Submitted:
27 August 2024
Posted:
29 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
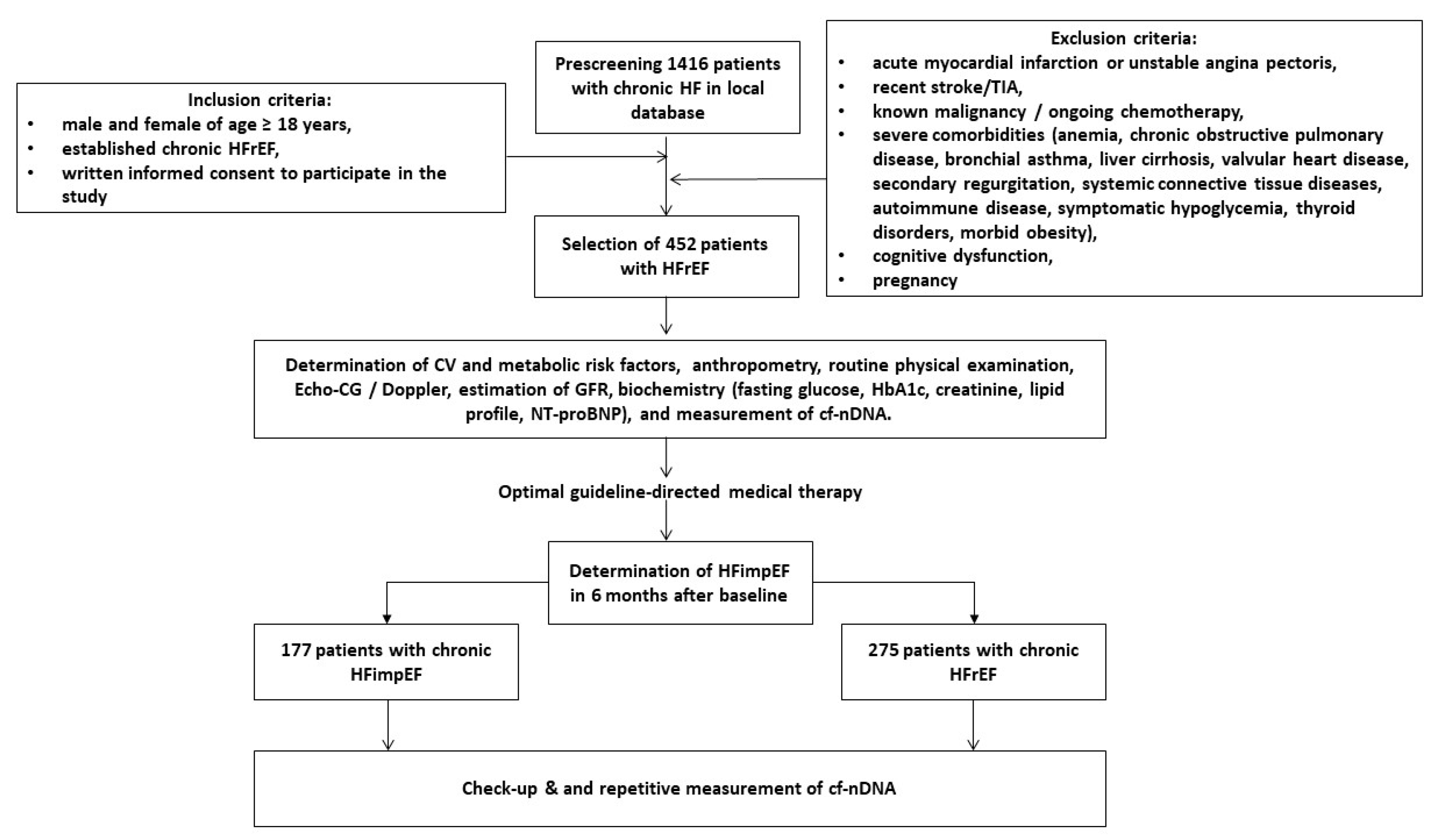
2.1. Study Patients
2.2. Determination of LVEF Improvement
2.3. Medical Information Collection
2.3. Examination of Hemodynamics
2.4. Glomerular Filtration Rate Calculation
2.5. Blood Sampling
2.6. Biomarker Evaluation
2.7. Cell-Free DNA Extraction
2.6. Measurement of Cell-Free DNAs in Plasma Samples
2.7. Statistical Analysis
3. Results
3.1. General Clinical Characteristics of the Patients
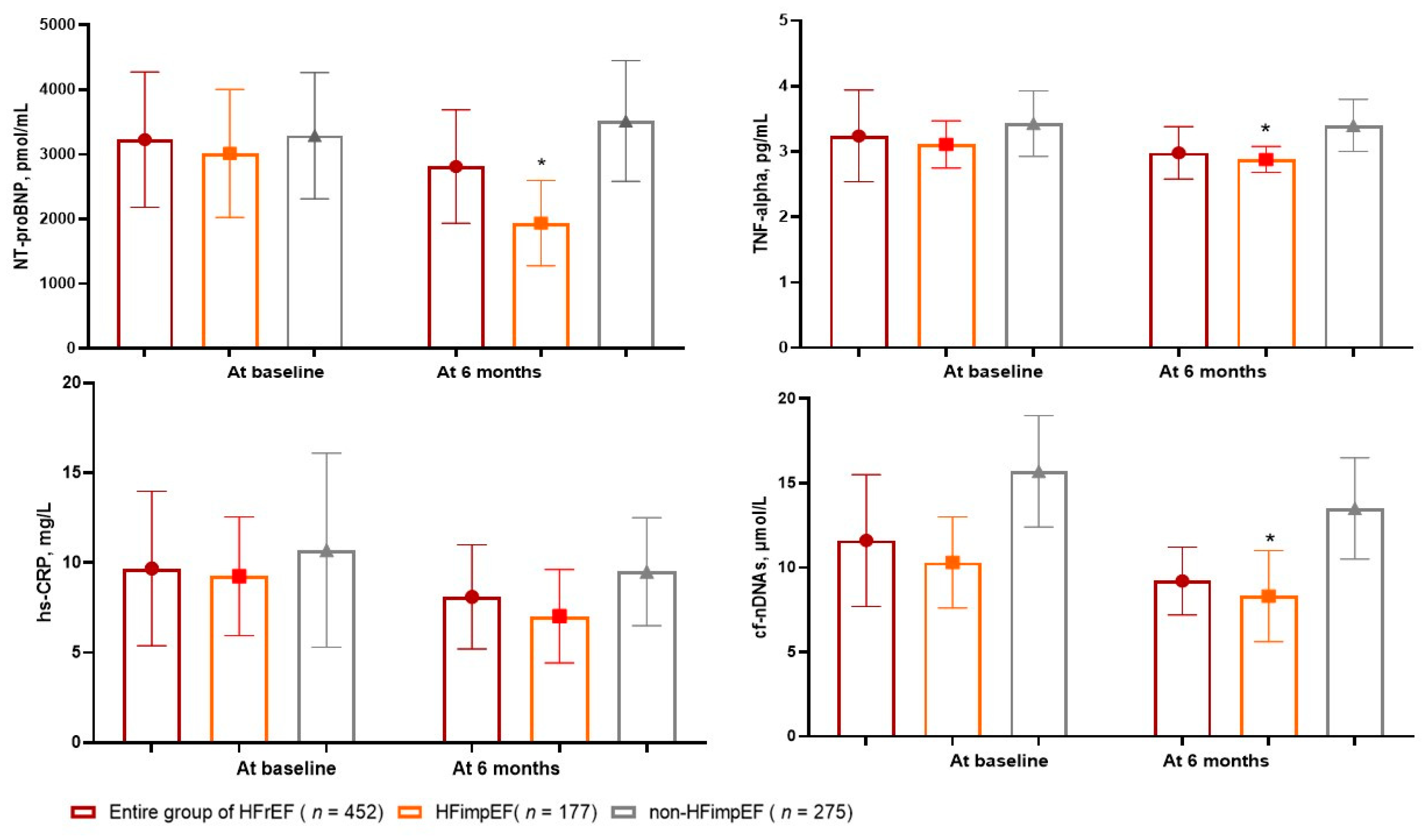
3.2. The Dynamics of Circulating Biomarkers Levels
3.3. Spearman’s Correlation between Circulating Levels of Cell-Free DNA and Other Parameters
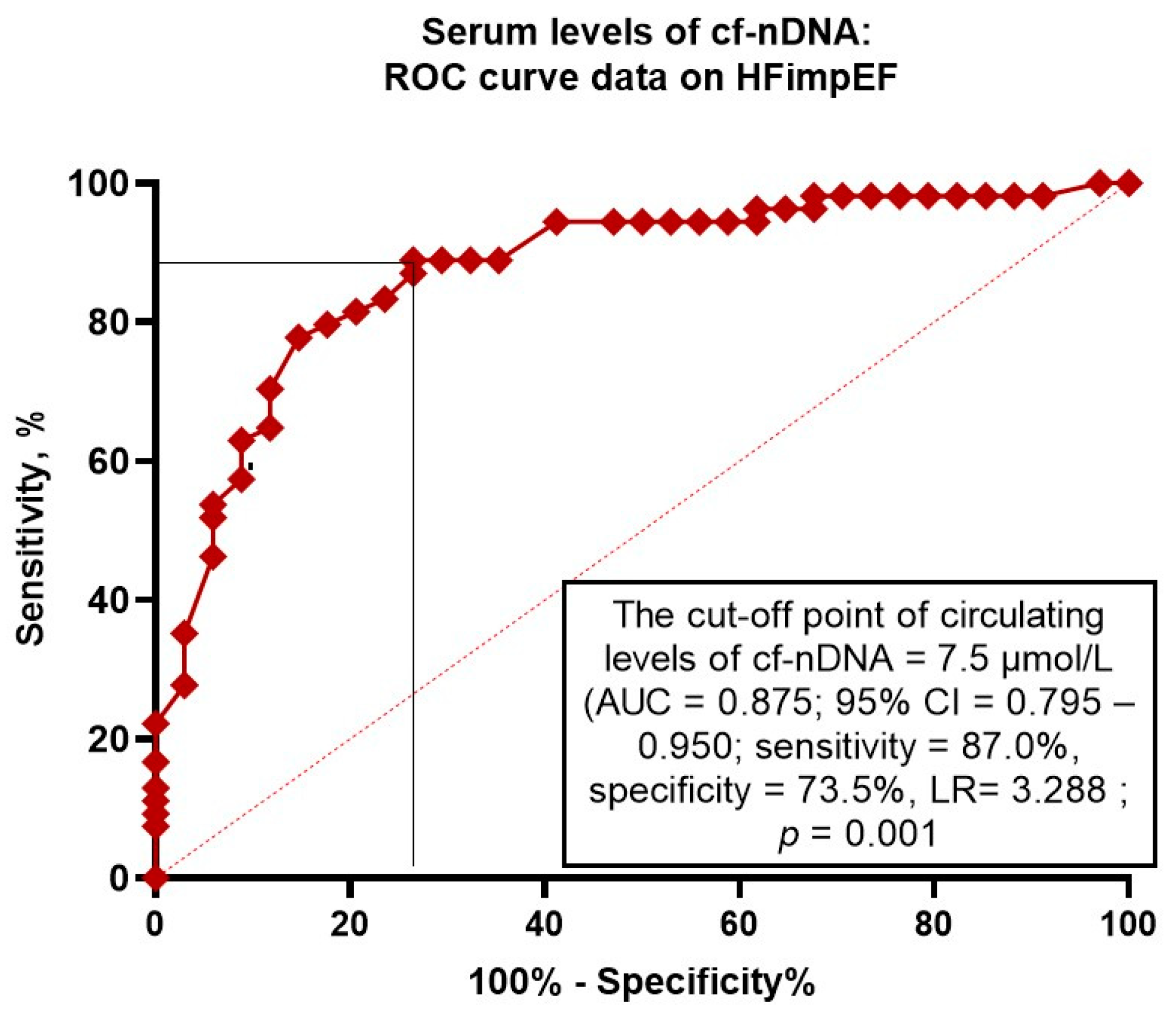
3.4. The Reliability of Circulating Levels of cf-nDNA: The Results of the ROC Curve Analysis
3.5. The Predictors of HFimpEF: The Univariate and Multivariate Logistic Regression
3.5. Comparison of the Models for HFimpEF
4. Discussion
5. Study Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bozkurt B, Coats AJS, Tsutsui H, Abdelhamid CM, Adamopoulos S, Albert N, Anker SD, Atherton J, Böhm M, Butler J, Drazner MH, Michael Felker G, Filippatos G, Fiuzat M, Fonarow GC, Gomez-Mesa JE, Heidenreich P, Imamura T, Jankowska EA, Januzzi J, Khazanie P, Kinugawa K, Lam CSP, Matsue Y, Metra M, Ohtani T, Francesco Piepoli M, Ponikowski P, Rosano GMC, Sakata Y, Seferović P, Starling RC, Teerlink JR, Vardeny O, Yamamoto K, Yancy C, Zhang J, Zieroth S. Universal definition and classification of heart failure: a report of the Heart Failure Society of America, Heart Failure Association of the European Society of Cardiology, Japanese Heart Failure Society and Writing Committee of the Universal Definition of Heart Failure: Endorsed by the Canadian Heart Failure Society, Heart Failure Association of India, Cardiac Society of Australia and New Zealand, and Chinese Heart Failure Association. Eur J Heart Fail. 2021; 23(3):352-380. [CrossRef]
- Keshvani N, Shah S, Ayodele I, Chiswell K, Alhanti B, Allen LA, Greene SJ, Yancy CW, Alonso WW, Van Spall HG, Fonarow GC, Heidenreich PA, Pandey A. Sex differences in long-term outcomes following acute heart failure hospitalization: Findings from the Get With The Guidelines-Heart Failure registry. Eur J Heart Fail. 2023 Sep;25(9):1544-1554. [CrossRef]
- Chimed S, Stassen J, Galloo X, Meucci MC, van der Bijl P, Knuuti J, Delgado V, Marsan NA, Bax JJ. Impact of Worsening Heart Failure on Long-Term Prognosis in Patients With Heart Failure With Reduced Ejection Fraction. Am J Cardiol. 2022 Dec 1;184:63-71. [CrossRef]
- Chen S, Huang Z, Liang Y, Zhao X, Aobuliksimu X, Wang B, He Y, Kang Y, Huang H, Li Q, Yao Y, Lu X, Qian X, Xie X, Liu J, Liu Y. Five-year mortality of heart failure with preserved, mildly reduced, and reduced ejection fraction in a 4880 Chinese cohort. ESC Heart Fail. 2022;9(4):2336-2347. [CrossRef]
