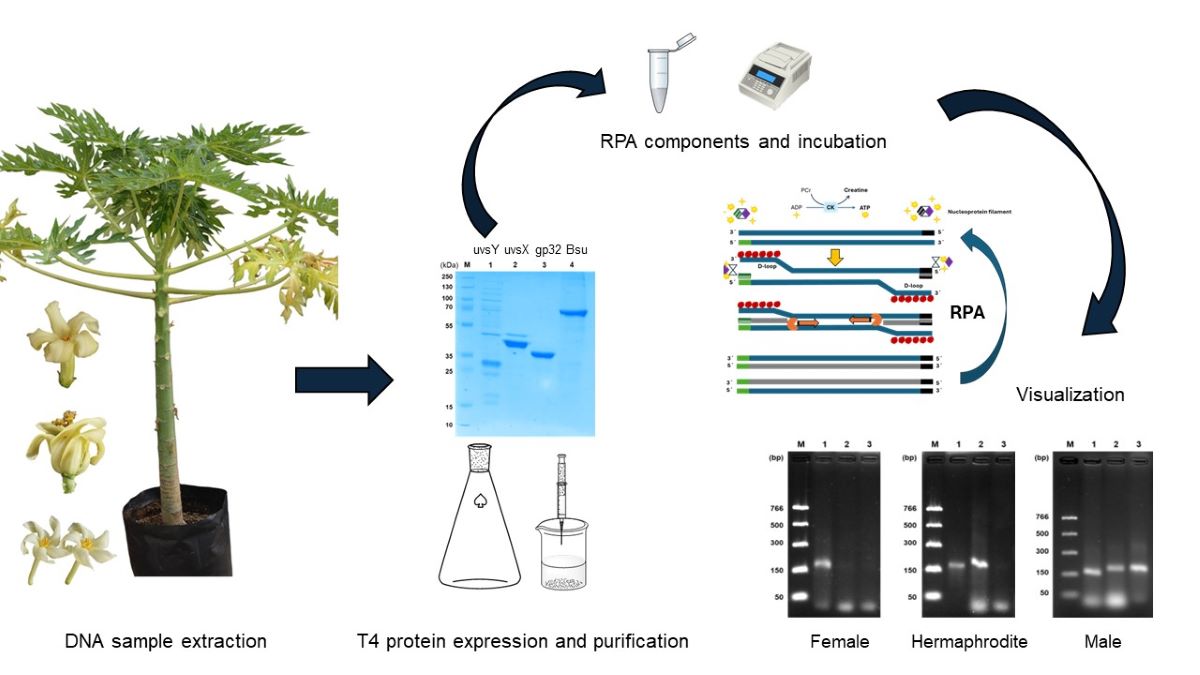
1. Introduction
Sex identification in papaya has been a common practice among growers since this plant can produce male, female, or hermaphrodite flowers almost four months after planting. However, molecular sex determination at the vegetative stage has been reported using molecular markers to save time and resources by PCR-based techniques [
1,
2,
3,
4,
5]. These approaches limit applicability in the field as a point of need strategies, e.g., in greenhouses; instead, isothermal amplification techniques have been used for such purposes. Isothermal methods are widely used to amplify nucleic acids with the premise of not performing denaturation steps, thus eliminating the need for thermocyclers. Some of these methods include loop-mediated isothermal amplification (LAMP) [
6], nucleic acid sequence-based amplification (NASBA) [
7], strand displacement amplification (SDA) [
8], rolling circle amplification (RCA) [
9], recombinase polymerase amplification (RPA) [
10], and so on. Among these methods, LAMP and, recently, RPA have been the most widely used for clinical and plant diagnosis of bacteria, fungi, and viruses, including the SARS-CoV-2 virus, even coupled to lateral flow biosensors. These approaches have in common that they amplify nucleic acids at a constant temperature (37 to 65°C) and a time that can vary according to the technique, from 20 minutes to 1 hour [
11,
12,
13,
14,
15,
16,
17].
