Submitted:
27 August 2024
Posted:
28 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
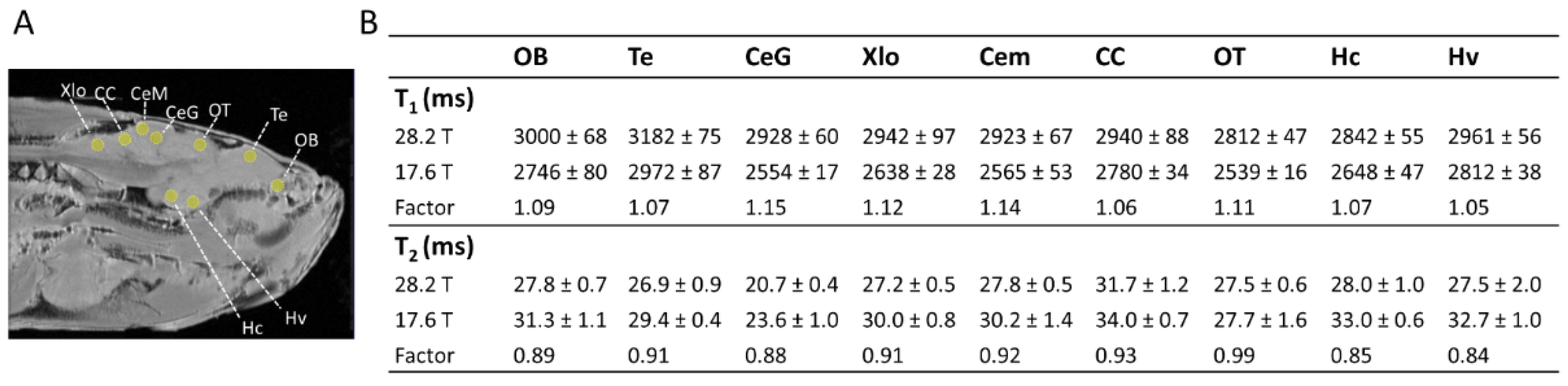
2.1. Relaxation Times
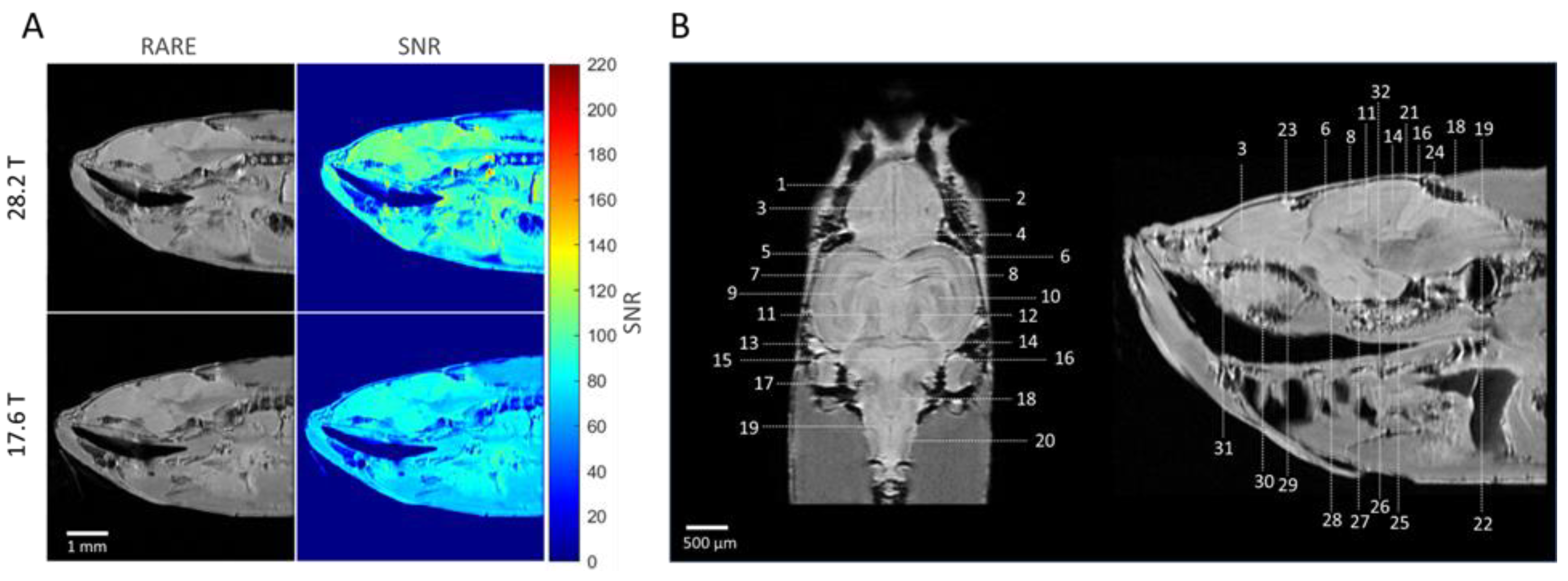
2.2. Anatomical Imaging
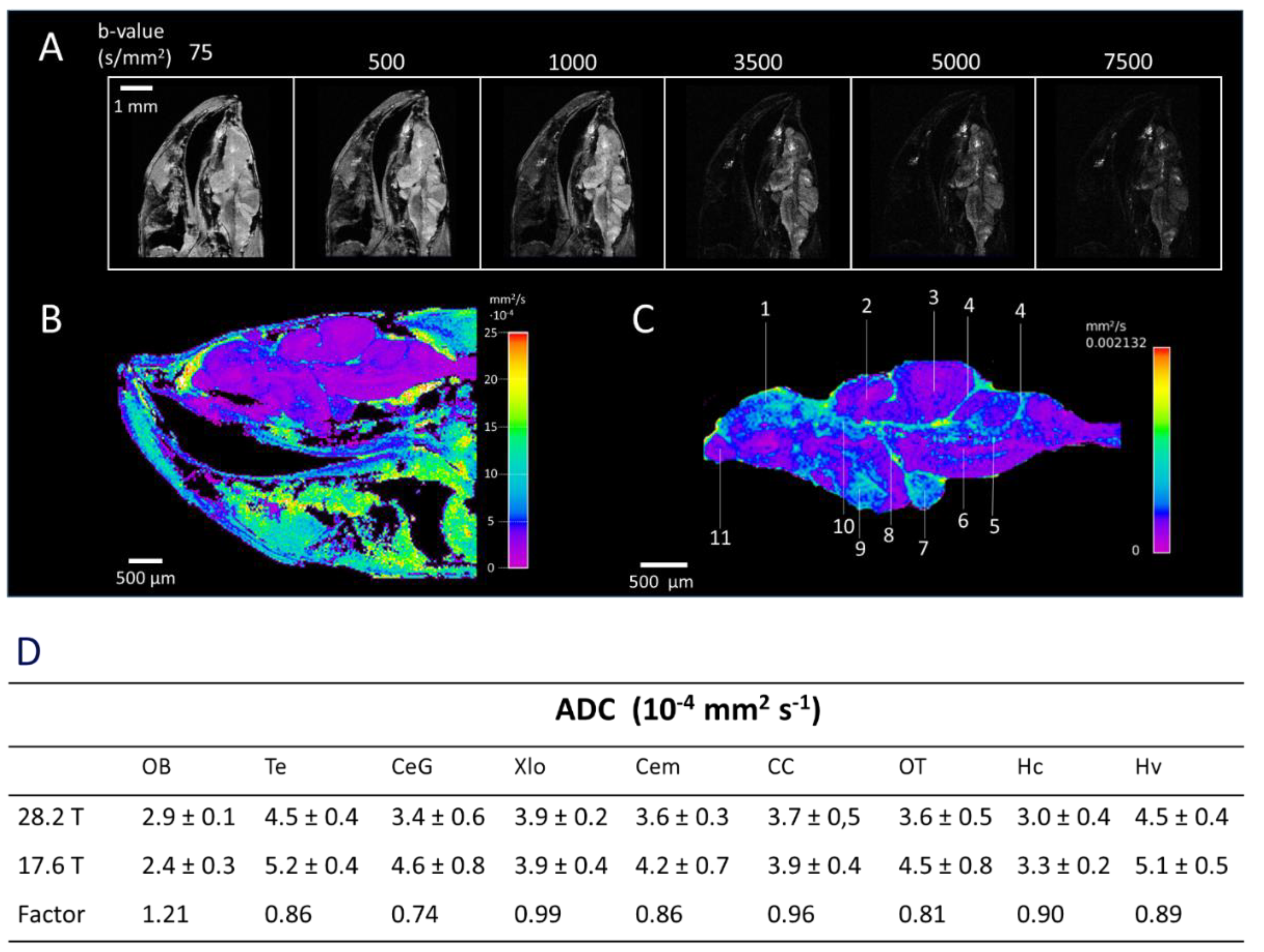
2.3. Diffusion Weighted Imaging (DWI)
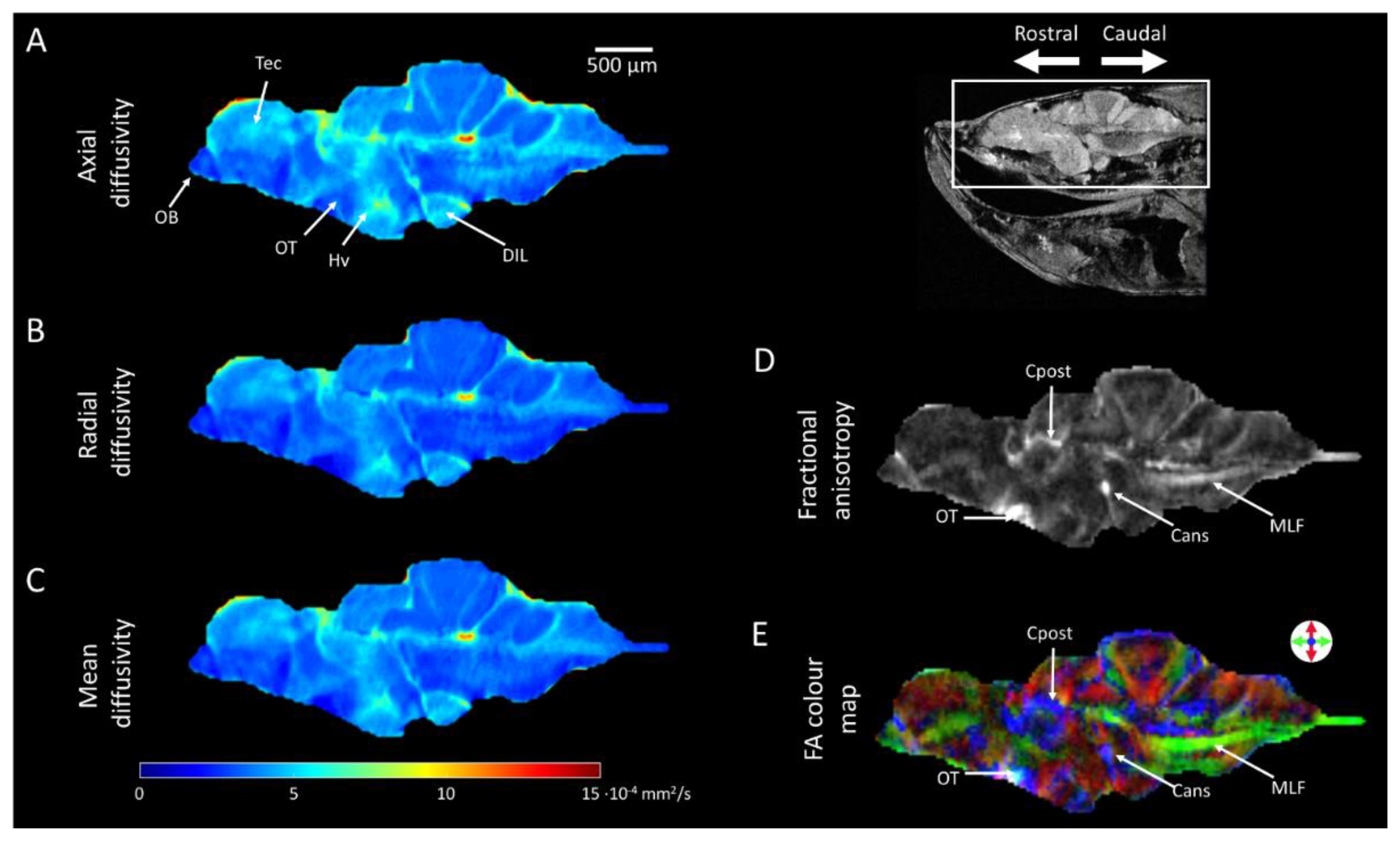
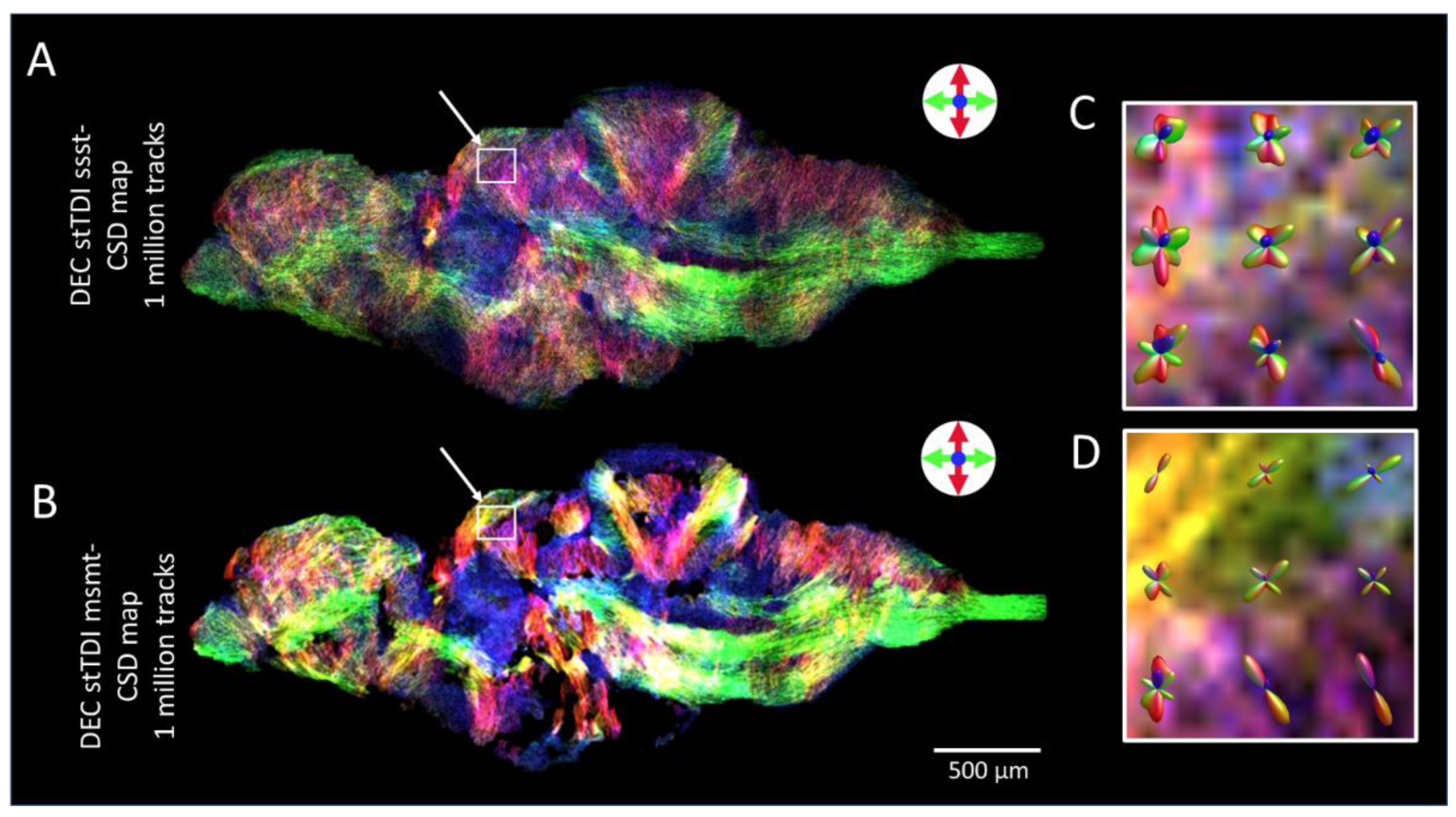
