1. Introduction
The characteristic analysis is a significantly concerning aspect in various fields, such as geology, medicine, business, or engineering systems, including image and data processing [
1,
2]. To be seen as a vital tool for characteristic analysis, clustering explores the data structure and distills meaningful information via analyzing, separating, and merging similar data points into distinct groups. There have been many clustering approaches, each one owning its specific techniques [
3,
4,
5]. Among them, density-based clustering is one of the most famous paradigms [
6,
7,
8,
9].
In [
6], Martin Ester et al. presented Density-Based Spatial Clustering of Applications with Noise (DBSCAN). It finds data points in a circle of radius
ε and adds them into a cluster of data points with the same characteristic. If the number of neighbors of an unassigned point is less than a predefined threshold
minPts, the point is seen as noise. Its considerable benefit is the ability to merge data points into arbitrary-shaped clusters without any assumption about the number of data clusters. This aspect is necessary and vital to face an inevitable characteristic of large data sets in real applications [
10,
11]. It is the advantage of most density-based methods. However, using Euclidean distance in most algorithms results in computational complexity, especially for large datasets [
12]. Inefficiencies when dealing with severely noisy datasets are also a common problem of density-based clustering, including the method shown in [
6]. Over recent years, many different methods have been proposed deriving from efforts to extend the ability of density-based methods [
13,
14,
15]. Also, several combinations of private approaches have been considered to form more efficient methods, such as the collaboration of density-based and grid-based clustering solutions [
13,
16,
17,
18,
19,
20,
21,
22]. The GRIDBSCAN [
13] is a development of the DBSCAN based on the grid approach to provide a solution to different densities that DBSCAN cannot overcome. It follows three phases. The first one selects appropriate grids for homogeneous density in each grid. The second one merges cells with similar densities and recognizes the most suitable values as
ε and
minPts to run the DBSCAN with these identified constants in the third phase. Even though getting the compared advantages in terms of accuracy of the generated CDS, the high time-consuming and matching with small-sized databases only are its considerable disadvantages.
To solve the above issues partly, the HDBSCAN [
16,
17], DPC [
18], and DPCSA [
19] considered sophisticated density-based approaches. As an extension of the DBSCAN, the HDBSCAN [
16] presented a multiple clustering in a hierarchy to get the best possible solution. It took
minPts as input and employed a non-parametric density estimation method followed by Hartigan’s notion of hard clusters to create a simplified cluster tree to obtain the final solution for an unlimited range of density thresholds. For the DPC [
18], it was a density-based clustering via the density-peak clustering approach to determine the optimal number of clusters in a dataset. The idea behind this strategy is that the cluster centers have larger densities and that the distances between them are significant. The method, however, is dependent on user input due to the manually selected center points from the decision graph. Yu et al. expanded the DPC by proposing the DPCSA [
19]. It used a two-stage cluster label assignment to data points to reduce error propagation. By using a dynamic closest neighbor table update approach, the DPCSA overcomes the constraints of DPC and improves clustering performance. However, a user still needs to select the cluster centers from the decision graph.
Recently, a fast-density grid-based method (DGB) was presented in [
20] with the improvement of computational time. This method proposes a type of density computation that has rarely been found in convenient algorithms before. Instead of calculating the density of grid cells as the famous algorithms like ENCLUS [
21], CLIQUE [
22], or DSTREAM [
23], it would compute the density of grid nodes only without considering that at each data point in the cells. There is no need to calculate the Euclidean distances between mutual data points to improve the computational complexity. However, like to the traditional density grid-based clustering methods, the challenge of misclassification caused by incorrectly recognizing the number of clusters may usually exist.
It can observe that density grid-based clustering owns promising advantages. However, the limitations mentioned above blur its effectiveness. Especially, the existence of noise may cause weak links between some grids that make the obtained results unstable and inaccurate. Focusing on this aspect, we propose a Density Grid-Based Clustering algorithm named SWCG-DGB using a strategy of Weak-Connected Grid search. First, we suggest a concept called Weak-Connected Grid Cell (WCG), by which we describe outside grids, neighboring grids, and a buffer. A fuzzy type approximation [
24,
25] is then employed to calculate the density of grid nodes for establishing initial clusters. A search strategy is carried out to seek and remove WCG existing in the initial clusters. As a result, Density Grid-Based data clusters are finally generated in the filtered data space. As usual, simulation surveys are performed to evaluate the SWCG-DGB. Our main contributions are as follows:
We point out a common drawback often appearing in the density grid-based clustering approach related to the WCG. Accordingly, we propose a solution for improving the clustering validity, including filtering noise and fine-tuning the generated CDS.
We propose a buffer for storing features of data points and the grid-cell’s coordinates. It allows detecting WCGs more effectively with a lower computational cost. The reason is that only the data points marked in the buffer are indicated as border or noise points instead of the entire dataset. This enhancement is very efficient for large datasets.
The rest of the paper is organized as follows. The related concepts are presented in
Section 2 to depict the SWCG-DGB in
Section 3. The following section shows surveys and evaluations via benchmark datasets. Finally,
Section 5 gives the vital conclusions of this approach.
2. Related Concepts
Definition 1.
(Characteristic Vector). The characteristic vector is a tuple (pos, C, mark) created by a structure type table. Here, pos is the position of grid cells (or cells for short), C is a set of data points in the same cell, and mark is the number of clusters marked for all cells in the cluster matrix.
Definition 2.
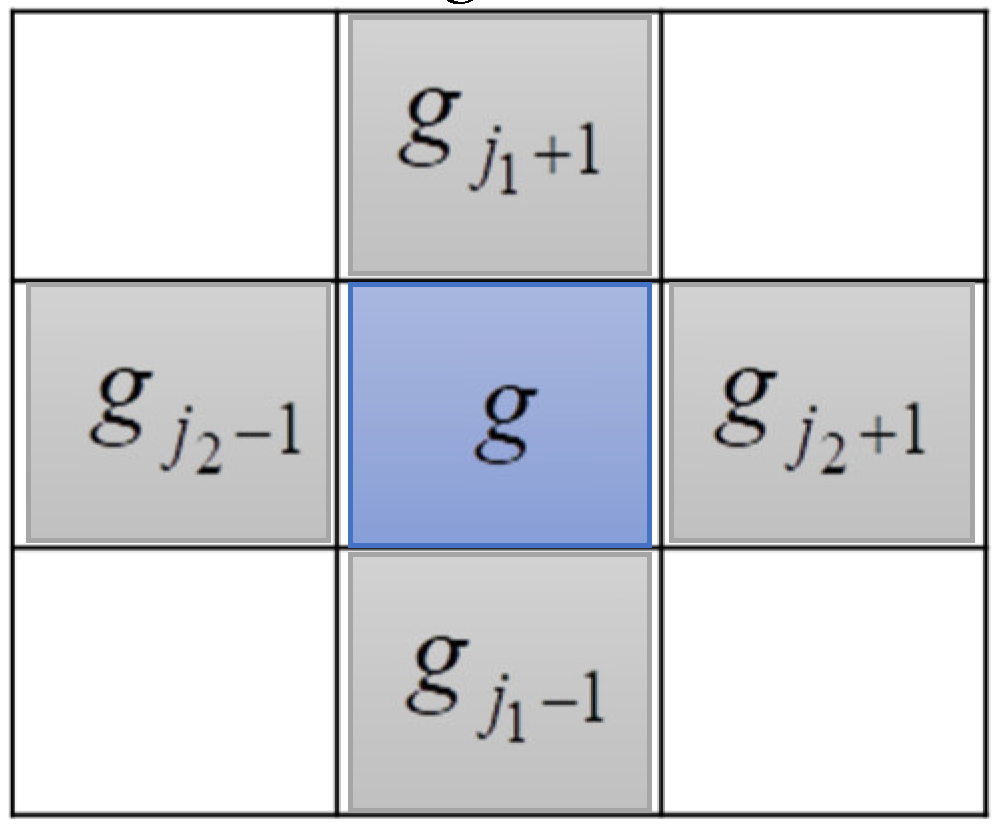
(Neighboring Cells). A cell is a neighbor of another one if both have at least one common boundary.
Let’s consider two cells
and
in a
k-dimension data space. Where
and
are the ordinal numbers on two different data dimensions. We follow the standard grid concept, to which the distance between any two neighboring nodes on an arbitrary data dimension is one. Accordingly, in general,
g1 and
g2 are neighbors (or neighboring cells, denoted
g1 ~ g2) if the two conditions in (1) must be satisfied:
Figure 1 shows the neighboring cells of cell
g in the two-dimension data space
Definition 3.
(Grid Cluster and Outside Grid) [
23]. A set of cells
is a grid cluster (or a cluster for short) if for any two cells
, there exist a sequence of cells
such that
,
, and
,
,…, and
. Consider
, if
gk has at least one neighbor not belonging to
G, then
gk is outside grid in
G.
Definition 4.
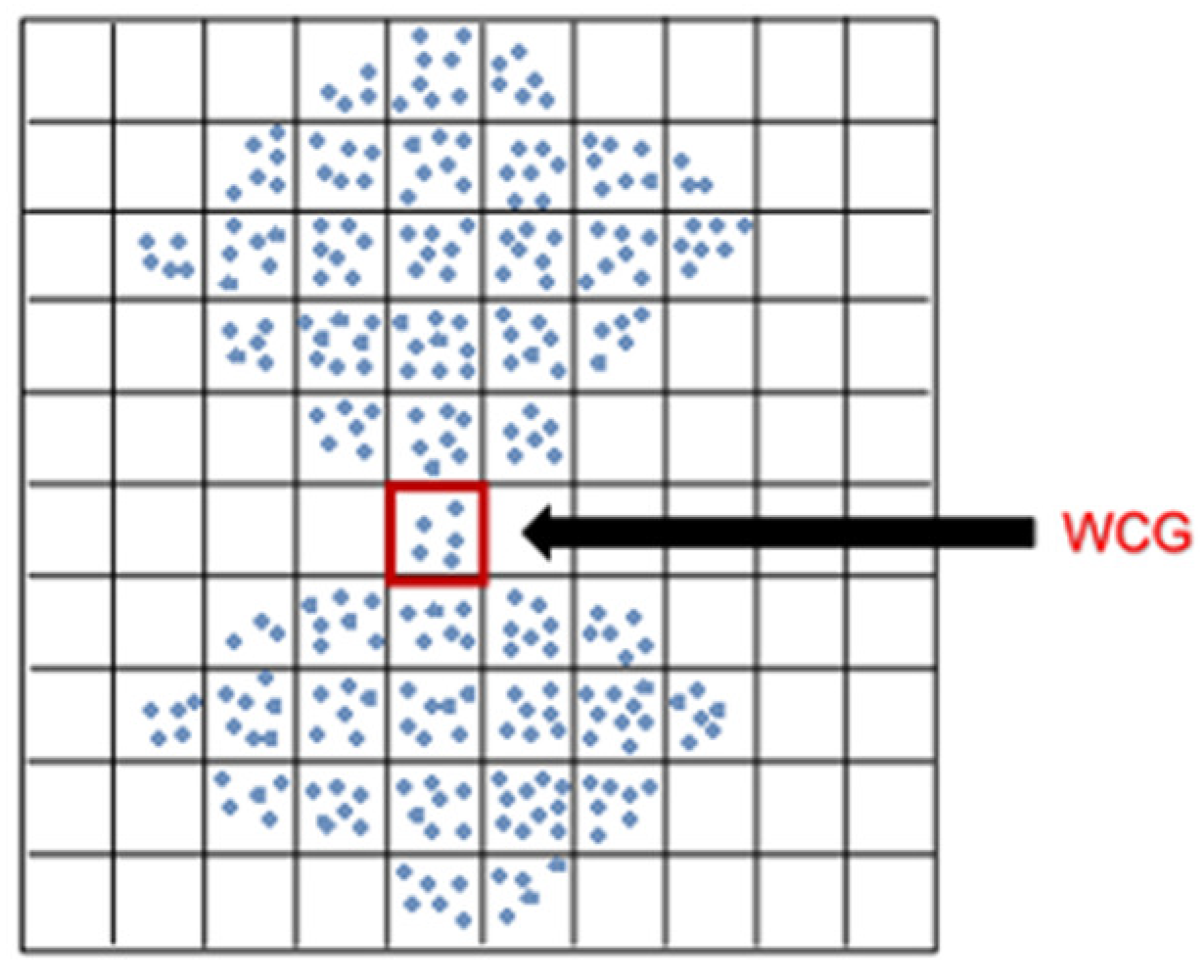
(Weak-Connected Grid Cell, WCG). Let’s consider a cell in a certain cluster. The cell is a WCG if both of the following statements are satisfied. 1) On one arbitrary data dimension, two neighbors of this cell belong to this cluster; and 2) Two of its neighbors on any of the remaining data dimensions do not belong to this cluster.
