ultimate goal of users. A more robust evaluation should consider the ability of these methods to capture true particles representing the diverse orientations of protein structures that are important for building better 3D density maps. Therefore, in this work, we used the resolution of 3D density maps reconstructed from particles picked by each method to evaluate its performance. We also considered the distribution of viewing directions of the picked particles and the local resolution of the resulting density maps in the evaluation.
8. Evaluation in Terms of the Resolution of 3D Density Maps Reconstructed from Picked Particles
For each protein (identified by EMPIAR ID) in the CryoPPP test dataset, we generated star files that contain protein particles picked by each method. The files were then imported into CryoSPARC [
46] for 3D
ab initio reconstruction of density maps and homogeneous refinement. During the reconstruction, a 3D density map was generated solely from the set of particles. During the process, the homogeneous refinement was used to correct higher-order aberrations and refine particle defocus caused by beam tilt, spherical aberration, and other optical challenges. The 3D resolution of the density maps constructed from the picked particles by Deep Picker, crYOLO, Topaz, CASSPER, CryoTransformer, and CryoSegNet was compared. The comparative evaluation was carried out across three trials with random seed initialization for CryoSPARC, and the best resolution among the three trials was used for comparison.
In addition to evaluating the methods on the CryoPPP test dataset, which includes approximately 300 micrographs per protein, we expanded our assessment to use the complete set of micrographs for each protein available on the EMPIAR website to benchmark them. This extended evaluation aimed to gauge the resolution these methods can achieve in a real-world setting where many micrographs are usually generated for a protein. The results of these methods in the two settings are summarized in
Table 3.
Table 3 shows that as the number of micrographs increases, as indicated in the CryoPPP and EMPIAR columns, the total number of particles picked by each method also increases. This increase in particles leads to an improvement in the 3D resolution of the constructed 3D density maps for all methods. CryoSegNet demonstrates superior performance for three out of four proteins (EMPIAR-10028, EMPIAR-10345, and EMPIAR-10532) for both CryoPPP and EMPIAR datasets, while CryoTransformer performs the best on EMPIAR-10093 in CryoPPP. Topaz shows the best result for EMPIAR-10093 in the EMPIAR dataset.
According to the average resolution across the four test proteins in the two datasets, CryoSegNet, Topaz and CryoTransformer exhibit better performance than the other three methods, among which CryoSegNet performs best. For instance, the average resolution of CryoSegNet over the four test proteins in the EMPIAR dataset is 3.28 Å, which is the highest among the six methods. CrYOLO and CASSPER yielded similar average resolutions, whereas Deep Picker had the poorest resolution.
9. Evaluation in Terms of the Viewing Directions of Picked Particles
Picking particles representing a broad range of particle orientations/views, particularly rare ones, is critical for achieving high-resolution reconstruction of 3D density maps. Here, we evaluated the quality of viewing direction of the particles picked by each method. Our assessment involved comparing the visual orientation of selected particles, focusing on elevation versus azimuth plots for each test EMPIAR ID as shown in
Figure 2.
Deep Picker picked a relatively fewer number of particles and hence the distribution of the particles representing various orientations is also less intense for EMPIAR 10028, EMPIAR 10345 and EMPIAR 10093. CASSPER struggled to pick particles of diverse orientations for EMPIAR 10028. For all other methods, the distribution of particles with various orientations has a similar pattern. An analysis of
Figure 2 reveals that EMPIAR 10028 and EMPIAR 10093 present a greater challenge for the methods to pick particles than EMPIAR 10345 and EMPIAR 10532, indicated by the abundance of blueish color in the plots of the latter.
10. Evaluation in Terms of the Visualized Reconstructed 3D Maps and Their GSFSC Curves
The 3D density map reconstructed by each method for each protein was visualized in
Figure 3. Moreover, in
Figure 4, the Fourier Shell Correlation (FSC) curves are used to evaluate the resolution of the 3D density maps. Two different versions of FSC plots, one based on a 'loose mask' curve generated automatically with a 15 Å falloff, and another using a 'tight mask' curve with a 6 Å falloff for all FSC plots, are presented.