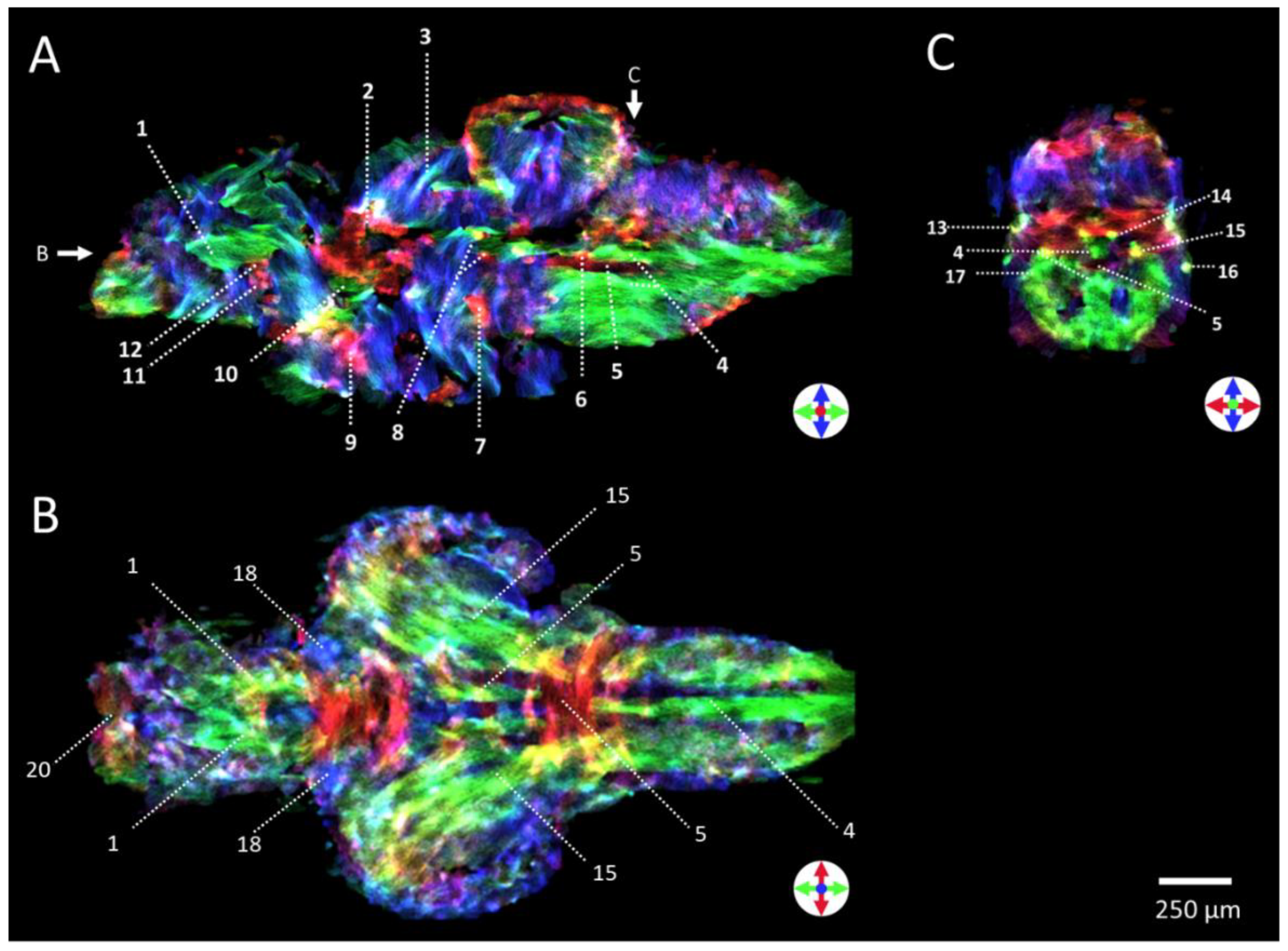
2.4. Diffusion Tensor Imaging (DTI)
3. Materials and Methods
3.1. Zebrafish Husbandry
3.2. Magnetic Resonance Imaging
3.3. Data Processing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bailone RL, Aguiar LKd, Roca RdO, Borra RC, Corrêa T, Janke H, Fukushima HCS (2019) “Zebrafish as an animal model for food safety