Figure 2 illustrates a WCG existing in a cluster. In this case, the cluster may be split easily into two smaller ones at this WCG. For our algorithm, the first separation step makes a rough space of initial clusters. This observation takes the role of a direction for the next phase: fine separation. The next section details these aspects.
Definition 5.
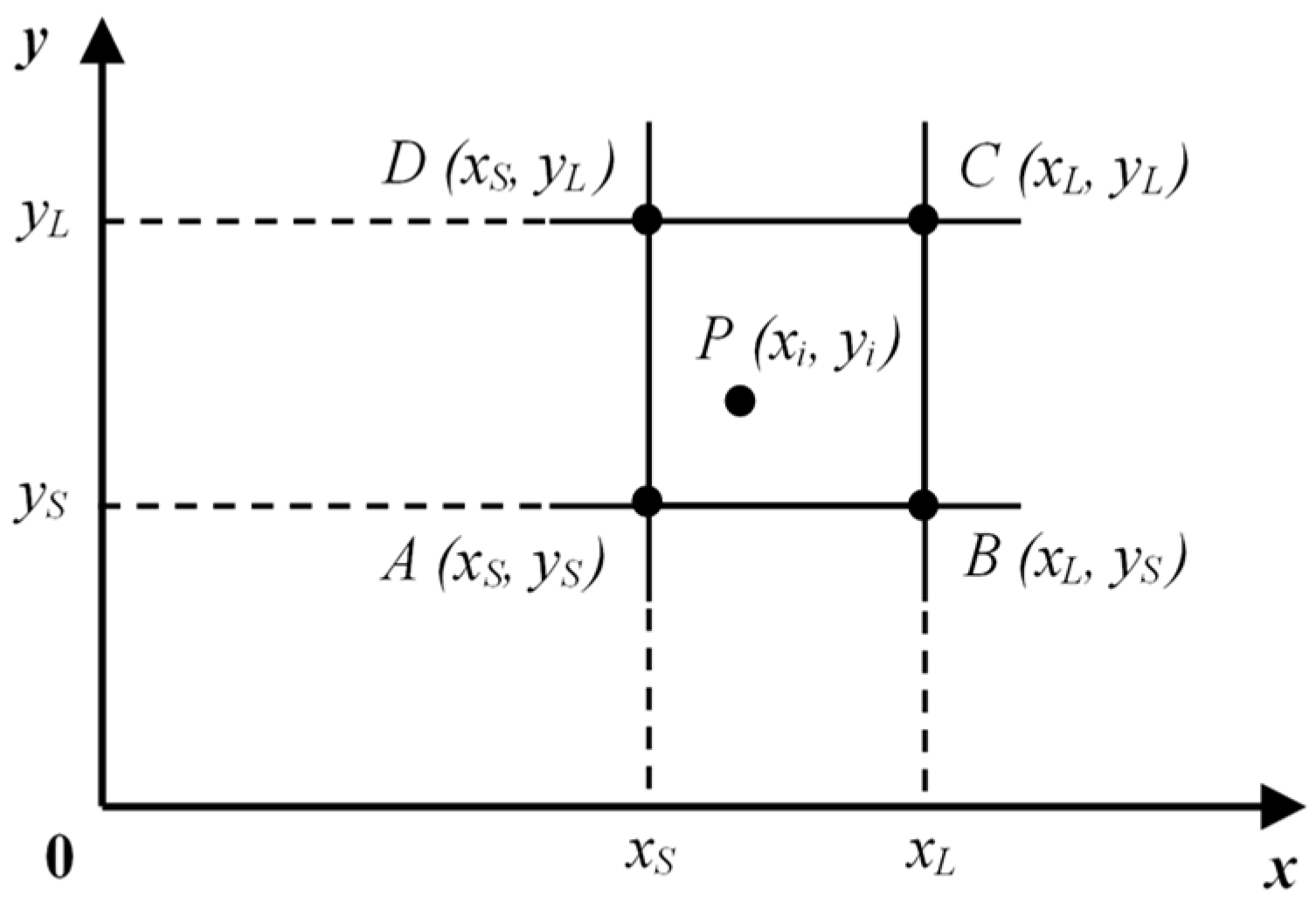
(Fuzzy Type Approximation). Let’s consider data point
P on the
d-th data dimension of a normalized
k-dimension data space. A fuzzy membership is defined as follows:
where,
λ is the length of each cell,
,
Pd is the coordinate of
P, and
Nd is the node’s coordinate in
d-th dimension (see
Figure 3).
In
Figure 3,
xS and
xL (
yS and
yL) are two coordinates on the
x-axis (
y-axis), signed
xSmall and
xLarge (
ySmall and
yLarge), respectively. Points
A,
B,
C, and
D are four nodes of the cell containing
P. The density of these nodes is calculated, and node
C (xL, yL), which is the representation of data point
P, is merged into the
struct_table type buffer.
Be noted that the distance between any two neighbor nodes in the standard grid (
λ) equals one. Equation (2) shows the contribution of each data point to its surrounding nodes and is utilized to calculate the local density of the nodes according to one data dimension. In the
n-dimensional data space, the node density is therefore calculated:
For example, for data point
P(
xi,
yi) in
Figure 3, the weighted fuzzy densities to four nodes A, B, C, and D are
The node’s local density is applied by summing up the density of all the variables according to (Equations 4-7):
dp is the density of data point
P, which is set to 1 in the general case to simplify the defuzzification process (8)-(11) because the denominator in these equations would always be one
Definition 6.
(Density Matrix). The density_matrix keeps the density of nodes. It has the size to be square of no_grid, which is the size of the grid in each dimension.
In the beginning, the density of all nodes is computed via equations (2-11) and stored in the density_matrix. It is then used for the mountain ridge searching process by finding the nodes with a density higher than a density threshold. The nodes owning a density value less than this threshold are marked as zero-density nodes. Hence, instead of processing all the nodes, one only needs to operate on a small number of nodes with non-zero density to accelerate the speed of the clustering process.
3. Proposed Method
Section 1 mentioned some typically density-based clustering methods, such as the DBSCAN [
6], HDBSCAN [
16], DPC [
18], DPCSA [
19], and DGB [
20]. Despite owning advantages in fast computation, they have a misclassification when finding the number of data clusters in many cases related to WCGs. Here, we present the algorithm SWCG-DGB for density-based clustering with a proposed solution for the above difficulty. It is an extension of the DGB [
20] with a supplemented vital tool for finding WCGs to filter noise, make an initial CDS, and perform fine-tuning of the CDS. Let’s consider a given source dataset. The main steps of the SWCG-DGB are as follows.
3.1. Initialization
It consists of normalizing the data space, finding the corner coordinates of each data point to map the data point into the buffer, and calculating the local density of nodes.
We normalize the original dataset to [0, 1], and then each data point is scaled into a grid range of [1,
no_grid]. Where,
no_grid is the size of the grid in each dimension. In other words, it is the number of cells in each data dimension. Here, we select
for all the data dimensions. As an example, with any data point
P(
xi,
yi) in
Figure 3, the normalization takes place as:
where
xmin (
ymin) and
xmax (
ymax) respectively are the minimum and maximum values of the dataset in
x-dimension (
y-dimension).
With each normalized data point, calculating local density for four nodes is done by Equations (8-12) using the fuzzy type approximation mentioned in Definition 5. These densities are saved in different local matrices, and then the sum of local density matrices is stored in a matrix named density_matrix with an initial setting to be zero.
3.2. Finding the Number of Clusters
Finding mountain ridges (or initial clusters) is a core process for the SWCG-DGB. In this progress, we map data points onto individual data clusters and store the information in a cell array named
CL_set. Algorithm 1 depicts this process. First, from the density of nodes sorted in descending order, the highest density is chosen for the mountain ridge searching task. Then, for each mountain ridge, its neighboring nodes whose density is higher than a threshold set up prior are merged into it. The mapping of data points into suited clusters happens regularly.
|
Algorithm 1. Searching mountain ridges |
Inputs: All data points.
Outputs: The updating position of the node in the buffer and CL set.
1: Calculating density of nodes and add them into density_matrix
2: Reshaping density_matrix, finding the nodei has max_density
3: while (max_density > edge_factor )
4: for i =1 to length(k) of high_density_nodes set
5: (xi, yi)=get_position(rowi, coli)
6: cluster_matrix(rowi, coli)=n (no_cluster)
7: updating node(xj, yj) into the buffer and CL_set
8: for m =k+1 to length of (no_grid*no_grid)
9: if (density of nodem > edge_factor)
10: (xm, ym)=get_position(rowm, colm)
11: if (node(xm, ym) is neighbor of node(xi, yi) )
12: cluster_matrix(rowm, colm) = cluster_matrix(rowi, coli)
13: density_matrix(rowm, colm)=max_density
14: high_density_nodes=[high_density_nodes m]
15: updating node(xm, ym) into the buffer and CL_set
16: end (if 11)
17: end (if 9)
18: end (for 8)
19: end (for 4)
20: high_density_nodes=[1]; n=n+1
21: end (while 3) |
In this algorithm we select a density threshold for finding the first mountain ridge. It is the maximal density of random grid cells arranged in density_vector. Then we get the position of this cell and assign it the corresponding location in cluster_matrix equals no_cluster (n), which is the name of the cluster and was set one in the first stage. After that, we examine the position of the next cell in the density_matrix and get the cells’ position. It is done if it has a high enough density. As a result, the neighbors of those cells contain nodes satisfying the edge_factor threshold condition, which is a ratio of the prior maximum peak density value. The value of the edge_factor parameter is inferred in the initial step based on the characteristics of each dataset. Simultaneously, we tune the grid ratio value to obtain clustering results. Besides, merging and marking these cells’ data points into the buffer and mapping them onto CL_set are conducted. This process establishes core clusters containing most of the source data points in cells that are the high-density areas.