In
Figure 3, some notable differences between the results of Deep Picker and CASSPER with other methods can be observed. For instance, in the case of EMPIAR 10093, the tips of the spiral shaped protein were not reconstructed from the particles picked by Deep Picker and CASSPER but were built from those picked by all other methods. In the case of EMPIAR 10532, CASSPER failed to reconstruct a segment of the rod-like protein structure, while all other methods reconstructed it. For EMPIAR 10532, the 3D maps constructed from Deep Picker, CrYOLO, Topaz and CASSPR picked particles contains a lot of dust (noisy) particles, while the 3D maps of CryoTransformer and CryoSegNet constructs are smooth and solid with few dusts.
11. Evaluation in Terms of the Local Resolution of 3D Maps Reconstructed from Picked Particles
The 3D density maps generated from the picked particles of the six methods are further analyzed using local resolution maps to interpret their structural details. These maps reveal how resolution varies across different regions of the density maps. A high local resolution for a region indicates well-defined structural details being constructed for the region. Conversely, a low local resolution points to less detailed and less reliable structural information in the region. This analysis helps us gain an in-depth understanding of the performance of the particle picking methods.
We utilized CryoSPARC's Local Refine job to generate the local resolution maps for the reconstructed density maps. Each local resolution map was overlaid onto the corresponding original density map using Chimera X [
47]. A color scale was applied to indicate resolution. High-resolution areas are depicted in gray and low-resolution areas in red, as illustrated in
Figure 5.
For EMPIAR 10028 and EMPIAR 10345, the density maps of CryoSegNet have high resolution indicated by gray color. CryoSegNet and CryoTransformer yielded similar local resolution for EMPIAR 10532. For EMPIAR 10093, the density map of CryoTransformer has the highest resolution in the open tip region. The better performance is due to a method’s ability to capture a wider range of particle orientations. The density maps of Deep Picker and CASSPER have generally lower local resolution than those of the other methods.
12. Remaining Challenges in Particle Picking.
As discussed in the previous sections, AI has substantially advanced the state of the art of single-protein cryo-EM particle picking. However, there are still the following significant hurdles to be overcome to move the particle picking to the next level.
13. Complexity within Cryo-EM Micrographs
The complex nature of micrographs affects the accuracy and reliability of particle identification. The low SNR, stemming from the weak scattering of electrons, complicates the distinction of particles from background noise. This difficulty is compounded by complex noisy background objects caused by ice contamination, support film artifacts, and other non-particle features, which can lead to false positives. Additionally, biological samples are often significantly heterogeneous, with particles in various conformations, orientations, and states, making consistent and accurate identification of the particles challenging. The frequent overlap and crowding of particles further complicate the accurate identification and separation of individual particles, necessitating the development of more advanced, robust computational techniques that can reliably deal with the background noise and particle heterogeneity.
The large size of cryo-EM micrographs also presents a significant computational challenge. Individual micrographs, often exceeding 7000 x 7000 pixels and comprising up to 3.0 GB in size, demand substantial computational resources for loading and processing them, particularly when training AI models with them. Resizing or compressing large micrographs to manage computational load risks the loss of critical information, potentially degrading the quality and accuracy of particle picking.
14. Lack of Benchmarking Data
The scarcity of labeled cryo-EM micrograph datasets poses a significant challenge in cryo-EM particle picking. Unlike some other domains such as protein tertiary structure prediction where large, annotated datasets are readily available for training machine learning models, annotated cryo-EM micrograph datasets are often limited in size and diversity. This scarcity of data makes it difficult to develop robust and generalizable particle picking methods, as the data may not capture the full spectrum of particle variability present in real-world samples. As a result, researchers often face difficulties in achieving high performance and accuracy, especially when dealing with novel or rare biological structures. The recent development of the expert-curated CryoPPP dataset used in this work is one step forward to address this issue. It has helped train several AI-based picking methods, including SAM-based picking [
48], CryoTransformer [
41], CryoMAE [
49], CryoSegNet [
40], and Cryo-EMMAE [
50]. However, the size of CryoPPP is still rather small compared to the image datasets available in the other field. Larger datasets that commensurate with the fast growth of the unannotated cryo-EM image data in the EMPIAR database need to be created by the community. Furthermore, the subjectivity in defining true and false particles is also an issue to be addressed in the data annotation and performance evaluation.
In addition to increasing the amount of the annotated cryo-EM micrograph data, machine learning techniques dealing with few data such as data augmentation and transfer learning can be applied to improve the performance of AI methods under the existing data constraints.