- McDonagh TA, Metra M, Adamo M, Gardner RS, Baumbach A, Bohm M, et al; ESC Scientific Document Group. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J. 2021; 42(36):3599-3726. [CrossRef]
- Shah KS, Xu H, Matsouaka RA, Bhatt DL, Heidenreich PA, Hernandez AF, Devore AD, Yancy CW, Fonarow GC. Heart Failure With Preserved, Borderline, and Reduced Ejection Fraction: 5-Year Outcomes. J Am Coll Cardiol. 2017;70(20):2476-2486. [CrossRef]
- Tang J, Wang P, Liu C, Peng J, Liu Y, Ma Q. Pharmacotherapy in patients with heart failure with reduced ejection fraction: A systematic review and meta-analysis. Chin Med J (Engl). 2024. Epub ahead of print. [CrossRef] [PubMed]
- Mo X, Lu P, Yang X. Efficacy of sacubitril-valsartan and SGLT2 inhibitors in heart failure with reduced ejection fraction: A systematic review and meta-analysis. Clin Cardiol. 2023;46(10):1137-1145. [CrossRef]
- Rosano GMC, Vitale C, Spoletini I. Precision Cardiology: Phenotype-targeted Therapies for HFmrEF and HFpEF. Int J Heart Fail. 2024;6(2):47-55. [CrossRef]
- Romero E, Baltodano AF, Rocha P, Sellers-Porter C, Patel DJ, Soroya S, Bidwell J, Ebong I, Gibson M, Liem DA, Jimenez S, Bang H, Sirish P, Chiamvimonvat N, Lopez JE, Cadeiras M. Clinical, Echocardiographic, and Longitudinal Characteristics Associated With Heart Failure With Improved Ejection Fraction. Am J Cardiol. 2024; 211:143-152. [CrossRef]
- Solymossi B, Muk B, Sepp R, Habon T, Borbély A, Heltai K, Majoros Z, Járai Z, Vágány D, Szatmári Á, Sziliczei E, Bánfi-Bacsárdi F, Nyolczas N. Incidence and predictors of heart failure with improved ejection fraction category in a HFrEF patient population. ESC Heart Fail. 2024; 11(2):783-794. [CrossRef]
- Su K, Li M, Wang L, Tian S, Su J, Gu J, Chen S. Clinical characteristics, predictors, and outcomes of heart failure with improved ejection fraction. Int J Cardiol. 2022;357:72-80. [CrossRef]
- Ho LT, Juang JJ, Chen YH, Chen YS, Hsu RB, Huang CC, Lee CM, Chien KL. Predictors of Left Ventricular Ejection Fraction Improvement in Patients with Early-Stage Heart Failure with Reduced Ejection Fraction. Acta Cardiol Sin. 2023;39(6):854-861. [CrossRef]
- Segev A, Avrahamy B, Fardman A, Matetzky S, Freimark D, Regev O, Kuperstein R, Grupper A. Heart failure with improved ejection fraction: patient characteristics, clinical outcomes and predictors for improvement. Front Cardiovasc Med. 2024;11:1378955. [CrossRef]
- Si J, Ding Z, Hu Y, Zhang X, Zhang Y, Cao H, Liu Y. Predictors and prognostic implications of left ventricular ejection fraction trajectory improvement in the spectrum of heart failure with reduced and mildly reduced ejection fraction. J Cardiol. 2024;83(4):250-257. [CrossRef]
- Lo YMD, Han DSC, Jiang P, Chiu RWK. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science. 2021;372(6538):eaaw3616. [CrossRef]