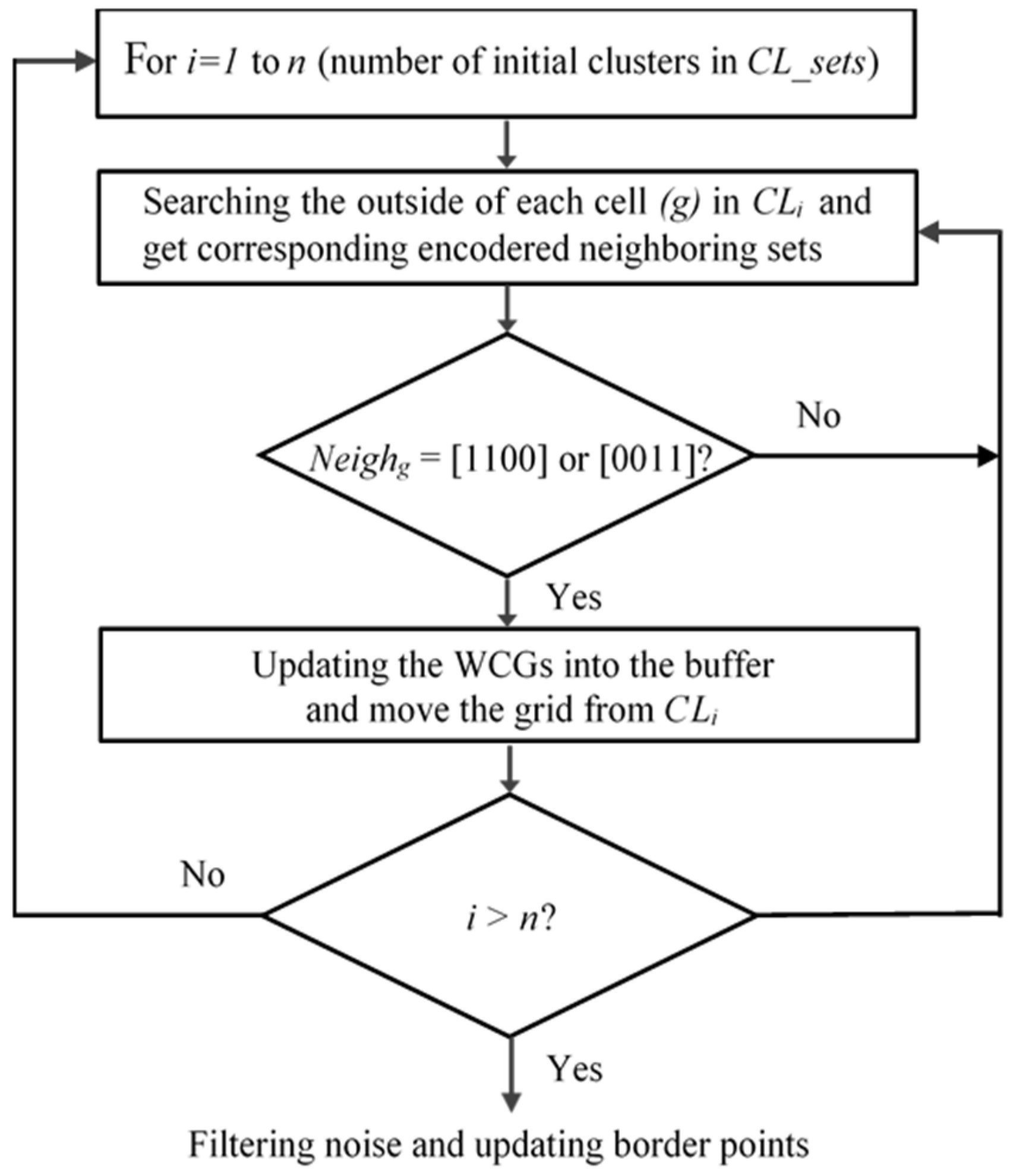
3.3. Searching the Weak-Connected Grids
Searching for WCGs is a meaningful development in our algorithm. With every cluster formed and located in
CL_set, we search for the WCGs inside it. One thing noted in this process is that after finding out the WCGs, we do not delete data points inside but just move and mark them into the buffer as a zero-number, which are called
zero_marked_data. These data will be reclassified and returned to those clusters. This essential work ensures the maximum avoidance of data loss. The workflow for searching the WCGs is in
Figure 4.
In the above process, outside grids are searched in each cluster and then we check the order of neighboring grids in terms of binary code. An outside grid is a WCG if its encoded neighboring set is [1 1 0 0] or [0 0 1 1]. In other words, an outside grid is a WCG if it has two neighbors on the same dimension located in the same cluster and the other neighbors are not in this cluster. When the WCG is found, data points in it are moved from the cluster and marked with a zero-number in the buffer for filtering noise and updating border points in the next part. The following work is to get the position of WCGs and mark them in the
density_matrix for splitting big clusters into smaller clusters separately. The search for the WCGs is depicted in Algorithm 2 below.
|
Algorithm 2. Searching WCGs |
Input: All grids on the CL set.
Output: Detecting and moving the WCGs out of each CL set.
1: for i =1 to length_of CL_set
2: for m =1 to length_of CLi
3: if gridm is outside of CLi
4: [Neighm]= get_neighbors of gridm
5: if Neighm =[0; 0; 1; 1] or [1; 1; 0; 0]
6: marking data points of (gridm) as noise in buffer
7: moving data points of (gridm) out of CLi
8: (xm, ym)=get position of (gridm) from buffer
9: density_matrix (rowm, colm) = 0
10: end (if 5)
11: end (if 3)
12: end (for 2)
13: end (for 1) |
3.4. Filtering Noise and Updating Border Points
Algorithm 3 below describes the progress for filtering noise and updating border points. In the finding of mountain ridges presented in the above section, the border nodes are detected via setting a threshold that is the
edge_factor parameter for identification of mountain ridges and border points. From the
noise_threshold set up via the initial parameter function, nodes whose local density is smaller than the threshold are considered noise.
|
Algorithm 3. Filtering noise and updating border points |
Input: All points of zero-number_grid set in buffer.
Output: Removing noise and updated border points in obtained clusters.
1: for i =1 to length of zero-number_grid set in buffer
2: get_position(rowi, coli);
3: if ( cluster_matrix(rowi, coli)=0 and
density_matrix (rowi, coli) > noise_threshold )
4: for k=1 to 4
5: if cluster_matrix (nodek) > 0
6: satisfactory_node = satisfactory_node+1;
7: sum = sum(satisfactory_node);
8: end (if 5)
9: end (for 4)
10: end (if 3)
11: if sum = 1, get density of satisfactory node
12: if node_ density <noise_threshold
13: cluster_matrix (rowi, coli)=0;
14: gridi = noise; break;
15: else
16: move gridi to corresponding cluster;
17: end (if 12)
18: end (if 11)
19: if sum > 1, get total density of satisfactory nodes
20: if total node_ density <noise_threshold
21: cluster_matrix (rowi, coli)=0;
22: gridi = noise; break;
23: else
24: move gridi to corresponding cluster;
25: end (if 20)
26: end (if 19)
27: end (for 1) |
Be noted that the buffer for storing and marking the original data set above-mentioned makes finding noise and updating border points convenient. Instead of searching for all data points in the entire data set, we only handle data points marked zero that does not belong to any arbitrary cluster. It allows for speeding up the process, which is vital for real datasets in real-world applications, especially data streams changing over time.
The entire progress of SWCG-DGB is presented by Algorithm 4.
|
Algorithm 4. The SWCG-DGB |
1: Normalizing each data point P(xi,yi) following equations (13) and (14).
2: Calculating corner coordinates of each data point P(xi,yi) and update C(xL, yL) into the buffer.
3: Calculating the local density of nodes following equations (4)-(7) and add them into the density_matrix.
4: Finding the mountain ridges (or initial clusters) and map data points corresponding to these clusters into CL_sets (Algorithm 1).
5: Searching the Weak-Connected Grids (Algorithm 2).
6: Filtering Noise and Updating Border Points (Algorithm 3). |
4. Experimental Simulation
4.1. Approach
As usual, the accuracy of the quantified cluster number (QCN) is the coinciding degree between the QCN and the actual (‘right’) number of clusters (RNC). The reasonableness in the distribution of samples in the created cluster space reflects clustering validity. In addition, one often considers calculating time (CT) in actual applications. As a result, the higher the accuracy of QCN and the clustering validity degree along with the lower the CT, the higher the clustering effectiveness. In this section:
Based on the above direction in evaluating, we compare the proposed SWCG-DGB method with the five other density-based clustering methods discussed in
Section 1, namely DBSCAN [
6], HDBSCAN [
16], DPC [
18], DPCSA [
19], and the density grid-based clustering method, DGB [
20]. The two HDBSCAN and DBSCAN need user input parameters. Hence, we try to specify the best values of their parameters before the surveys. To benefit DPC and DPCSA, the detected number of clusters is adjusted to the correct number of classes in accordance with the decision graph. Due to the DGB and SWCG-DGB not needing any input parameter, we carry out surveys with similar no_grid values. Accordingly, we set up their parameters as in
Table 2.
We employ six 2-dimensional synthetic datasets with the pre-observed RNC detailed in
Table 1 and
Figure 5 to evaluate the SWCG-DGB. These datasets are commonly used as benchmarks to test (
https://github.com/deric/clustering-benchmark). The surveys are on MATLAB R2019a under the Windows 10, 64-bit operating system, Intel (R) Core (TM) i5-1135G7 @ 2.40 GHz, 16GB RAM.
4.2. Evaluation Measures
The accuracy of the quantified cluster number (QCN) is evaluated via visualization and the calculating time (CT) are compared. For a given synthetic dataset, the target is to seek QCN such that QCN is equal to its RNC with a CT as low as possible. Besides, Rand Index (RI), Adjusted Rand Index (ARI) and Normalized Mutual Information (NMI) are employed to estimate the performance of comparing algorithms. Both of their values are in [0, 1]. And better clustering performance is indicated by higher values for them.
The RI [
27] is a measure to compare the output of a clustering algorithm with actual clusters. This measure may also be used for comparing the results of two clustering methods. RI is described as follows:
In (15), n is the total number of samples, X and C represent two distinct sets of clusters, and a and b represent the number of pairs of samples that belong to (non-similar) distinct groups in X and C, respectively.
Another measure is the ARI [
28] that is used to assesses the clustering results via assuming a generalized hypergeometric distribution as the model of randomness. This measure is presented as follows:
where
X is the results and
C denotes the actual labels.
nij is the number of samples same in both clusters
xi and
,
ai and
bi are respectively the number of the same samples of
xi and
ci clusters.
Finally, NMI [
29] is a widely employed metric to evaluate clustering approaches. The following definition, this metric is applied information theory to quantify the differences between two clustering partitions, is:
In the above, Mutual Information (MI) achieves the degree of information between the two random variables and is defined in (18).
where
p(
xi,
ci) is the probability of belonging an instance to clusters
xi and
ci at the same time. Additionally,
p(
xi) and
p(
ci) are the probabilities that an instance will belong to associated clusters
xi and
ci, respectively. Take note that NMI takes its values between 0 and 1. It takes the value of 1 if two clusters are precisely similar, and it takes the value of 0 if two clusters are independent.