research: trends in the animal research”. Food Biotechnology 33, 283-302.
- Cassar S, Adatto I, Freeman JL, Gamse JT, Iturria I, Lawrence C, Muriana A, Peterson RT, Van Cruchten S, Zon LI (2020) Use of Zebrafish in Drug Discovery Toxicology. Chem Res Toxicol 33, 95-118.
- Choi TY, Choi TI, Lee YR, Choe SK, Kim CH (2021) Zebrafish as an animal model for biomedical research. Exp Mol Med 53, 310-317.
- Ding Y, Haks MC, Forn-Cuni G, He J, Nowik N, Harms AC, Hankemeier T, Eeza MNH, Matysik J, Alia A, Spaink HP (2021) Metabolomic and transcriptomic profiling of adult mice and larval zebrafish leptin mutants reveal a common pattern of changes in metabolites and signaling pathways. Cell Biosci 11, 126.
- Bashirzade AA, Zabegalov KN, Volgin AD, Belova AS, Demin KA, de Abreu MS, Babchenko VY, Bashirzade KA, Yenkoyan KB, Tikhonova MA, Amstislavskaya TG, Kalueff AV (2022) Modeling neurodegenerative disorders in zebrafish. Neurosci Biobehav Rev 138, 104679.
- Wang X, Zhang JB, He KJ, Wang F, Liu CF (2021) Advances of Zebrafish in Neurodegenerative Disease: From Models to Drug Discovery. Front Pharmacol 12, 713963.