- Cahilog Z, Zhao H, Wu L, Alam A, Eguchi S, Weng H, Ma D. The Role of Neutrophil NETosis in Organ Injury: Novel Inflammatory Cell Death Mechanisms. Inflammation. 2020;43(6):2021-2032. [CrossRef]
- Stanley KE, Jatsenko T, Tuveri S, Sudhakaran D, Lannoo L, Van Calsteren K, de Borre M, Van Parijs I, Van Coillie L, Van Den Bogaert K, De Almeida Toledo R, Lenaerts L, Tejpar S, Punie K, Rengifo LY, Vandenberghe P, Thienpont B, Vermeesch JR. Cell type signatures in cell-free DNA fragmentation profiles reveal disease biology. Nat Commun. 2024;15(1):2220. [CrossRef]
- Oellerich M, Sherwood K, Keown P, Schütz E, Beck J, Stegbauer J, Rump LC, Walson PD. Liquid biopsies: donor-derived cell-free DNA for the detection of kidney allograft injury. Nat Rev Nephrol. 2021;17(9):591-603. [CrossRef]
- Tan E, Liu D, Perry L, Zhu J, Cid-Serra X, Deane A, Yeo C, Ajani A. Cell-free DNA as a potential biomarker for acute myocardial infarction: A systematic review and meta-analysis. Int J Cardiol Heart Vasc. 2023;47:101246. [CrossRef]
- Antonatos D, Patsilinakos S, Spanodimos S, Korkonikitas P, Tsigas D. Cell-free DNA levels as a prognostic marker in acute myocardial infarction. Ann N Y Acad Sci. 2006; 1075(1):278–281. [CrossRef]
- Medina JE, Dracopoli NC, Bach PB, Lau A, Scharpf RB, Meijer GA, Andersen CL, Velculescu VE. Cell-free DNA approaches for cancer early detection and interception. J Immunother Cancer. 2023; 11(9):e006013. [CrossRef]
- Berezina TA, Berezin AE. Cell-free DNA as a plausible biomarker of chronic kidney disease. Epigenomics. 2023;15(17):879-890. [CrossRef]
- Mansueto G, Benincasa G, Della Mura N, Nicoletti GF, Napoli C. Epigenetic-sensitive liquid biomarkers and personalised therapy in advanced heart failure: a focus on cell-free DNA and microRNAs. J Clin Pathol. 2020;73(9):535-543. [CrossRef]
- Yokokawa T, Misaka T, Kimishima Y, Shimizu T, Kaneshiro T, Takeishi Y. Clinical Significance of Circulating Cardiomyocyte-Specific Cell-Free DNA in Patients With Heart Failure: A Proof-of-Concept Study. Can J Cardiol, 2020; 36(6):931-935. [CrossRef]
- Berezina, T.A.; Kopytsya, M.P.; Petyunina, O.V.; Berezin, A.A.; Obradovic, Z.; Schmidbauer, L.; Lichtenauer, M.; Berezin, A.E. Lower Circulating Cell-Free Mitochondrial DNA Is Associated with Heart Failure in Type 2 Diabetes Mellitus Patients. Cardiogenetics 2023, 13, 15–30. [Google Scholar] [CrossRef]
- American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2021. Diabetes Care. 2021; 44(Suppl 1):S15-S33. [CrossRef]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J. 2021, 42, 3599–3726. [Google Scholar] [CrossRef] [PubMed]
- Williams B, Mancia G, Spiering W, Agabiti Rosei E, Azizi M, Burnier M, et al; ESC Scientific Document Group. 2018 ESC/ESH Guidelines for the management of arterial hypertension. Eur Heart J. 2018; 39(33):3021-3104. [CrossRef]
- Mach F, Baigent C, Catapano AL, Koskinas KC, Casula M, Badimon L, et al; ESC Scientific Document Group. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur Heart J. 2020; 41(1):111-188. [CrossRef]