15. Lack of Standard Evaluation Metrics for Particle Picking
The lack of standardized metrics in this relatively young field is a challenge for evaluating the performance of particle picking methods and identifying the key areas that need improvement. Traditionally, the number of picked particles, precision, recall, F1 score, and Dice score are often used in evaluation. However, these metrics do not provide a comprehensive evaluation of the usefulness of picked particles, i.e., the quality of 3D density maps built from them. Directly evaluating the quality of the density maps built from picked particles such as the resolution used in this work can be added into the evaluation of new methods. Finally, it is important to evaluate AI methods on a diverse set of cryo-EM micrographs data containing diverse proteins of different size and shapes, heterogeneous particles, and noisy background to realistically estimate their performance. Evaluating methods only on relatively simple and idealized datasets such as Keyhole Limpet Hemocyanin (KHL) [
18] and Apoferritin [
21] may not be sufficient.
16. Potential Future Development
Addressing Data Scarcity
EMPIAR, the largest Cryo-EM image database in the field, provides a vast repository of raw cryo-EM image data. Annotating many cryo-EM micrographs in EMPIAR by one person or by one group may be unrealistic. However, a collaborative particle annotation effort by the community may be able to create large datasets of labeled cryo-EM image data for particle picking. To facilitate the effort, a standard protocol of reducing conflicts and subjectivity in human particle annotations may be needed.
17. Preprocessing and Efficient Representation of Cryo-EM Micrographs
Denoising micrographs is crucial for effective cryo-EM particle detection. However, standard image denoising techniques are often ineffective for cryo-EM micrographs due to the significant variability between micrographs. For instance, some micrographs may require saturation correction, others may need contrast enhancement, and for some micrographs, additional techniques such as noise reduction and edge preservation may be necessary. Therefore, adaptive denoising methods, tailored to the specific characteristics of each micrograph, are important for improving particle detection accuracy.
Moreover, it may be useful to develop micrograph cleansing tools to eliminate artifacts like ice contamination and carbon edges. Tools for resolving beam-induced movement and sample drift for motion correction, and methods that can estimate the contrast transfer function (CTF) parameters, which are crucial for correcting phase reversals and improving image contrast, may also help improve the accuracy of particle picking.
Using a more efficient representation of cryo-EM micrographs can facilitate the training and development of AI methods for particle picking. Typically, micrographs are stored as 32-bit float images of MRC format, which imposes significant computational burdens for loading and training machine learning models. By compressing the micrographs to 8-bit representation and normalizing pixels ranging from 0 to 255 as JPEG images can substantially reduce the requirement of the GPU/CPU memory and disk space for processing them.
18. Adoption of Comprehensive Performance Evaluation Metrics
Applying a comprehensive set of complimentary metrics to evaluate particle picking methods is important to objectively assess their strengths and weaknesses. In addition to the generic machine learning evaluation metrics such as precision, recall, and F1-measure, the following domain-specific metrics that directly measure the usefulness of picked particles can be applied to evaluate the performance of particle picking methods.
-
a)
2D Class Resolution of Picked Particles
Picked particles are often grouped together to form 2D particle classes whose resolution is evaluated before they are used to build 3D density maps. Better resolution of 2D classes indicates a more effective picking. Visualizing different 2D particle classes can show the breadth of the orientations of particles captured by a particle picking methods. The Initial Classification Uncertainty Factor (ICUF) and maximum alignment resolution are specified to align particles into classes (typically around 50) for this analysis.
Three diagnostic measures in this analysis are useful: the resolution (Å) of a class, the number of particles in a class (higher numbers are generally better), and the visual appearance of a class, which provides distinct views of particle structures. Solely considering the number of particles in a class is insufficient, as some classes with fewer particles may represent unique views of the protein. For a more detailed evaluation of viewing directions, analyzing the azimuthal curve is necessary, which is discussed below.
-
b)
Elevation vs Azimuthal Plot
It is worth pointing out that simply having a large number of particles does not guarantee the high resolution of 3D density maps. It's crucial to select a sufficient number of high-quality particles that cover a wide range of viewing angles.
The elevation versus azimuth plot can assess the view directions of picked particles. The plot provides a visual representation of the distribution of picked particle orientations in 3D space as represented in
Figure 2. By analyzing the elevation vs azimuthal angles, one can assess whether a particle picking method effectively samples particles from a wide range of viewing directions. A well-distributed plot with uniform coverage across all angles indicates robust particle picking. Discrepancies or gaps in the plot highlight regions in the particle orientation space that are underrepresented or not picked by the method.