- Wang J, Cao H (2021) Zebrafish and Medaka: Important Animal Models for Human Neurodegenerative Diseases. Int J Mol Sci 22.
- Kabli S, Alia A, Spaink HP, Verbeek FJ, De Groot HJ (2006) Magnetic resonance microscopy of the adult zebrafish. Zebrafish 3, 431-439.
- Haud N, Kara F, Diekmann S, Henneke M, Willer JR, Hillwig MS, Gregg RG, Macintosh GC, Gartner J, Alia A, Hurlstone AF (2011) rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc Natl Acad Sci U S A 108, 1099-1103.
- Ramirez IB, Pietka G, Jones DR, Divecha N, Alia A, Baraban SC, Hurlstone AF, Lowe M (2012) Impaired neural development in a zebrafish model for Lowe syndrome. Hum Mol Genet 21, 1744-1759.
- Kabli S, Spaink HP, De Groot HJ, Alia A (2009) In vivo metabolite profile of adult zebrafish brain obtained by high-resolution localized magnetic resonance spectroscopy. J Magn Reson Imaging 29, 275-281.
- Eeza MNH, Singer R, Ding Y, He J, Zuberi Z, Baelde HJ, de Groot HJM, Matysik J, Spaink HP, Alia A (2023) Probing microstructural changes in muscles of leptin-deficient zebrafish by non-invasive ex-vivo magnetic resonance microimaging. PLoS One 18, e0284215.
- Kabli S, He S, Spaink HP, Hurlstone A, Jagalska ES, De Groot HJ, Alia A (2010) In vivo magnetic resonance imaging to detect malignant melanoma in adult zebrafish. Zebrafish 7, 143-148.
- Alexander AL, Lee JE, Lazar M, Field AS (2007) Diffusion tensor imaging of the brain. Neurotherapeutics 4, 316-329.
- Auning E, Kjaervik VK, Selnes P, Aarsland D, Haram A, Bjornerud A, Hessen E, Esnaashari A, Fladby T (2014) White matter integrity and cognition in Parkinson’s disease: a cross-sectional study. BMJ Open 4, e003976.
- Bede P, Elamin M, Byrne S, McLaughlin RL, Kenna K, Vajda A, A. F, Bradley DG, Hardiman O (2014) Patterns of cerebral and cerebellar white matter degeneration in ALS. J Neurol Neurosurg Psychiatry 86.
- de Schipper LJ, Hafkemeijer A, Bouts M, van der Grond J, Marinus J, Henselmans JML, van Hilten JJ (2019) Age- and disease-related cerebral white matter changes in patients with Parkinson’s disease. Neurobiol Aging 80, 203-209.
- Gold BT, Johnson NF, Powell DK, Smith CD (2012) White matter integrity and vulnerability to Alzheimer’s disease: preliminary findings and future directions. Biochim Biophys Acta 1822, 416-422.
- Huang P, Xu X, Gu Q, Xuan M, Yu X, Luo W, Zhang M (2014) Disrupted white matter integrity in depressed versus non-depressed Parkinson’s disease patients: a tract-based spatial statistics study. J Neurol Sci 346, 145-148.
- Kantarci K, Murray ME, Schwarz CG, Reid RI, Przybelski SA, Lesnick T, Zuk SM, Raman MR, Senjem ML, Gunter JL, Boeve BF, Knopman DS, Parisi JE, Petersen RC, Jack CR, Jr., Dickson DW (2017) White-matter integrity on DTI and the pathologic staging of Alzheimer’s disease. Neurobiol Aging 56, 172-179.
- Novak MJ, Seunarine KK, Gibbard CR, Hobbs NZ, Scahill RI, Clark CA, Tabrizi SJ (2014) White matter integrity in premanifest and early Huntington’s disease is related to caudate loss and disease progression. Cortex 52, 98-112.
- Reading SA, Yassa MA, Bakker A, Dziorny AC, Gourley LM, Yallapragada V, Rosenblatt A, Margolis RL, Aylward EH, Brandt J, Mori S, van Zijl P, Bassett SS, Ross CA (2005) Regional white matter change in pre-symptomatic Huntington’s disease: a diffusion tensor imaging study. Psychiatry Res 140, 55-62.
- Stricker NH, Schweinsburg BC, Delano-Wood L, Wierenga CE, Bangen KJ, Haaland KY, Frank LR, Salmon DP, Bondi MW (2009) Decreased white matter integrity in late-myelinating fiber pathways in Alzheimer’s disease supports retrogenesis. Neuroimage 45, 10-16.