- Knuuti J, Wijns W, Saraste A, Capodanno D, Barbato E, Funck-Brentano C, Prescott E, Storey RF, Deaton C, Cuisset T, Agewall S, Dickstein K, Edvardsen T, Escaned J, Gersh BJ, Svitil P, Gilard M, Hasdai D, Hatala R, Mahfoud F, Masip J, Muneretto C, Valgimigli M, Achenbach S, Bax JJ; ESC Scientific Document Group. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur Heart J. 2020;41(3):407-477. Erratum in: Eur Heart J. 2020 Nov 21;41(44):4242. 10.1093/eurheartj/ehz825. [CrossRef]
- Inker LA, Astor BC, Fox CH, Isakova T, Lash JP, Peralta CA, Kurella Tamura M, Feldman HI. KDOQI US commentary on the 2012 KDIGO clinical practice guideline for the evaluation and management of CKD. Am J Kidney Dis. 2014;63(5):713-35. [CrossRef]
- Mitchell, C.; Rahko, P.S.; Blauwet, L.A.; Canaday, B.; Finstuen, J.A.; Foster, M.C.; Horton, K.; Ogunyankin, K.O.; Palma, R.A.; Velazquez, E.J. Guidelines for Performing a Comprehensive Transthoracic Echocardiographic Examination in Adults: Recommendations from the American Society of Echocardiography. J. Am. Soc. Echocardiogr. 2018, 32, 1–64. [Google Scholar] [CrossRef] [PubMed]
- Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, et al; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration). A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009; 150(9):604-12. [CrossRef]
- Hasenleithner SO, Speicher MR. A clinician's handbook for using ctDNA throughout the patient journey. Mol Cancer. 2022; 21(1):81. [CrossRef]
- He Y, Ling Y, Guo W, Li Q, Yu S, Huang H, Zhang R, Gong Z, Liu J, Mo L, Yi S, Lai D, Yao Y, Liu J, Chen J, Liu Y, Chen S. Prevalence and Prognosis of HFimpEF Developed From Patients With Heart Failure With Reduced Ejection Fraction: Systematic Review and Meta-Analysis. Front Cardiovasc Med. 2021; 8:757596. [CrossRef]
- Heidenreich PA, Bozkurt B, Aguilar D, Allen LA, Byun JJ, Colvin MM, Deswal A, Drazner MH, Dunlay SM, Evers LR, Fang JC, Fedson SE, Fonarow GC, Hayek SS, Hernandez AF, Khazanie P, Kittleson MM, Lee CS, Link MS, Milano CA, Nnacheta LC, Sandhu AT, Stevenson LW, Vardeny O, Vest AR, Yancy CW. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation. 2022;145(18):e895-e1032. [CrossRef]
- Zamora E, González B, Lupón J, Borrellas A, Domingo M, Santiago-Vacas E, Cediel G, Codina P, Rivas C, Pulido A, Crespo E, Velayos P, Diaz V, Bayes-Genis A. Quality of life in patients with heart failure and improved ejection fraction: one-year changes and prognosis. ESC Heart Fail. 2022;9(6):3804-3813. [CrossRef]
- Yoshimura R, Hayashi O, Horio T, Fujiwara R, Matsuoka Y, Yokouchi G, Sakamoto Y, Matsumoto N, Fukuda K, Shimizu M, Izumiya Y, Yoshiyama M, Fukuda D, Fujimoto K, Kasayuki N. The E/e' ratio on echocardiography as an independent predictor of the improvement of left ventricular contraction in patients with heart failure with reduced ejection fraction. J Clin Ultrasound. 2023;51(7):1131-1138. [CrossRef]