Besides visual inspection, quantitative metrics such as the angular coverage and distribution uniformity can be derived from the elevation vs azimuthal plot. These metrics provide objective measures of a method’s ability to capture particles from diverse orientations.
-
c)
3D Resolution of Density Maps with Multiple Trails
The 3D resolution of density maps reconstructed from picked particles reflects the overall level of structural detail obtained from the particles. High resolution indicates that the picked particles accurately represent the underlying protein structure, whereas lower resolution may high false positives or few true positives in the picked particles. The star file of the picked particles can be used for ab initio density map reconstruction. To avoid any random bias in the 3D resolution, multiple ab initio 3D reconstruction trials with different random seeds (typically three trails) can be used, followed by reporting the average or best resolution. Then the average or the best resolution can be used in the evaluation. The variations of Fourier Shell Correlation (FSC) plots, such as those with tight or loose masks, can further assess the range of the resolution of the 3D density maps, offering comprehensive insights into the performance of particle picking methods.
-
d)
Local Resolution of Density Maps
The local resolution of 3D density maps provides a full picture of the variability of resolution across different regions of the 3D structure of a protein. By examining local resolution, one can identify specific regions where a particle picking method may struggle, such as regions with high conformational flexibility or insufficient coverage. Regions of the 3D structure with consistently high local resolution suggest that the picked particles in those areas are accurate and consistent. Conversely, regions with variable or low local resolution may indicate the presence of heterogeneous or incorrectly picked particles, highlighting the need for better particle picking or better cryo-EM image data in the first place. Furthermore, the quantitative metrics derived from local resolution maps, such as the mean local resolution and the distribution of resolution values, can be used to statistically compare different particle picking methods.
19. Exploration of Advanced AI Architectures and Ensemble Methods
Continuing to explore and refine advanced AI architectures is important for further improving the robustness and accuracy of particle picking. For instance, utilizing large-scale image models, such as Swin Transformer [
51], Segment Anything Model (SAM) [
44], and DETR [
52], can be useful to improve cryo-EM picking particles. Even though these models were initially trained on natural image datasets not containing cryo-EM micrographs for particle detection, they can be fine-tuned with cryo-EM data or directly used as a component in a cryo-EM particle picking pipeline, as exemplified by CryoSegNet.
Another possibility is to develop ensemble learning techniques to combine multiple particle picking methods to obtain consensus predictions. This approach may leverage the strengths of multiple methods, enhancing reliability of particle picking.
20. Conclusions
Particle picking methods have evolved from manual and semi-automated techniques to fully automated AI approaches. Early AI methods built on small, simple cryo-EM datasets lack robustness needed in practical applications. Recent AI methods leveraging advanced AI architectures and larger datasets have significantly improved the accuracy and reliability of particle picking. This work provides a rather complete review of the strengths, novelty, and weaknesses of many AI particle picking methods developed in the last two decades. We also conducted a comparative evaluation of several state-of-the-art deep learning particle picking methods using the metrics most relevant to the need of end users, such as the resolution of density maps reconstructed from picked particles. Through an in-depth review of the existing methods and the comparative evaluation, we identified several remaining challenges in the field, including improving accuracy of picking particles for small proteins with complicated micrographs, picking particles representing a complete, diverse range of view directions, and the lack of large, diverse, labeled particle datasets. Finally, we suggest some future developments to tackle the challenges.
Key Points
1. This review provides a rather complete, deep analysis of the existing AI-based methods for single-protein cryo-EM particle picking.
2. A comprehensive benchmarking of six state-of-the-art deep learning particle picking methods that can help users apply them in
practice is conducted.
3. Several key remaining challenges in cryo-EM protein particle picking are identified and the potential future developments to address them are discussed.