- van der Burgh HK, Westeneng HJ, Walhout R, van Veenhuijzen K, Tan HHG, Meier JM, Bakker LA, Hendrikse J, van Es MA, Veldink JH, van den Heuvel MP, van den Berg LH (2020) Multimodal longitudinal study of structural brain involvement in amyotrophic lateral sclerosis. Neurology 94, e2592-e2604.
- Zhang J, Yin X, Zhao L, Evans AC, Song L, Xie B, Li H, Luo C, Wang J (2014) Regional alterations in cortical thickness and white matter integrity in amyotrophic lateral sclerosis. J Neurol 261, 412-421.
- Abe O, Yamada H, Masutani Y, Aoki S, Kunimatsu A, Yamasue H, Fukuda R, Kasai K, Hayashi N, Masumoto T, Mori H, Soma T, Ohtomo K (2004) Amyotrophic lateral sclerosis: diffusion tensor tractography and voxel-based analysis. NMR Biomed 17, 411-416.
- Agosta F, Pagani E, Petrolini M, Caputo D, Perini M, Prelle A, Salvi F, Filippi M (2010) Assessment of white matter tract damage in patients with amyotrophic lateral sclerosis: a diffusion tensor MR imaging tractography study. AJNR Am J Neuroradiol 31, 1457-1461.
- Kitamura S, Kiuchi K, Taoka T, Hashimoto K, Ueda S, Yasuno F, Morikawa M, Kichikawa K, Kishimoto T (2013) Longitudinal white matter changes in Alzheimer’s disease: a tractography-based analysis study. Brain Res 1515, 12-18.
- Kok JG, Leemans A, Teune LK, Leenders KL, McKeown MJ, Appel-Cresswell S, Kremer HPH, de Jong BM (2020) Structural Network Analysis Using Diffusion MRI Tractography in Parkinson’s Disease and Correlations With Motor Impairment. Front Neurol 11, 841.
- Lo CY, Wang PN, Chou KH, Wang J, He Y, Lin CP (2010) Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer’s disease. J Neurosci 30, 16876-16885.
- Phillips O, Sanchez-Castaneda C, Elifani F, Maglione V, Di Pardo A, Caltagirone C, Squitieri F, Sabatini U, Di Paola M (2013) Tractography of the corpus callosum in Huntington’s disease. PLoS One 8, e73280.
- Zhang Y, Wu IW, Buckley S, Coffey CS, Foster E, Mendick S, Seibyl J, Schuff N (2015) Diffusion tensor imaging of the nigrostriatal fibers in Parkinson’s disease. Mov Disord 30, 1229-1236.
- Freidlin RZ, Morris HD, Horkay F, Pierpaoli C, Toyama R, Dawid IB, Basser PJ (2004) Diffusion Tensor MR Microscopy of Adult Zebrafish. Proc. Intl. Soc. Mag. Reson. Med. 11.
- Ullmann JF, Calamante F, Collin SP, Reutens DC, Kurniawan ND (2015) Enhanced characterization of the zebrafish brain as revealed by super-resolution track-density imaging. Brain Struct Funct 220, 457-468.
- Dhollander T, Raffelt D, Connelly A (2016) Unsupervised 3-tissue response function estimation from single-shell or multi-shell diffusion MR data without a co-registered T1 image.
- Dhollander T, Raffelt D, Mito R, Connelly A (2019) Improved white matter response function estimation for 3-tissue constrained spherical deconvolution. Conference Paper.
- Jeurissen B, Tournier JD, Dhollander T, Connelly A, Sijbers J (2014) Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 103, 411-426.
- Singer, R, Oganezova, I, Hu W, Liu L, Ding Y, de Groot HJ, Spaink HP, Alia A (2024) Ultrahigh field diffusion magnetic resonance imaging uncovers intriguing microstructural changes in the adult zebrafish brain caused by Toll-like receptor 2 genomic deletion. NMR Biomed, p. e5170.
- Bottomley PA, Foster TH, Argersinger RE, Pfeifer LM (1984) A review of normal tissue hydrogen NMR relaxation times and relaxation mechanisms from 1-100 MHz: dependence on tissue type, NMR frequency, temperature, species, excision, and age. Med Phys 11, 425-448.
- de Graaf RA, Brown PB, McIntyre S, Nixon TW, Behar KL, Rothman DL (2006) High magnetic field water and metabolite proton T1 and T2 relaxation in rat brain in vivo. Magn Reson Med 56, 386-394.
- Korb JP, Bryant RG (2002) Magnetic field dependence of proton spin-lattice relaxation times. Magn Reson Med 48, 21-26.