- Cao TH, Tay WT, Jones DJL, Cleland JGF, Tromp J, Emmens JE, Teng TK, Chandramouli C, Slingsby OC, Anker SD, Dickstein K, Filippatos G, Lang CC, Metra M, Ponikowski P, Samani NJ, Van Veldhuisen DJ, Zannad F, Anand IS, Lam CSP, Voors AA, Ng LL. Heart failure with improved versus persistently reduced left ventricular ejection fraction: A comparison of the BIOSTAT-CHF (European) study with the ASIAN-HF registry. Eur J Heart Fail. 2024. Epub ahead of print. [CrossRef] [PubMed]
- Lam CSP, Li YH, Bayes-Genis A, Ariyachaipanich A, Huan DQ, Sato N, Kahale P, Cuong TM, Dong Y, Li X, Zhou Y. The role of N-terminal pro-B-type natriuretic peptide in prognostic evaluation of heart failure. J Chin Med Assoc. 2019; 82(6):447-451. [CrossRef]
- Liu D, Hu K, Schregelmann L, Hammel C, Lengenfelder BD, Ertl G, Frantz S, Nordbeck P. Determinants of ejection fraction improvement in heart failure patients with reduced ejection fraction. ESC Heart Fail. 2023;10(2):1358-1371. [CrossRef]
- Yamamoto M, Ishizu T, Sato K, Minami K, Terauchi T, Nakatsukasa T, Kawamatsu N, Machino-Ohtsuka T, Ieda M. Longitudinal Changes in Natriuretic Peptides and Reverse Cardiac Remodeling in Patients with Heart Failure Treated with Sacubitril/Valsartan Across the Left Ventricular Ejection Traction Spectrum. Int Heart J. 2023 Nov 30;64(6):1071-1078. [CrossRef]
- Butt JH, Adamson C, Docherty KF, de Boer RA, Petrie MC, Inzucchi SE, Kosiborod MN, Maria Langkilde A, Lindholm D, Martinez FA, Bengtsson O, Schou M, O'Meara E, Ponikowski P, Sabatine MS, Sjöstrand M, Solomon SD, Jhund PS, McMurray JJV, Køber L. Efficacy and Safety of Dapagliflozin in Heart Failure With Reduced Ejection Fraction According to N-Terminal Pro-B-Type Natriuretic Peptide: Insights From the DAPA-HF Trial. Circ Heart Fail. 2021;14(12):e008837. [CrossRef]
- Nassif ME, Windsor SL, Tang F, Khariton Y, Husain M, Inzucchi SE, McGuire DK, Pitt B, Scirica BM, Austin B, Drazner MH, Fong MW, Givertz MM, Gordon RA, Jermyn R, Katz SD, Lamba S, Lanfear DE, LaRue SJ, Lindenfeld J, Malone M, Margulies K, Mentz RJ, Mutharasan RK, Pursley M, Umpierrez G, Kosiborod M. Dapagliflozin Effects on Biomarkers, Symptoms, and Functional Status in Patients With Heart Failure With Reduced Ejection Fraction: The DEFINE-HF Trial. Circulation. 2019 Oct 29;140(18):1463-1476. [CrossRef]
- Martinsson A, Oest P, Wiborg MB, Reitan Ö, Smith JG. Longitudinal evaluation of ventricular ejection fraction and NT-proBNP across heart failure subgroups. Scand Cardiovasc J. 2018;52(4):205-210. [CrossRef]
- Dutta A, Das M, Ghosh A, Rana S. Molecular and cellular pathophysiology of circulating cardiomyocyte-specific cell free DNA (cfDNA): Biomarkers of heart failure and potential therapeutic targets. Genes Dis. 2022 Aug 31;10(3):948-959. PMCID: PMC10308167. [CrossRef] [PubMed]