4.3. Simulating Results on Synthetic Datasets
First, we employed the five methods to cluster Dataset 1, which contains 3300 two-dimensional data points with 5% noise and RNC=9 (see
Figure 5 and
Table 1). The results in
Figure 6 and
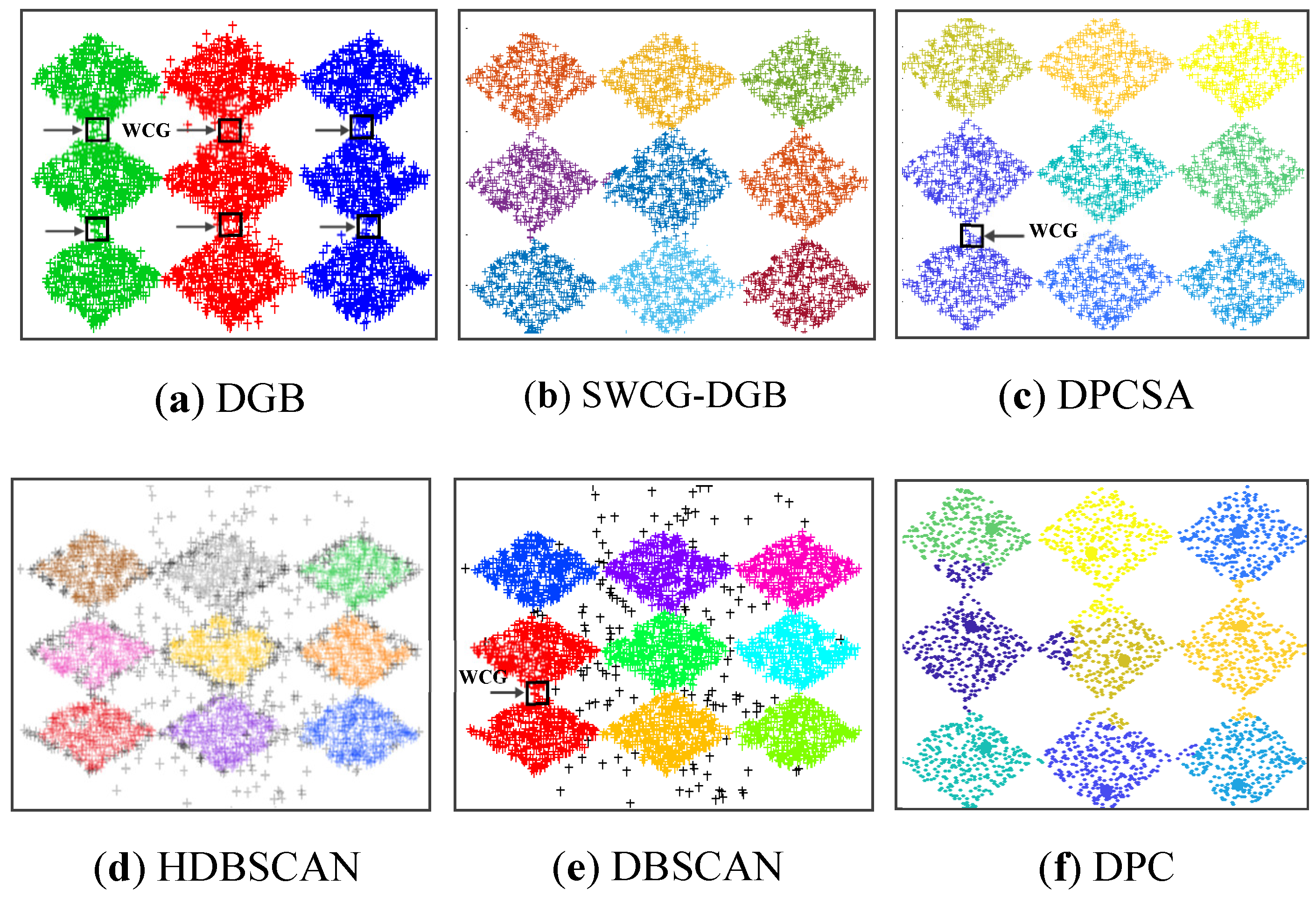
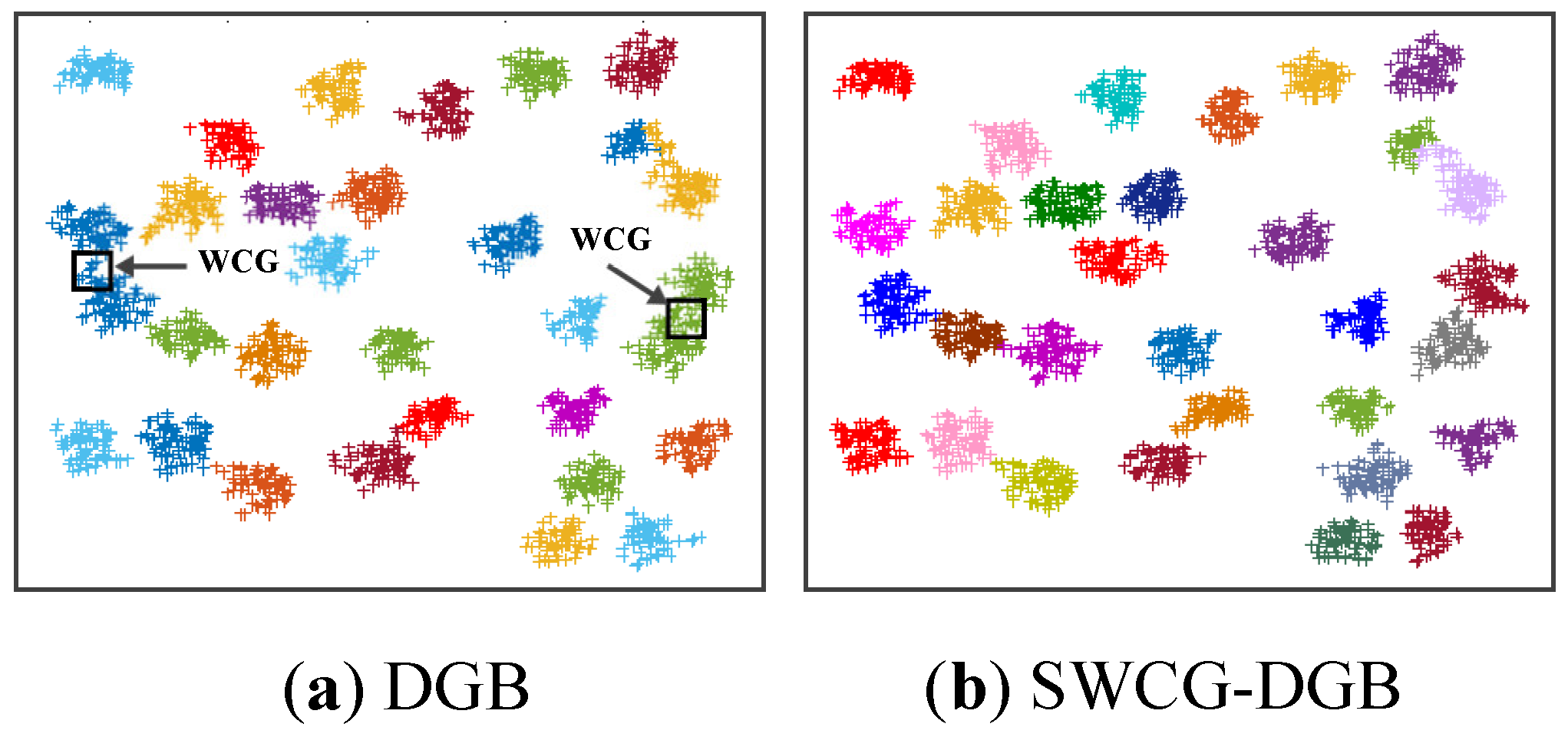
Table 3 reflect that the SWCG-DGB could cluster the database correctly with QNC= RNC=9. The other methods are inferior to the SWCG-DGB. Namely, QNC=3 for the DGB, QNC=8 for the DBSCAN, and QNC=10 for the DPCSA. For the DPC and HDBSCAN, despite the accurate number of clusters (QNC=9=RNC), some confusion between border points and noise is present in imperfect data clusters.