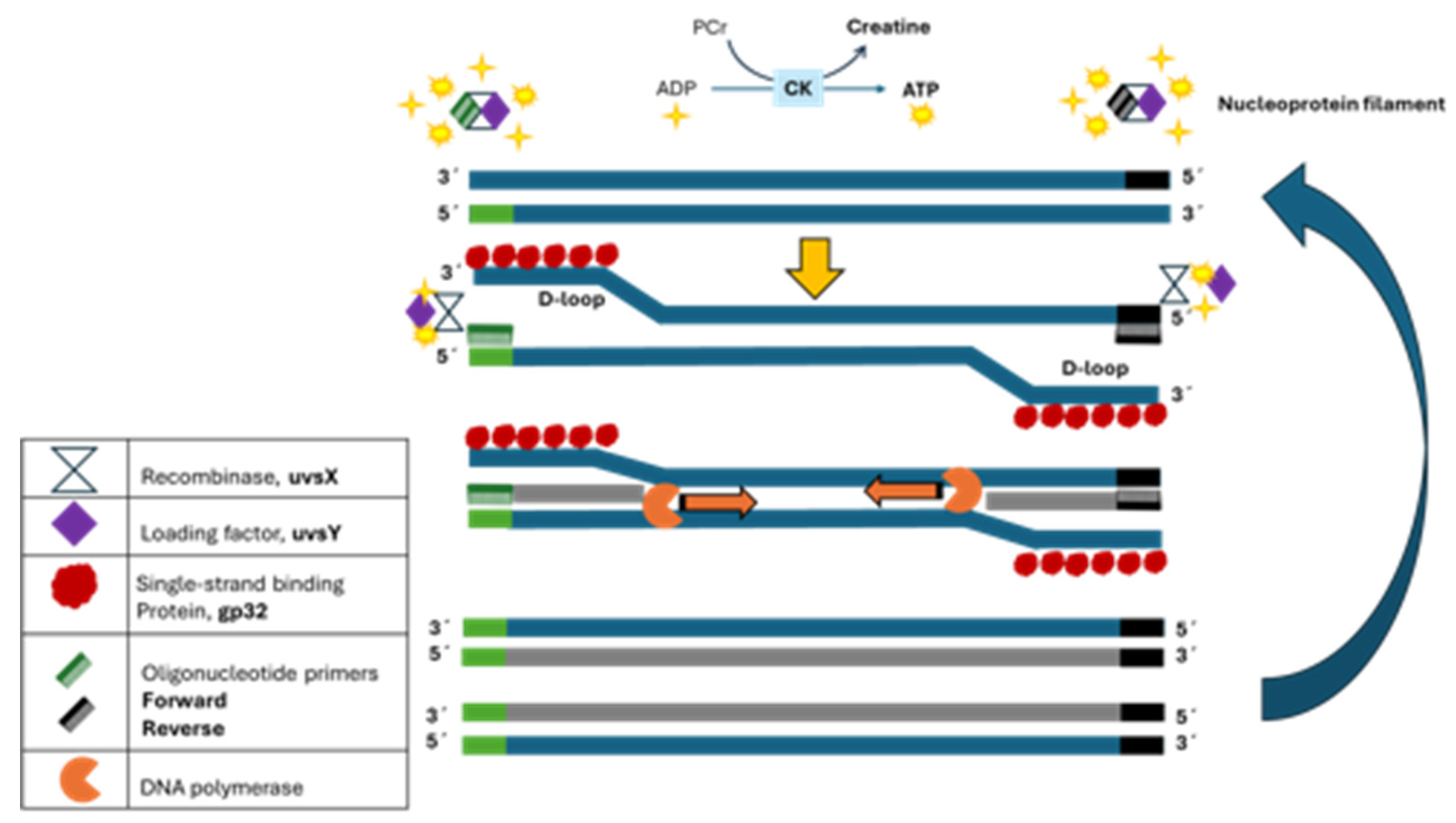
Recombinase polymerase amplification, or RPA, was developed in 2006 by Piepenburg and colleagues [
10] as a promising isothermal technique for nucleic acid amplification. This technology requires a set of enzymes based on the T4 bacteriophage and other components. The reaction starts with the recombinase uvsX, which binds to DNA primer (single-stranded DNA) assisted by uvsY and, in the presence of ATP, forms a presynaptic filament (nucleoprotein filament). The recombinase-usvY-primers complex actively hydrolyzes ATP to ADP as the driving force to search homologous sequences on the double-stranded DNA, and once homology is found, the complex invades it, and a D-loop is formed. The unwounded DNA strand is stabilized by aligned multimers of the single-stranded DNA-binding protein (gp32). The disassembly of the complex allows strand-displacement DNA polymerase access to the 3’-end of the two opposite primers to initiate primer extension and exponential DNA amplification. Thus, the recombinase can bind with a new primer and initiate several strand displacement processes to enhance DNA amplification in minutes and at a constant temperature (
Figure 1).