- Ren J, Jiang L, Liu X, Liao Y, Zhao X, Tang F, et al. Heart-specific DNA methylation analysis in plasma for the investigation of myocardial damage. J Transl Med. 2022; 20(1):36. [CrossRef]
- Berezin A. Neutrophil extracellular traps: The core player in vascular complications of diabetes mellitus. Diabetes Metab Syndr. 2019; 13(5):3017-3023. [CrossRef]
- Thorsen SU, Moseholm KF, Clausen FB. Circulating cell-free DNA and its association with cardiovascular disease: what we know and future perspectives. Curr Opin Lipidol. 2024;35(1):14-19. [CrossRef]
- Vulesevic B, Lavoie SS, Neagoe PE, Dumas E, Räkel A, White M, et al. CRP Induces NETosis in Heart Failure Patients with or without Diabetes. Immunohorizons. 2019;3(8):378-388. [CrossRef]
- Liu LP, Cheng K, Ning MA, Li HH, Wang HC, Li F, et al. Association between peripheral blood cells mitochondrial DNA content and severity of coronary heart disease. Atherosclerosis. 2017; 261:105-110. [CrossRef]
- Frangogiannis, NG. The inflammatory response in myocardial injury, repair, and remodelling. Nat Rev Cardiol. 2014; 11(5):255-65. [CrossRef]
- Liu Q, Wu J, Zhang X, Li X, Wu X, Zhao Y, Ren J. Circulating mitochondrial DNA-triggered autophagy dysfunction via STING underlies sepsis-related acute lung injury. Cell Death Dis. 2021;12(7):673. [CrossRef]
- Oommen SG, Man RK, Talluri K, Nizam M, Kohir T, Aviles MA, Nino M, Jaisankar LG, Jaura J, Wannakuwatte RA, Tom L, Abraham J, Siddiqui HF. Heart Failure With Improved Ejection Fraction: Prevalence, Predictors, and Guideline-Directed Medical Therapy. Cureus. 2024;16(6):e61790. [CrossRef]
- Kim KA, Kim SH, Lee KY, Yoon AH, Hwang BH, Choo EH, Kim JJ, Choi IJ, Kim CJ, Lim S, Park MW, Yoo KD, Jeon DS, Ahn Y, Jeong MH, Chang K. Predictors and Long-Term Clinical Impact of Heart Failure With Improved Ejection Fraction After Acute Myocardial Infarction. J Am Heart Assoc. 2024;13(16):e034920. [CrossRef]
| Variables | Entire patient cohort ( n = 452) |
Patients with HFimpEF ( n = 177) |
Patients with persistent HFrEF ( n = 275) |
P value |
|---|---|---|---|---|
| Demographics and anthropomorphic parameters | ||||
| Age, year | 59 (50-68) | 59 (52-65) | 60 (49-72) | 0.48 |
| Male / female n (%) | 266 (58.9) / 186 (41.2) | 102 (57.6) / 75 (42.3) | 164 (59.6) / 111 (40.4) | 0.36 |
| BMI, kg/m2 | 25.8±3.5 | 25.1±2.9 | 26.1±2.7 | 0.44 |
| Comorbidities and CV risk factors | ||||
| Dyslipidemia, n (%) | 286 (63.2) | 115 (64.5) | 171 (62.2) | 0.77 |
| Hypertension, n (%) | 71 (15.7) | 28 (15.8) | 43 (15.6) | 0.88 |
| Ischemia-induced cardiomyopathy, n (%) | 141 (31.2) | 44 (24.9) | 97 (35.3) | 0.04 |
| Dilated cardiomyopathy, n (%) | 68 (15.0) | 21 (11.9) | 47 (17.1) | 0.52 |
| AF, n (%) | 137 (30.3) | 47 (26.6) | 90 (32.7) | 0.28 |
| Smoking, n (%) | 168 (37.2) | 65 (36.7) | 103 (37.5) | 0.88 |
| Abdominal obesity, n (%) | 112 (24.8) | 46 (26.0) | 66 (24.0) | 0.87 |