For Dataset 2,
Figure 7 and
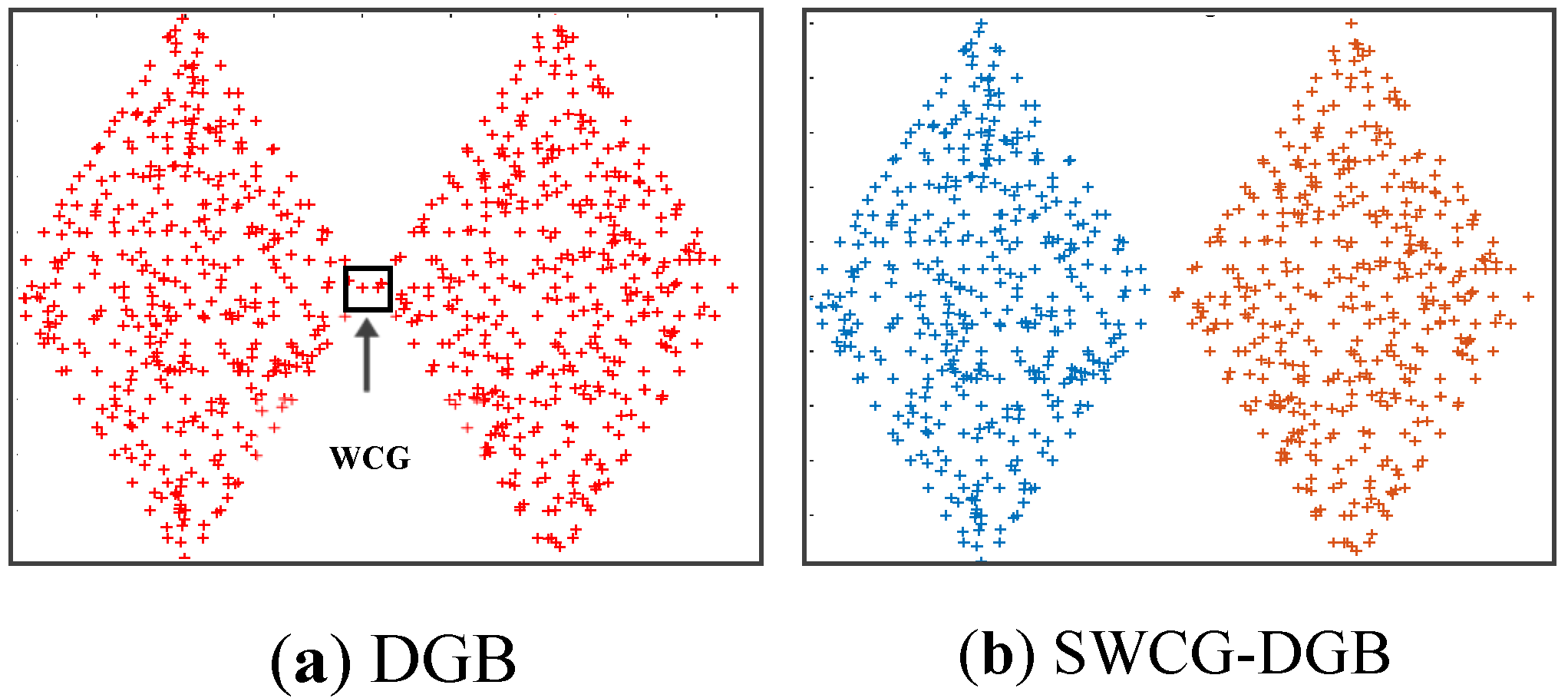
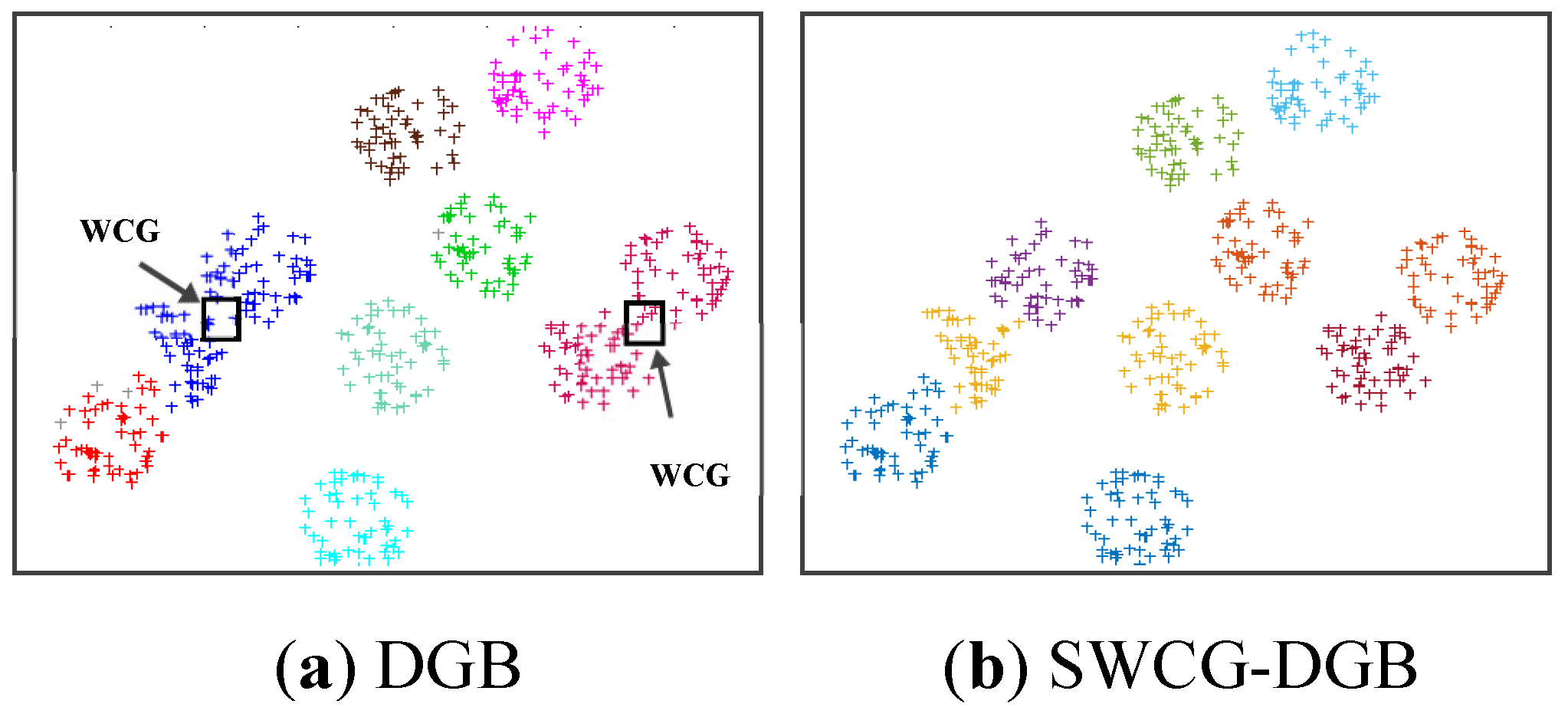
Table 3 show the SWCG-DGB could find the correct number of clusters (QNC= RNC=2), while the DGB is incorrect with QNC=1.
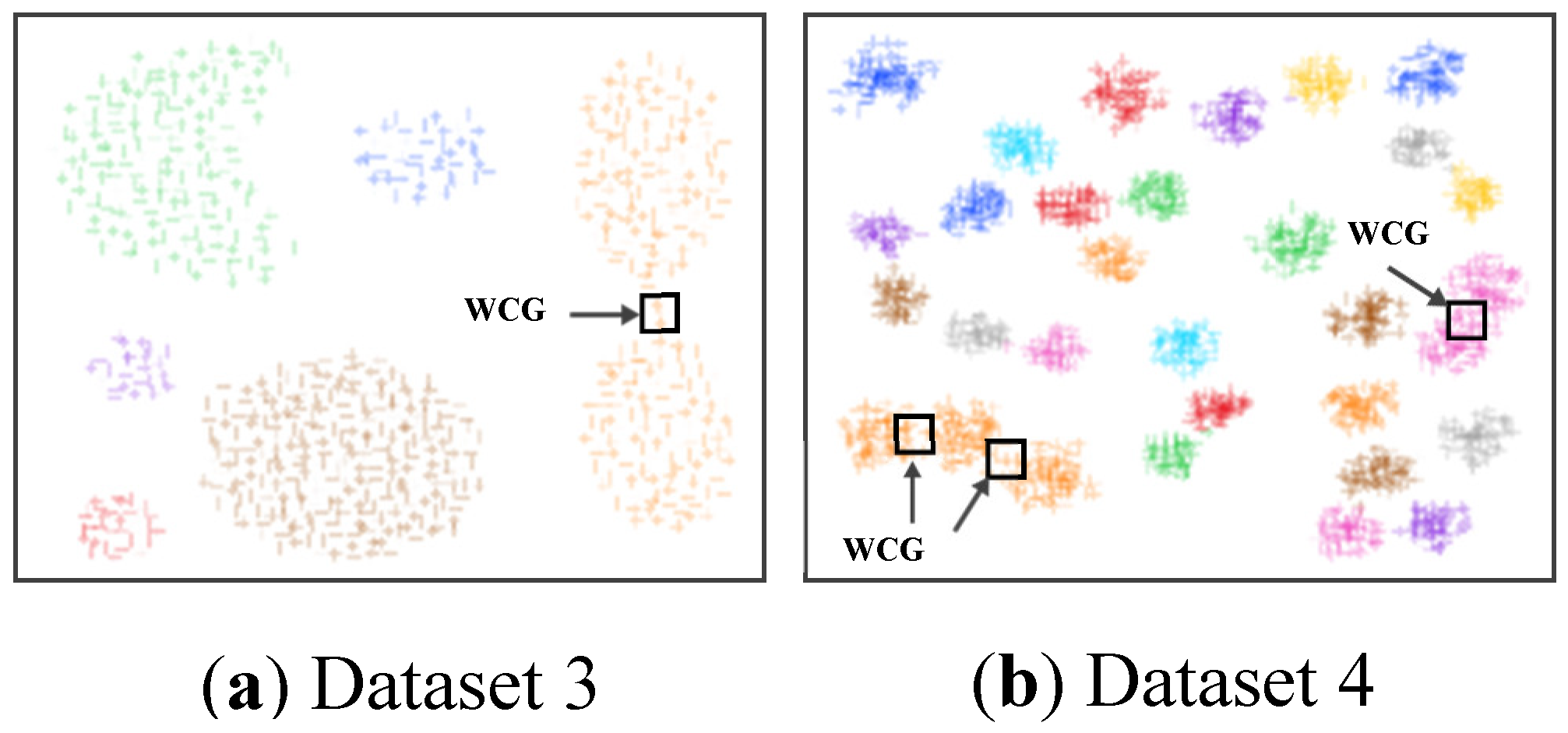
For Dataset 3 and Dataset 4, the compared effectiveness of the SWCG-DGB is much the same.
Figure 8 and
Figure 9 and
Table 3 depict QNC=RNC=7 for Dataset 3 and QNC= RNC=31 for Dataset 4 based on the proposed method.
Similarly, for Dataset 5 and Dataset 6 in
Figure 10 and
Figure 11 and
Table 3, they show the accuracy of the SWCG-DGB better than DGB with QNC=RNC=10, and DPCSA with QNC=RNC= 20 regarding Dataset 5 and 6, respectively.
In other words, the SWCG-DGB could always accurately quantify the cluster data space of the datasets in
Table 2, even in the presence of noise in the case of Dataset 1. Its advantage is the capability to recognize WCGs to adjust finely and re-establish the created CDS more reasonably. It is easy to see from
Figure 6,
Figure 7,
Figure 8,
Figure 9,
Figure 10,
Figure 11,
Figure 12 and
Figure 13 that the WCG generated a bridge between two adjacent clusters to form either an inappropriately larger one or data clusters with unsuitable data distribution. This aspect is the main reason for the incorrect results of the method DGB.