References
- Frank J. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu Rev Biophys Biomol Struct 2002; 31:303–319. [CrossRef]
- Dhakal A, Gyawali R, Wang L, et al. A large expert-curated cryo-EM image dataset for machine learning protein particle picking. Sci Data 2023; 10. [CrossRef]
- Frank J, Radermacher M, Penczek P, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 1996; 116:190–199. [CrossRef]
- Dhakal A, Gyawali R, Wang L, et al. CryoPPP: A Large Expert-Labelled Cryo-EM Image Dataset for Machine Learning Protein Particle Picking. bioRxiv 2023; 2023.02.21.529443. [CrossRef]
- Dhakal A, McKay C, Tanner JJ, et al. Artificial intelligence in the prediction of protein-ligand interactions: recent advances and future directions. Brief Bioinform 2022; 23. [CrossRef]
- Corso G, Stärk H, Jing B, et al. Diffdock: Diffusion steps, twists, and turns for molecular docking. arXiv preprint arXiv:2210.01776 2022. [CrossRef]
- Stärk H, Ganea O, Pattanaik L, et al. Equibind: Geometric deep learning for drug binding structure prediction. International conference on machine learning 2022; 20503–20521.
- Dhakal A, Gyawali R, Cheng J. Predicting Protein-Ligand Binding Structure Using E(n) Equivariant Graph Neural Networks. bioRxiv 2023; 2023.08.06.552202. [CrossRef]
- Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012; 180:519–530. [CrossRef]
- Tang G, Peng L, Baldwin PR, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol 2007; 157:38–46. [CrossRef]
- Gyawali R, Dhakal A, Wang L, et al. CryoVirusDB: A Labeled Cryo-EM Image Dataset for AI-Driven Virus Particle Picking. bioRxiv 2023. [CrossRef]
- Iudin A, Korir PK, Somasundharam S, et al. EMPIAR: the Electron Microscopy Public Image Archive. Nucleic Acids Res 2023; 51:D1503–D1511. [CrossRef]
- Bradski G. The opencv library. Dr. Dobb’s Journal: Software Tools for the Professional Programmer 2000; 25:120–123.
- Mallick SP, Zhu Y, Kriegman D. Detecting particles in cryo-EM micrographs using learned features. J Struct Biol 2004; 145:52–62. [CrossRef]
- Hoang T V, Cavin X, Schultz P, et al. gEMpicker: a highly parallel GPU-accelerated particle picking tool for cryo-electron microscopy. BMC Struct Biol 2013; 13:25. [CrossRef]
- Langlois R, Pallesen J, Ash JT, et al. Automated particle picking for low-contrast macromolecules in cryo-electron microscopy. J Struct Biol 2014; 186:1–7. [CrossRef]
- Heimowitz A, Andén J, Singer A. APPLE picker: Automatic particle picking, a low-effort cryo-EM framework. J Struct Biol 2018; 204:215–227. [CrossRef]
- Zhu Y, Carragher B, Glaeser RM, et al. Automatic particle selection: Results of a comparative study. J Struct Biol 2004; 145:3–14. [CrossRef]
- Azzawi A Al, Ouadou A, Tanner JJ, et al. A super-clustering approach for fully automated single particle picking in cryo-em. Genes (Basel) 2019; 10. [CrossRef]
- Al-Azzawi A, Ouadou A, Tanner JJ, et al. Autocryopicker: An unsupervised learning approach for fully automated single particle picking in cryo-em images. BMC Bioinformatics 2019; 20. [CrossRef]
- Rona G, Wynder EL, Helman P, et al. Plasma Hormone Profiles in Populations at Different Risk for Breast Cancer. Elife 2018; 36:1883–1885.
- Li S, Li H, Zhang C, et al. A Segmentation-aware Synergy Network for Single Particle Recognition in Cryo-EM. Proceedings - 2022 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2022 2022; 1066–1071.
- Wang F, Gong H, Liu G, et al. DeepPicker: A deep learning approach for fully automated particle picking in cryo-EM. J Struct Biol 2016; 195:325–336. [CrossRef]
- Zhu Y, Ouyang Q, Mao Y. A deep convolutional neural network approach to single-particle recognition in cryo-electron microscopy. BMC Bioinformatics 2017; 18. [CrossRef]
- Girshick R. Fast R-CNN. Proceedings of the IEEE International Conference on Computer Vision 2015; 2015 Inter:1440–1448.
- Xiao Y, Yang G. A fast method for particle picking in cryo-electron micrographs based on fast R-CNN. AIP Conf Proc 2017; 1836.