| T2DM, n (%) | 146 (32.3) | 54 (30.5) | 92 (33.5) | 0.26 |
| LVH, n (%) | 316 (69.9) | 120 (67.8) | 196 (71.3) | 0.44 |
| CKD 1-3 grades, n (%) | 132 (29.2) | 45 (25.4) | 87 (31.6) | 0.42 |
| Complete LBBB / RBBB on ECG, n (%) | 98 (21.7) | 35 (19.8) | 63 (22.9) | 0.18 |
| NYHA functional classification | ||||
| I/II HF NYHA class, n (%) | 144 (31.9) | 71 (40.1) | 73 (26.6) | 0.001 |
| III HF NYHA class, n (%) | 230 (50.8) | 85 (48.0) | 145 (52.7) | 0.06 |
| IV HF NYHA class, n (%) | 78 (17.3) | 21 (11.9) | 57 (20.7) | 0.036 |
| Hemodynamics performances | ||||
| SBP, mm Hg | 128±11 | 129±9 | 125±10 | 0.22 |
| DBP, mm Hg | 78±10 | 77±8 | 74±9 | 0.64 |
| LVEDV, mL | 171 (149-192) | 168 (136-188) | 181 (150-202) | 0.04 |
| LVESV, mL | 115 (89-127) | 109 (87-124) | 126 (90-131) | 0.01 |
| LVEF, % | 32 (29-39) | 35 (31-39) | 30 (27-34) | 0.02 |
| LVMMI, g/m2 | 226±15 | 218±15 | 234±13 | 0.46 |
| LAVI, mL/m2 | 46 (39-52) | 44 (35-51) | 47 (39-54) | 0.12 |
| E/e`, unit | 17.3±5.4 | 16.6±4.1 | 19.1±3.3 | 0.56 |
| Biochemistry parameters | ||||
| eGFR, mL/min/1.73 m2 | 72 ± 11 | 80 ± 9 | 65 ± 7 | 0.04 |
| Fasting glucose, mmol/L | 5.11 ± 0.77 | 5.06 ± 0.60 | 5.19 ± 1.1 | 0.66 |
| Creatinine, µmol/L | 99.6±12.8 | 78.9±9.1 | 115.2±8.2 | 0.04 |
| TC, mmol/L | 5.88±0.90 | 5.61±0.52 | 5.92±0.70 | 0.62 |
| HDL-C, mmol/L | 0.97±0.14 | 0.97±0.15 | 0.98±0.18 | 0.68 |
| LDL-C, mmol/L | 3.93±0.18 | 3.80±0.17 | 4.00±0.12 | 0.02 |
| TG, mmol/L | 1.98±0.17 | 1.90±0.12 | 2.03±0.15 | 0.64 |
| hs-CRP, mg/L | 9.68 (4.31 – 13.70) | 9.25 (3.45 – 12.70) | 10.70 (5.80 – 17.50) | 0.22 |
| TNF-alpha, pg/mL | 3.24 (2.70–3.98) | 3.11 (2.62–3.69) | 3.43 (2.95–4.12) | 0.04 |
| NT-proBNP, pmol/mL | 3228 (1910 – 5215) | 3015 (1780 – 5220) | 3290 (1820 – 5470) | 0.44 |
| cf-nDNA, μmol/L | 11.6 (7.68 - 15.7) | 9.8 (7.2 – 12.2) | 14.1 (11.8 – 16.5) | 0.02 |
| Concomitant medications | ||||
| ACEI, n (%) | 198 (43.8) | 79 (44.6) | 119 (43.3) | 0.88 |
| ARNI, n (%) | 134 (29.6) | 53 (29.9) | 81 (29.5) | 0.90 |
| ARB, n (%) | 86 (19.0) | 35 (19.7) | 51 (18.5) | 0.82 |
| Ivabradine, n (%) | 78 (17.3) | 28 (15.8) | 50 (18.2) | 0.56 |
| Calcium channel blocker, n (%) | 67 (14.8) | 23 (13.0) | 44 (16.0) | 0.44 |
| MRA, n (%) | 405 (89.6) | 161 (91.0) | 244 (88.7) | 0.86 |
| Digoxin, n (%) | 51 (11.3) | 14 (7.9) | 37 (13.5) | 0.010 |
| Loop diuretic, n (%) | 412 (91.2) | 159 (89.8) | 253 (92.0) | 0.46 |
| Antiplatelet, n (%) | 141 (31.2) | 54 (30.5) | 87 (31.6) | 0.84 |
| Anticoagulants, n (%) | 139 (30.8) | 55 (31.1) | 84 (30.5) | 0.82 |
| Metformin, n (%) | 138 (30.5) | 54 (30.5) | 84 (31.0) | 0.86 |
| SGLT2 inhibitors, n (%) | 434 (96.0) | 175 (98.9) | 259 (94.2) | 0.86 |
| Statins, n (%) | 350 (77.4) | 139 (78.5) | 211 (76.7) | 0.88 |
| Dependent variable: HFimpEF | ||||
| Variables | Univariate logistic regression | Multivariate logistic regression | ||
| OR (95% CI) | P-value | OR (95% CI) | P-value | |