Besides,
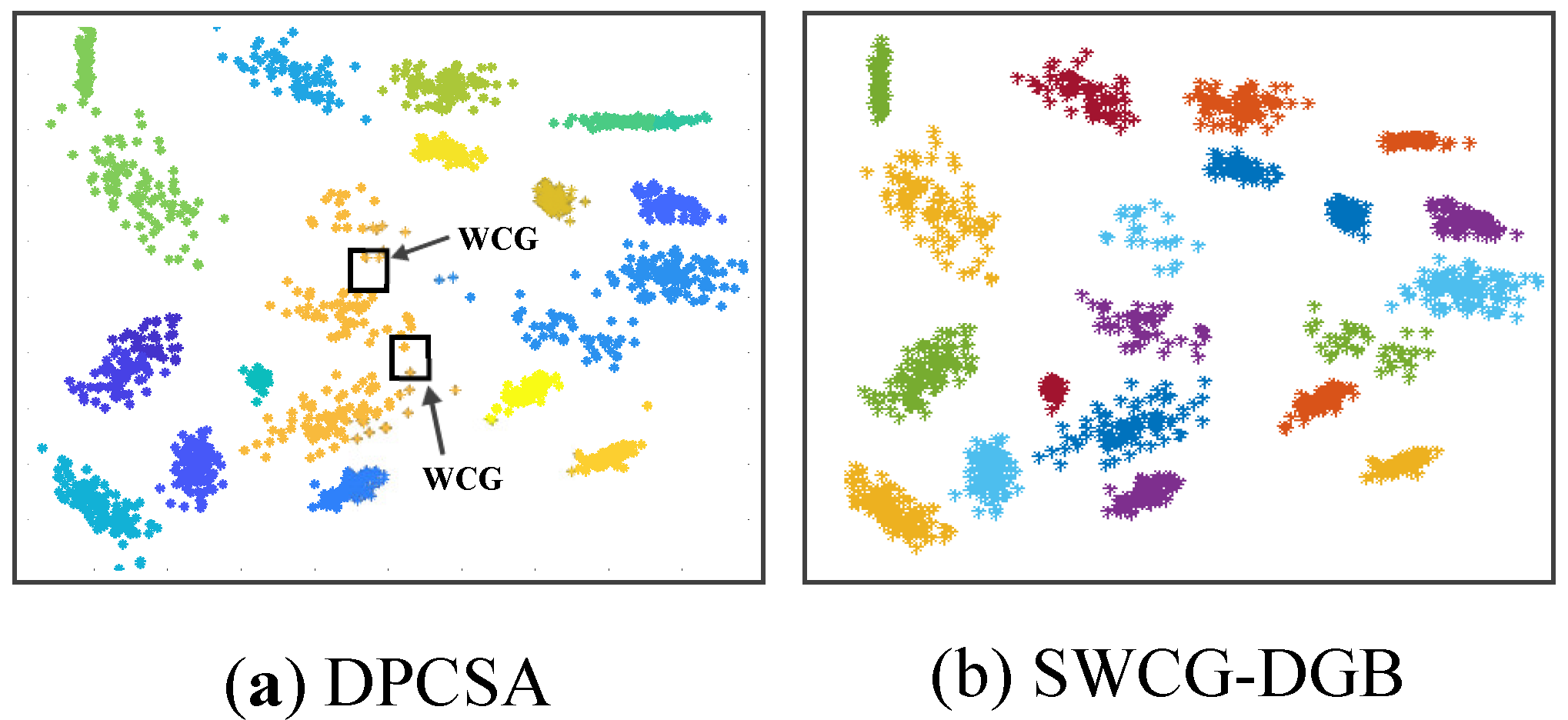
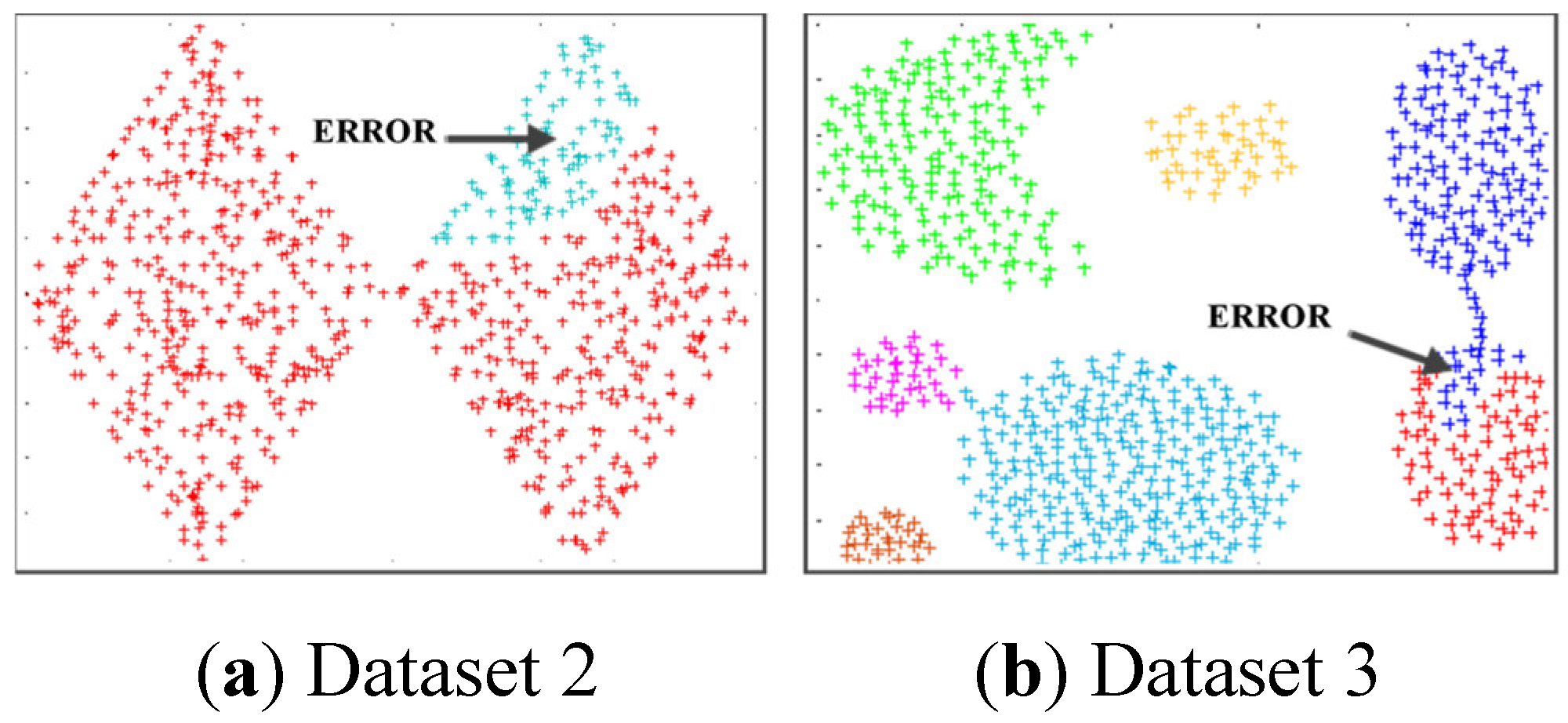
Figure 12 reflects the incorrect clustering results of the DPCSA when facing Dataset 2 and Dataset 3 as for Dataset 1 of the DPC in
Figure 6. In this case, the WCG causes the data classification error denoted in the figure. Similarly,
Figure 13 shows that WCG leads to the incorrect QNC of Dataset 3 and Dataset 4 with the HDBSCAN. It is much the same as the issue of the DGB abovementioned. Owning the ability to recognize WCGs, the proposed SWCG-DGB can overcome these difficulties to set up the fit cluster data spaces illustrated in
Table 3 and
Figure 6,
Figure 7,
Figure 8,
Figure 9,
Figure 10 and
Figure 11.
Together with evaluating the clustering validity via comparing the QNC and RNC and estimating the reasonableness of data distribution in each cluster, we also surveyed the running time or calculating time (CT) of the methods.
Table 3 and
Table 4 show that the SWCG-DGB is the best in the clustering validity but is not the fastest method. However, their difference is not much.
In addition, the performance of the clustering is then evaluated using three metrics, including RI, ARI, and NMI. We show the metrics on the six synthetic datasets in
Table 1. The results are shown in
Table 5,
Table 6 and
Table 7, and the best results are highlighted in boldface in each column.
Following the results reported, in most instances, the proposed method obtained the best results. From these results, we can see clearly that the performance of the SWCG-DGB is much better than the other clustering methods.
4.3. Simulating Results on Real Datasets
In this section, we compare the capacity of six clustering algorithms on three real datasets that described in
Table 1. Namely,
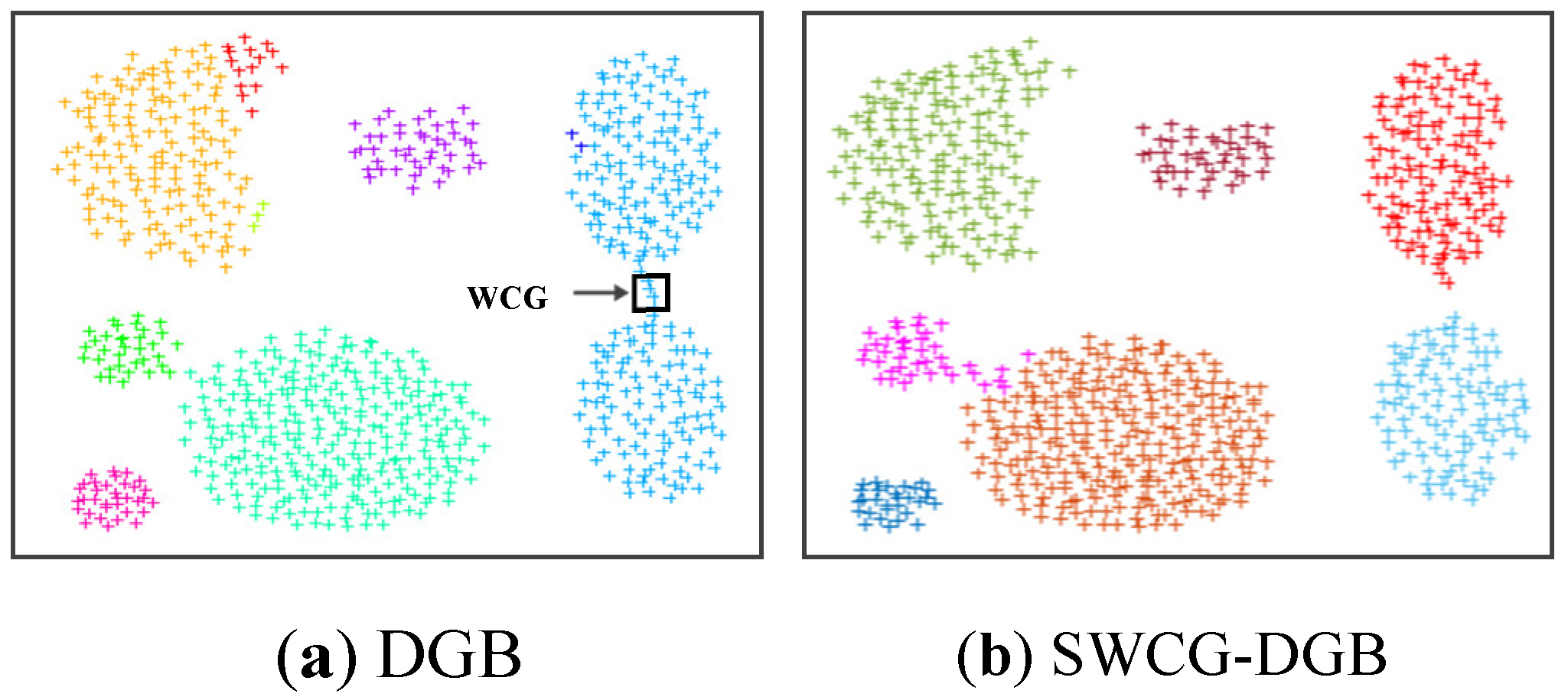
Figure 14 and
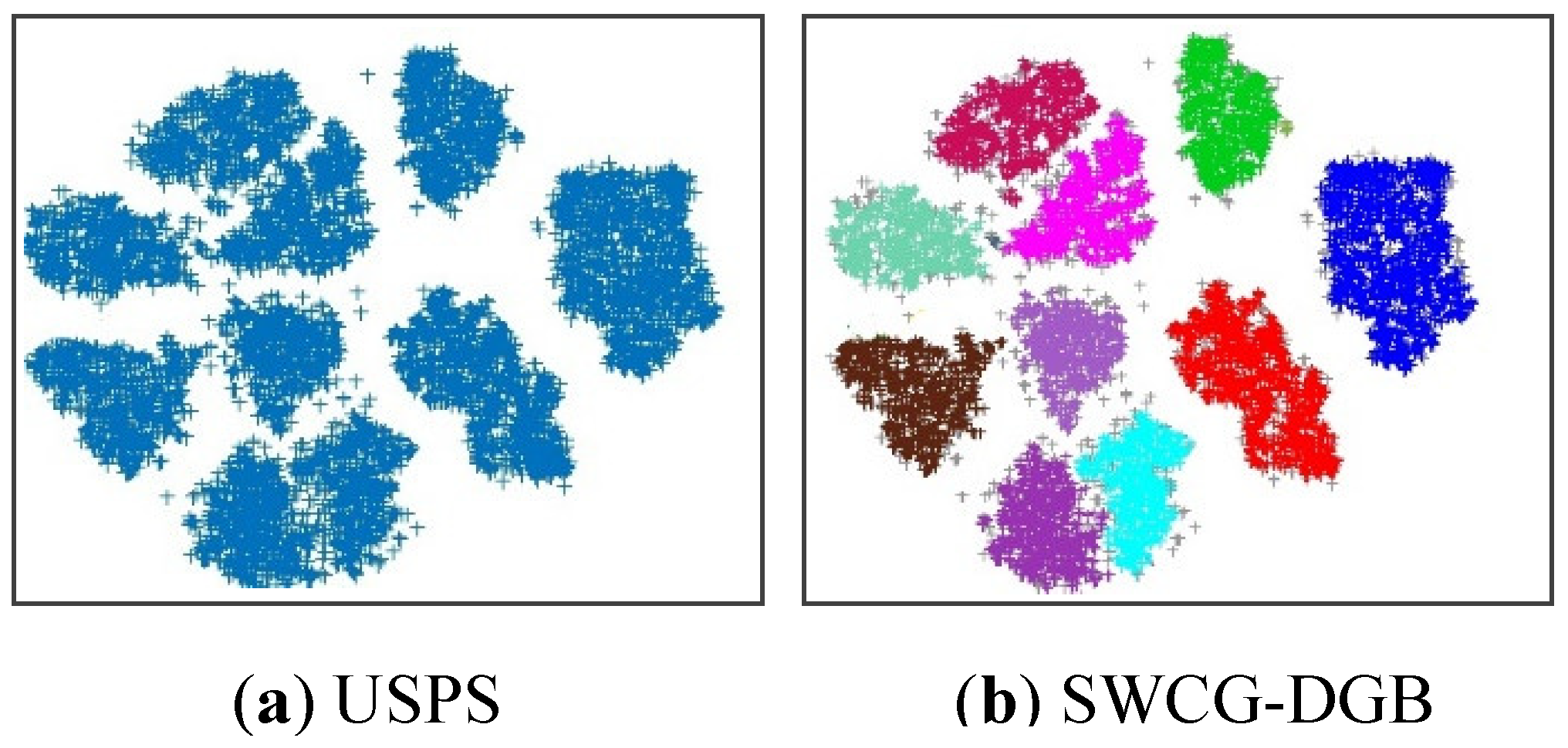
Figure 15 show one popular image data set (USPS) [
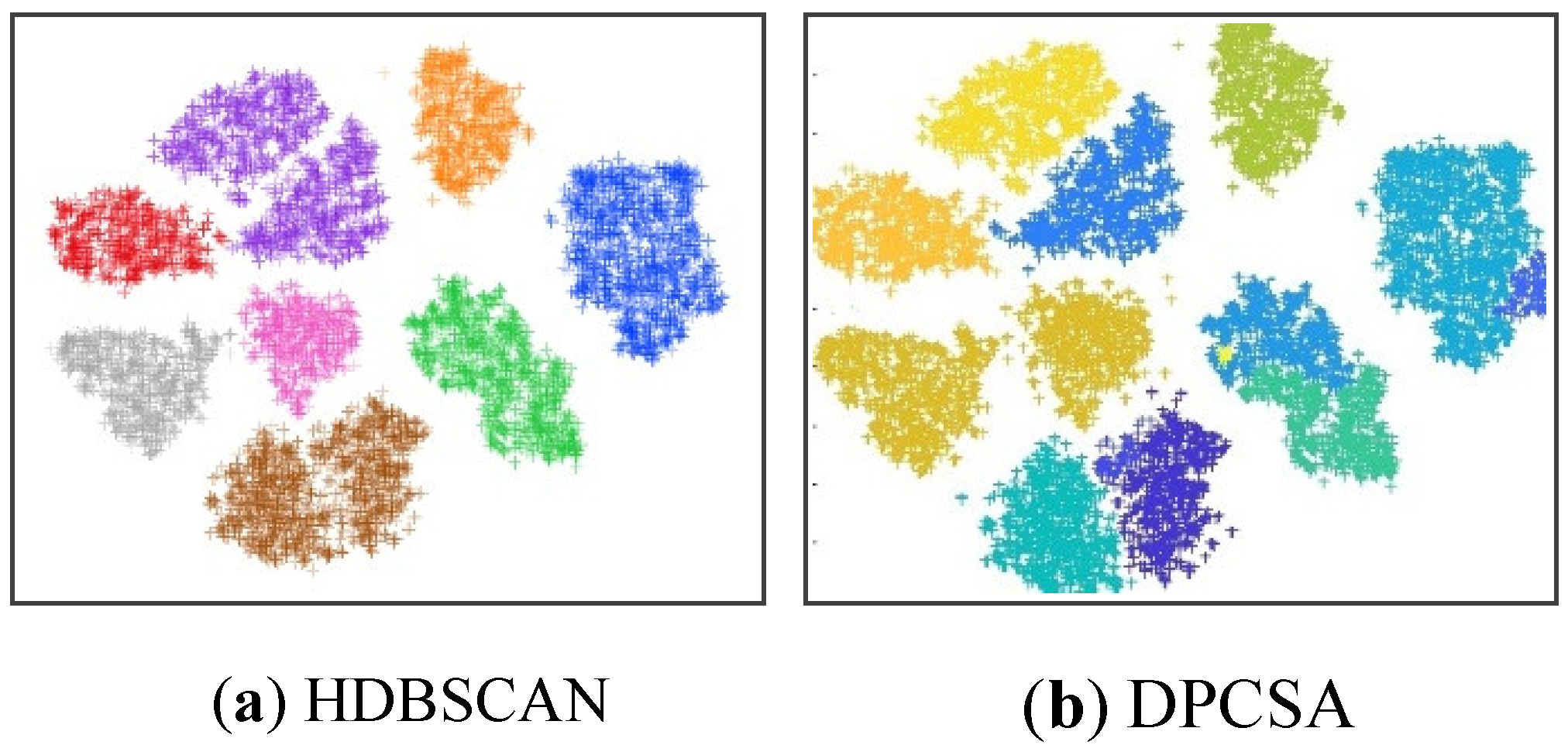
30], which the U.S. postal service collects from handwritten numbers on envelopes, along with the correctly clustered result of the proposed algorithm and the incorrectly clustered results of the HDBSCAN and DPCSA. All results on the number of clusters are shown in