- van de Ven RC, Hogers B, van den Maagdenberg AM, de Groot HJ, Ferrari MD, Frants RR, Poelmann RE, van der Weerd L, Kiihne SR (2007) T(1) relaxation in in vivo mouse brain at ultra-high field. Magn Reson Med 58, 390-395.
- Kara F, Chen F, Ronen I, de Groot HJ, Matysik J, Alia A (2013) In vivo measurement of transverse relaxation time in the mouse brain at 17.6 T. Magn Reson Med 70, 985-993.
- Hamilton N, Allen C, Reynolds S (2022) Longitudinal brain studies in adult zebrafish by MRI.
- Kenney JW, Steadman PE, Young O, Shi MT, Polanco M, Dubaishi S, Covert K, Mueller T, Frankland PW, A 3D adult zebrafish brain atlas (AZBA) for the digital age,eLife, http://azba.wayne.edu/, Accessed 09.
- Wullimann MF, Rupp B, Reichert H (1996) Neuroanatomy of the zebrafish brain A topological atlas, Basel Boston Berlin.
- Baliyan V, Das CJ, Sharma R, Gupta AK (2016) Diffusion weighted imaging: Technique and applications. World J Radiol 8, 785-798.
- Ogura A, Tamura T, Ozaki M, Doi T, Fujimoto K, Miyati T, Ito Y, Maeda F, Tarewaki H, Takahashi M (2015) Apparent Diffusion Coefficient Value Is Not Dependent on Magnetic Resonance Systems and Field Strength Under Fixed Imaging Parameters in Brain. J Comput Assist Tomogr 39, 760-765.
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A (2000) In vivo fiber tractography using DT-MRI data. Magnetic Resonance in Medicine 44, 625-632.
- Jeurissen B, Leemans A, Jones DK, Tournier JD, Sijbers J (2011) Probabilistic fiber tracking using the residual bootstrap with constrained spherical deconvolution. Hum Brain Mapp 32, 461-479.
- Jeurissen B, Leemans A, Tournier JD, Jones DK, Sijbers J (2013) Investigating the prevalence of complex fiber configurations in white matter tissue with diffusion magnetic resonance imaging. Hum Brain Mapp 34, 2747-2766.
- Jillings S, Morez J, Vidas-Guscic N, Van Audekerke J, Wuyts F, Verhoye M, Sijbers J, Jeurissen B (2020) Multi-tissue constrained spherical deconvolution in a murine brain. Proc. Intl. Soc. Mag. Reson. Med. 28.
- He J, Ding Y, Nowik N, Jager C, Eeza MNH, Alia A, Baelde HJ, Spaink HP (2021) Leptin deficiency affects glucose homeostasis and results in adiposity in zebrafish. J Endocrinol 249, 125-134.
- Carr HY, Purcell EM (1954) Effects of Diffusion on Free Precession in Nuclear Magnetic Resonance Experiments. Physical Review 94, 630-638.
- Milford D, Rosbach N, Bendszus M, Heiland S (2015) Mono-Exponential Fitting in T2-Relaxometry: Relevance of Offset and First Echo. PLoS One 10, e0145255.
- Tournier JD, Smith R, Raffelt D, Tabbara R, Dhollander T, Pietsch M, Christiaens D, Jeurissen B, Yeh CH, Connelly A (2019) MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage 202, 116137.
- Veraart J, Fieremans E, Novikov DS (2016) Diffusion MRI noise mapping using random matrix theory. Magn Reson Med 76, 1582-1593.
- Basser PJ, Mattiello J, Lebihan D (1994) Estimation of the Effective Self-Diffusion Tensor from the NMR Spin Echo. Journal of Magnetic Resonance B, 8.
- Veraart J, Sijbers J, Sunaert S, Leemans A, Jeurissen B (2013) Weighted linear least squares estimation of diffusion MRI parameters: strengths, limitations, and pitfalls. Neuroimage 81, 335-346.
- Tournier JD, Calamante F, Connelly A (2013) Determination of the appropriate b value and number of gradient directions for high-angular-resolution diffusion-weighted imaging. NMR Biomed 26, 1775-1786.
- Tournier JD, Calamante F, Connelly A (2007) Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage 35, 1459-1472.
- Tournier JD, Calamante F, Connelly A (2012) MRtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology 22, 53-66.
- Calamante F, Tournier JD, Jackson GD, Connelly A (2010) Track-density imaging (TDI): super-resolution white matter imaging using whole-brain track-density mapping. Neuroimage 53, 1233-1243.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