| Rough regression models | ||||
| Ischemia-induced cardiomyopathy (presence vs. absent) | 0.75 (0.62-0.88) | 0.044 | 0.77 (0.60-0.90) | 0.042 |
| IV HF NYHA class | 0.71 (0.57-0.92) | 0.001 | 0.76 (0.63-0.87) | 0.001 |
| T2DM (presence vs. absent) | 0.77 (0.71-0.82) | 0.040 | 0.84 (0.62-0.92) | 0.042 |
| CKD (presence vs. absent) | 0.89 (0.84-0.96) | 0.048 | 0.88 (0.80-0.10) | 0.050 |
| AF (presence vs. absent) | 0.94 (0.80-1.09) | 0.064 | - | |
| LVEDV | 0.93 (0.90-1.01) | 0.052 | - | |
| LAVI | 0.95 (0.92-0.98) | 0.042 | 0.96 (0.90-1.00) | 0.050 |
| E/e` | 0.92 (0.89-0.97) | 0.080 | - | |
| NT-proBNP (≤1940 pmol/mL vs. >1940 pmol/mL) | 1.42 (1.19-1.98) | 0.001 | 1.35 (1.12-1.76) | 0.001 |
| Relative decrease in NT-proBNP levels (>35% vs. ≤35%) from baseline | 1.67 (1.51-1.82) | 0.001 | 1.70 (1.61-1.83) | 0.001 |
| TNF-alpha (≤2.88 pg/mL vs. >2.88 pg/mL) | 1.06 (1.00-1.12) | 0.48 | - | |
| hs-CRP (≤7.02 mg/L vs. >7.02 mg/L) | 1.08 (1.00-1.17) | 0.60 | - | |
| cf-nDNA (≤7.5 μmol/L vs. >7.5 μmol/L) | 1.56 (1.07-2.94) | 0.001 | 1.64 (1.10-2.07) | 0.001 |
| Digoxin (presence vs. absent) | 0.85 (0.72-0.97) | 0.042 | 0.93 (0.86-1.00) | 0.052 |
| Adjustment to ischemia-induced cardiomyopathy, IV HF NYHA class, and digoxin use | ||||
| T2DM (presence vs. absent) | 0.84 (0.76-0.95) | 0.040 | 0.88 (0.74-1.01) | 0.12 |
| CKD (presence vs. absent) | 0.94 (0.90-1.02) | 0.46 | - | |
| AF (presence vs. absent) | 0.92 (0.80-1.12) | 0.52 | - | |
| LVEDV | 0.98 (0.92-1.14) | 0.66 | - | |
| LAVI | 0.97 (0.91-1.05) | 0.52 | - | |
| E/e` | 0.92 (0.84-1.03) | 0.14 | - | |
| NT-proBNP (≤1940 pmol/mL vs. >1940 pmol/mL) | 1.55 (1.23-2.06) | 0.001 | 1.43 (1.21-1.88) | 0.001 |
| Relative decrease in NT-proBNP levels (>35% vs. ≤35%) from baseline | 1.63 (1.53-0.74) | 0.001 | 1.52 (1.38-0.69) | 0.001 |
| TNF-alpha (≤2.88 pg/mL vs. >2.88 pg/mL) | 1.03 (1.00-1.07) | 0.56 | - | |
| hs-CRP (≤7.02 mg/L vs. >7.02 mg/L) | 1.06 (1.00-1.12) | 0.66 | - | |
| cf-nDNA (≤7.5 μmol/L vs. >7.5 μmol/L) | 1.64 (1.10-2.33) | 0.001 | 1.64 (1.19-2.15) | 0.001 |
| Predictive models | AUC | NRI | IDI | |||
| M (95% CI) | P-value | M (95% CI) | P-value | M (95% CI) | P-value | |
| Model 1 (NT-proBNP ≤1940 pmol/mL) | 0.783 (0.700–0.840) | - | Reference | - | Reference | - |
| Model 2 (relative decrease in NT-proBNP levels ≤35% from baseline) | 0.795 (0.745–0.861) | 0.06 | 0.23 (0.17–0.30) | 0.170 | 0.21 (0.18–0.25) | 0.240 |
| Model 3 (cf-nDNA ≤7.5 μmol/L) | 0.875 (0.795–0.950) | 0.001 | 0.54 (0.43–0.67) | 0.001 | 0.51 (0.45–0.58) | 0.001 |
| Model 4 (NT-proBNP levels ≤1940 pmol/mL + cf-nDNA) | 0.872 (0.820–0.941) | 0.001 | 0.48 (0.42–0.55) | 0.001 | 0.49 (0.41–0.56) | 0.001 |
| Model 5 (relative decrease in NT-proBNP levels ≤35% from baseline + cf-nDNA) | 0.893 (0.844–0.962) | 0.001 | 0.58 (0.45–0.72) | 0.001 | 0.55 (0.49–0.62) | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