Table 8 below. In general, for all three real datasets, the SWCG-DGB could find the correct number of clusters (QNC= RNC), similar to the DGB, while the remainder methods had some incorrect cases.
Besides, the clustering results obtained in
Table 9 evaluate the surveyed methods via the RI, ARI, and NMI using these real datasets. In each column, the best results are highlighted in boldface. It can see that the proposed method yields the best results in most cases.
5. Conclusions
The essential improvement to the density grid-based clustering to propose the method named SWCG-DGB has been presented. This approach has a remarkable advantage related to WCGs in solving the common issue of density grid-based clustering. The task of seeking WCGs to remove noise and adjust the created CDS takes a vital role in improving the clustering validity, including filtering noise and fine-tuning the CDS. In this phase, the calculating strategy without the Euclidean distance is to decrease the complexity of the clustering process. Besides, the buffer explored to store features of data points and coordinates of the grid cells is a fit technique solution. It allows for reducing considerable computational time in filtering noise and updating border points.
Many survey results show that the proposed SWCG-DGB can always better accurately quantify the cluster data space of the given datasets. Accordingly, clustering validity based on the SWCG-DGB is better than the other considered methods. However, the calculating cost is not a comparative advantage of this method. Improving the calculating time, hence, is our motivation in the following research.
Figure 1.
Neighbors of cell g in two-dimension data space.
Figure 1.
Neighbors of cell g in two-dimension data space.
Figure 2.
A WCG appearing between the two clusters in the two-dimension data space.
Figure 2.
A WCG appearing between the two clusters in the two-dimension data space.
Figure 3.
Node’s local density in the two-dimensional data space.
Figure 3.
Node’s local density in the two-dimensional data space.
Figure 4.
Workflow for searching the WCGs.
Figure 4.
Workflow for searching the WCGs.
Figure 5.
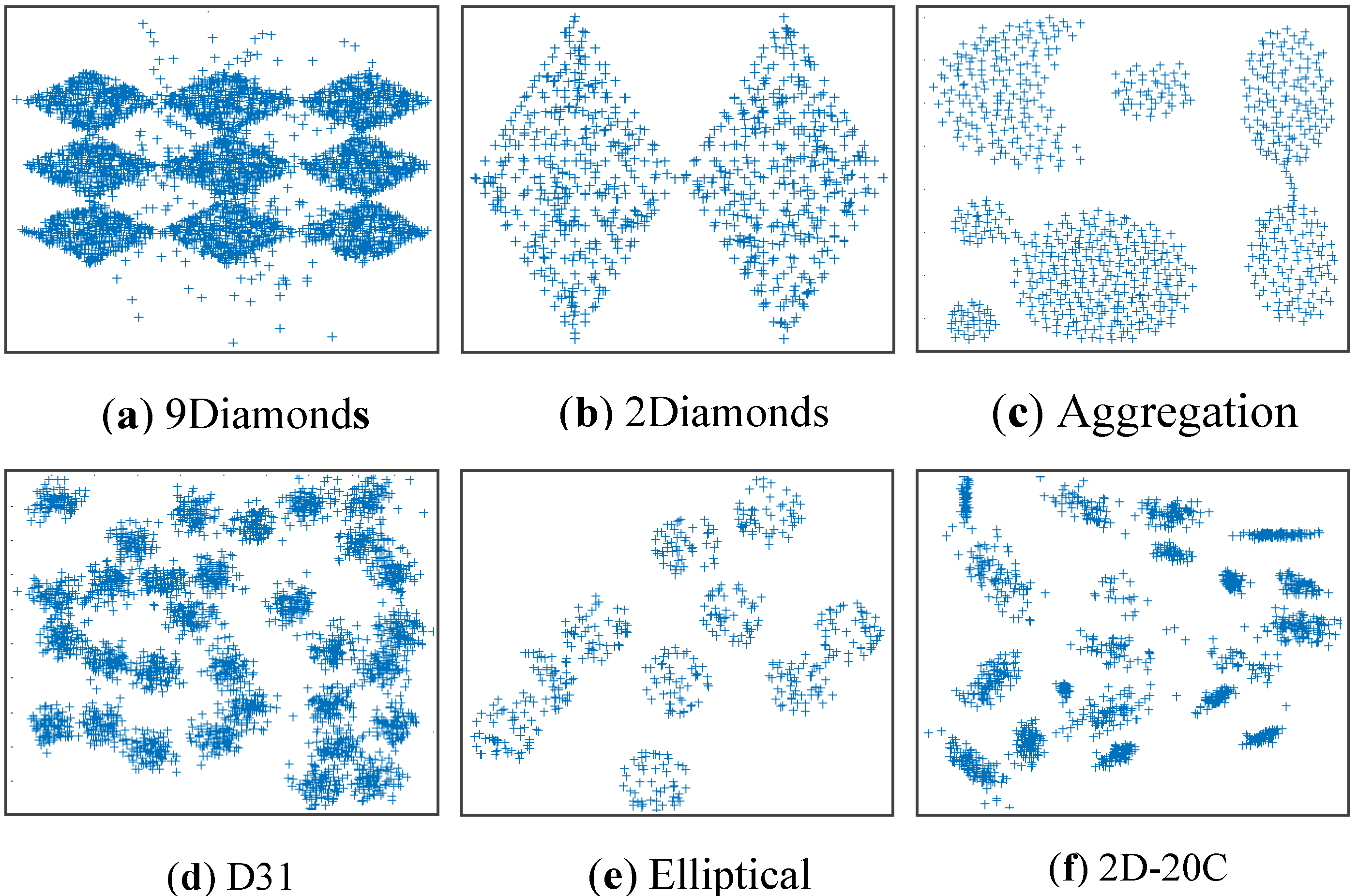
The six benchmark datasets: 9Diamonds (Dataset 1), 2Diamonds (Dataset 2), Aggregation (Dataset 3), D31 (Dataset 4), Elliptical (Dataset 5), and 2D-20C (Dataset 6).