- Zhang J, Wang Z, Chen Y, et al. PIXER: An automated particle-selection method based on segmentation using a deep neural network. BMC Bioinformatics 2019; 20. [CrossRef]
- Chen LC, Papandreou G, Kokkinos I, et al. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans Pattern Anal Mach Intell 2018; 40:834–848. [CrossRef]
- Masoumzadeh A, Brubaker M. HydraPicker: Fully automated particle picking in cryo-em by utilizing dataset bias in single shot detection. 30th British Machine Vision Conference 2019, BMVC 2019 2020.
- Wagner T, Merino F, Stabrin M, et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol 2019; 2. [CrossRef]
- Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods 2019; 16:1146–1152. [CrossRef]
- Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012; 180:519–530. [CrossRef]
- Bepler T, Morin A, Rapp M, et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat Methods 2019; 16:1153–1160. [CrossRef]
- Al-Azzawi A, Ouadou A, Max H, et al. DeepCryoPicker: fully automated deep neural network for single protein particle picking in cryo-EM. BMC Bioinformatics 2020; 21. [CrossRef]
- McSweeney DM, McSweeney SM, Liu Q. A self-supervised workflow for particle picking in cryo-EM. IUCrJ 2020; 7:719–727. [CrossRef]
- Nguyen NP, Ersoy I, Gotberg J, et al. DRPnet: automated particle picking in cryo-electron micrographs using deep regression. BMC Bioinformatics 2021; 22. [CrossRef]
- Zhang C, Li H, Wan X, et al. TransPicker: A Transformer-based Framework for Particle Picking in cryoEM Micrographs. Proceedings - 2021 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2021 2021; 1179–1184.
- George B, Assaiya A, Roy RJ, et al. CASSPER is a semantic segmentation-based particle picking algorithm for single-particle cryo-electron microscopy. Commun Biol 2021; 4. [CrossRef]
- Ouyang J, Zhang Y, Fang K, et al. Urdnet: A Cryo-EM Particle Automatic Picking Method. Computers, Materials and Continua 2022; 72:1593–1610. [CrossRef]
- Gyawali R, Dhakal A, Wang L, et al. CryoSegNet: accurate cryo-EM protein particle picking by integrating the foundational AI image segmentation model and attention-gated U-Net. Brief Bioinform 2024; 25:bbae282. [CrossRef]
- Dhakal A, Gyawali R, Wang L, et al. CryoTransformer: a transformer model for picking protein particles from cryo-EM micrographs. Bioinformatics 2024; 40. [CrossRef]
- Zhu X, Su W, Lu L, et al. Deformable detr: Deformable transformers for end-to-end object detection. arXiv preprint arXiv:2010.04159 2020. [CrossRef]
- Pratyush P, Bahmani S, Pokharel S, et al. LMCrot: an enhanced protein crotonylation site predictor by leveraging an interpretable window-level embedding from a transformer-based protein language model. Bioinformatics 2024; 40:btae290. [CrossRef]
- Kirillov A, Mintun E, Ravi N, et al. Segment Anything. Proceedings of the IEEE International Conference on Computer Vision 2023; 3992–4003.
- Gyawali R, Dhakal A, Wang L, et al. Accurate cryo-EM protein particle picking by integrating the foundational AI image segmentation model and specialized U-Net. bioRxiv 2024; 2023.10.02.560572. [CrossRef]
- Punjani A, Rubinstein JL, Fleet DJ, et al. CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 2017; 14:290–296. [CrossRef]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Science 2021; 30:70–82. [CrossRef]
- He F, Yang Z, Gao M, et al. Adapting Segment Anything Model (SAM) through Prompt-based Learning for Enhanced Protein Identification in Cryo-EM Micrographs. ArXiv 2023. [CrossRef]
- Xu C, Zhan X, Xu M. CryoMAE: Few-Shot Cryo-EM Particle Picking with Masked Autoencoders. ArXiv 2024. [CrossRef]
- Zamanos A, Koromilas P, Bouritsas G, et al. Towards generalizable particle picking in Cryo-EM images by leveraging Masked AutoEncoders. ICML 2024 Workshop on Efficient and Accessible Foundation Models for Biological Discovery 2024.
- Liu Z, Lin Y, Cao Y, et al. Swin transformer: Hierarchical vision transformer using shifted windows. Proceedings of the IEEE/CVF international conference on computer vision 2021; 10012–10022.
- Carion N, Massa F, Synnaeve G, et al. End-to-end object detection with transformers. European conference on computer vision 2020; 213–229.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).