Figure 5.
The six benchmark datasets: 9Diamonds (Dataset 1), 2Diamonds (Dataset 2), Aggregation (Dataset 3), D31 (Dataset 4), Elliptical (Dataset 5), and 2D-20C (Dataset 6).
Figure 6.
The comparative results deriving from Dataset 1 and the six methods consisting of DGB (no_grid=20), SWCG-DGB (no_grid=20), HDBSCAN (minPts=15), DBSCAN (minPts=14; ε=4), DPC, and DPCSA.
Figure 6.
The comparative results deriving from Dataset 1 and the six methods consisting of DGB (no_grid=20), SWCG-DGB (no_grid=20), HDBSCAN (minPts=15), DBSCAN (minPts=14; ε=4), DPC, and DPCSA.
Figure 7.
The comparative results between the SWCG-DGB and DGB (no_grid=22) with Dataset 2.
Figure 7.
The comparative results between the SWCG-DGB and DGB (no_grid=22) with Dataset 2.
Figure 8.
The comparative results between the SWCG-DGB and DGB (no_grid=16) with Dataset 3.
Figure 8.
The comparative results between the SWCG-DGB and DGB (no_grid=16) with Dataset 3.
Figure 9.
The comparative results between SWCG-DGB and DGB (no_grid=55) with Dataset 4.
Figure 9.
The comparative results between SWCG-DGB and DGB (no_grid=55) with Dataset 4.
Figure 10.
The comparative results between SWCG-DGB and DGB (no_grid=44) with Dataset 5.
Figure 10.
The comparative results between SWCG-DGB and DGB (no_grid=44) with Dataset 5.
Figure 11.
The comparative results between SWCG-DGB (no_grid=44) and DPCSA with Dataset 6.
Figure 11.
The comparative results between SWCG-DGB (no_grid=44) and DPCSA with Dataset 6.
Figure 12.
The incorrect clustering results of the method DPCSA with Dataset 2 and Dataset 3.
Figure 12.
The incorrect clustering results of the method DPCSA with Dataset 2 and Dataset 3.
Figure 13.
The incorrect clustering results of the method HDBSCAN with Dataset 3 and 4.
Figure 13.
The incorrect clustering results of the method HDBSCAN with Dataset 3 and 4.
Figure 14.
The original USPS dataset and correctly clustering results of the proposed method SWCG-DGB (no_grid=66) on it.
Figure 14.
The original USPS dataset and correctly clustering results of the proposed method SWCG-DGB (no_grid=66) on it.
Figure 15.
The incorrect clustering results of the methods HDBSCAN (minPts=15) and DPCSA with the number of clusters in the USPS dataset are 12 and 11, respectively.
Figure 15.
The incorrect clustering results of the methods HDBSCAN (minPts=15) and DPCSA with the number of clusters in the USPS dataset are 12 and 11, respectively.
Table 1.
Details of the datasets.
Table 1.
Details of the datasets.
| Datasets |
Samples |
Dimension |
RNC |
Noise |
Dataset 1 (9Diamonds)
Dataset 2 (2Diamonds)
Dataset 3 (Aggregation)
Dataset 4 (D31)
Dataset 5 (Elliptical)
Dataset 6 (2D-20C) |
3300
800
788
3100
500
1517 |
2
2
2
2
2
2 |
9
2
7
31
10
20 |
Yes (5%)
No
No
No
No
No |
Dataset 7 (Iris)
Dataset 8 (Seeds)
Dataset 9 (USPS) |
150
210
9298 |
4
7
256 |
3
3
10 |
No
No
No |
Table 2.
Input parameters of the methods in each survey.
Table 2.
Input parameters of the methods in each survey.
| Datasets |
HDBSCAN |
DBSCAN |
DGB |
SWCG-DGB |
| minPts |
minPts /ε
|
no_grid |
no_grid |
Dataset 1
Dataset 2
Dataset 3
Dataset 4
Dataset 5
Dataset 6 |
15
15
15
15
15
15 |
4 / 0.12
3 / 0.6
4 / 1.0
10 / 0.6
10 / 0.6
10 / 0.6 |
20
22
16
55
44
44 |
20
22
16
55
44
44 |
Dataset 7
Dataset 8
Dataset 9 |
15
15
15 |
6 / 0.16
10 / 0.2
4 / 0.12 |
60
34
66 |
60
34
66 |
Table 3.
The number of clusters corresponding to each dataset quantified by the methods (QNC), where the bold values reflect the correct quantified cases (QNC= RNC).
Table 3.
The number of clusters corresponding to each dataset quantified by the methods (QNC), where the bold values reflect the correct quantified cases (QNC= RNC).
| Methods |
|
Datasets |
|
|
| 1 |
2 |
3 |
4 |
5 |
6 |
DBSCAN
HDBSCAN
DPC
DPCSA
DGB
SWCG-DGB |
8
9
9
10
3
9
|
1
2
2
2
1
2
|
9
6
7
7
6
7
|
29
28
31
31
29
31
|
7
8
10
10
8
10
|
23
18
19
18
12
20
|
Table 4.
The calculating time (CT) of the algorithms.
Table 4.
The calculating time (CT) of the algorithms.
| Method |
|
Datasets |
|
|
| 1 |
2 |
3 |
4 |
5 |
6 |
DBSCAN
HDBSCAN
DPC
DPCSA
DGB
SWCG-DGB |
0.336
0.044
1.215
2.459
0.464
0.356 |
0.860
0.015
0.621
0.596
0.923
0.513 |
0.559
0.011
0.372
0.548
0.487
0.361 |
1.347
0.033
1.952
2.686
1.487
1.457 |
0.871
0.020
0.413
0.661
0.423
0.302 |
3.012
0.049
1.114
2.577
0.590
0.352 |
Table 5.
Comparison of results of the algorithms in terms of Rand Index (RI).
Table 5.
Comparison of results of the algorithms in terms of Rand Index (RI).
| Method |
|
Datasets |
|
|
| 1 |
2 |
3 |
4 |
5 |
6 |
DBSCAN
HDBSCAN
DPC
DPCSA
DGB
SWCG-DGB |
0.792
0.951
0.954
0.801
0.798
0.972
|
0.756
0.910
0.918
0.721
0.756
0.942
|
0.795
0.936
0.970
0.820
0.812
0.976
|
0.730
0.960
0.976
0.919
0.852
0.961 |
0.786
0.971
0.972
0.716
0.721
0.983
|
0.798
0.906
0.922
0.738
0.797
0.936
|
Table 6.
Comparison of results of the algorithms in terms of Adjusted Rand Index (ARI).
Table 6.
Comparison of results of the algorithms in terms of Adjusted Rand Index (ARI).
| Method |
|
Datasets |
|
|
| 1 |
2 |
3 |
4 |
5 |
6 |
DBSCAN
HDBSCAN
DPC
DPCSA
DGB
SWCG-DGB |
0.641
0.743
0.891
0.750
0.727
0.904
|
0.632
0.801
0.910
0.603
0.689
0.915
|
0.755
0.819
0.922
0.729
0.734
0.939
|
0.623
0.805
0.908
0.835
0.780
0.942
|
0.704
0.901
0.855
0.461
0.627
0.913
|
0.720
0.602
0.896
0.590
0.705
0.901
|
Table 7.
Comparison of results of the algorithms in terms of Normalized Mutual Information (NMI).
Table 7.
Comparison of results of the algorithms in terms of Normalized Mutual Information (NMI).
| Method |
|
Datasets |
|
|
| 1 |
2 |
3 |
4 |
5 |
6 |
DBSCAN
HDBSCAN
DPC
DPCSA
DGB
SWCG-DGB |
0.716
0.832
0.841
0.765
0.742
0.932
|
0.702
0.815
0.816
0.678
0.713
0.928
|
0.811
0.875
0.972
0.790
0.789
0.953 |
0.718
0.855
0.953
0.862
0.832
0.957
|
0.723
0.940
0.912
0.524
0.664
0.952
|
0.755
0.723
0.811
0.622
0.765
0.933
|
Table 8.
The number of clusters corresponding to each real dataset quantified by the methods (QNC), where the bold values reflect the correct quantified cases (QNC= RNC).
Table 8.
The number of clusters corresponding to each real dataset quantified by the methods (QNC), where the bold values reflect the correct quantified cases (QNC= RNC).
| Methods |
|
QNC |
|
|
| |
Dataset 7
(Iris) |
Dataset 8 (Seeds) |
|
Dataset 9 (USPS) |
|
DBSCAN
HDBSCAN
DPC
DPCSA
DGB
SWCG-DGB |
|
2
2
3
2
3
3
|
2
3
3
3
3
3
|
|
15
12
9
11
10
10
|
|
Table 9.
Comparison of results of the algorithms in terms of RI, ARI, and NMI for three real datasets 7-9.
Table 9.
Comparison of results of the algorithms in terms of RI, ARI, and NMI for three real datasets 7-9.
| Methods |
RI |
ARI |
NMI |
| Dataset 7 (Iris) |
|---|
| DBSCAN |
0.751 |
0.682 |
0.740 |
| HDBSCAN |
0.776 |
0.568 |
0.733 |
| DPC |
0.935 |
0.893 |
0.917 |
| DPCSA |
0.892 |
0.886 |
0.870 |
| DGB |
0.955 |
0.902 |
0.919 |
| SWCG-DGB |
0.962 |
0.931 |
0.948 |
| Dataset 8 (Seeds) |
| DBSCAN |
0.782 |
0.632 |
0.704 |
| HDBSCAN |
0.753 |
0.413 |
0.531 |
| DPC |
0.921 |
0.910 |
0.933 |
| DPCSA |
0.746 |
0.703 |
0.693 |
| DGB |
0.923 |
0.905 |
0.910 |
| SWCG-DGB |
0.934 |
0.887 |
0.895 |
| Dataset 9 (USPS) |
| DBSCAN |
0.675 |
0.596 |
0,638 |
| HDBSCAN |
0.973 |
0.868 |
0.882 |
| DPC |
0.927 |
0.903 |
0.916 |
| DPCSA |
0.851 |
0.783 |
0.843 |
| DGB |
0.945 |
0.901 |
0.916 |
| SWCG-DGB |
0.984 |
0.925 |
0.938 |