To date, the RPA technology has been patented and is sold as kits by companies such as TwistDx (UK), Agdia Inc. (USA), and Intact Genomics (USA) for a wide range of applications for research use only. The user-friendly nature of this technology has promoted its multiple applications in health, environment, and food, mainly for the detection of pathogens, either point-of-care or point-of-need, as it has demonstrated high sensitivity, specificity, and multiplexing. In addition, it does not require pre-denaturation of the sample and uses only one pair of primers, as compared to other methods where more than two pairs of primers are needed [
18]. Nevertheless, in some applications, it is not possible to optimize certain reaction conditions due to the nature of the commercial kit. Hence, Yasukawa and co-workers have optimized enzyme concentrations and buffer pH, defined optimal reaction temperatures and times, and evaluated the activity of novel polymerases and other additives for reaction efficiency for the detection of SARS-coV-2, rice yellow mottle virus and ureaplasma parvum serovar 3 [
19,
20,
21,
22,
23,
24].
In the present work, we established an isothermal DNA amplification system for papaya sex determination. For this purpose, we used purified recombinant enzymes uvsX, uvsY, gp32, and Bsu DNA polymerase. Reaction conditions were evaluated for reaction efficiency using specific primers to detect female, hermaphrodite, and male sexes. To our knowledge, this is the first report of a recombinase polymerase amplification assay for sex identification in papaya. Our results indicate a valuable potential for use in point-of-need strategies.
2. Materials and Methods
2.1. The T4 Enzymes uvsX, gp32, uvsY y Bsu DNA Polymerase
Escherichia coli chemically competent cells were obtained by the calcium chloride method [
25] and transformed with the plasmid coding for T4 uvsX, uvsY, gp32 proteins, and Bsu DNA polymerase in
E.
coli BL21(DE3). The genes, expression, and purification of proteins were according to Cordoba-Andrade et al. [
26], from where Dr. Brieba gently provided us with the plasmids and methodologies to purify the proteins. All proteins were stored at -20°C after snap freezing using liquid nitrogen. Protein quantification was carried out by the Bradford microassay (Bio-Rad, Hercules, CA, USA) method at 595 nm by triplicate, using bovine serum albumin (BSA) as a standard (Sigma Co., Burlington, MA, USA).
2.2. RPA Primer Design
Primers were designed manually and validated by OligoAnalyzer
® Tool (Integrated DNA Technologies), according to Strayer-Sherer et al. [
27] specification. For female, hermaphrodite, and male sex identification, DNA markers CpTrnL (603 bp), W11 (832 bp), and PMSM2 (548 bp) previously reported [
28] were used to design the RPA primers (
Table 1).
2.3. Preparation of the DNA Used as Standard
Standard DNA was prepared as follows: the DNA fragments were amplified by PCR according to specific primers (
Table 1) from papaya leaf genomic DNA, previously isolated [
28], under the following conditions: 50 µL reaction volume containing 1X buffer, 2.5 mM MgCl
2, 0.2 mM dNTPs, 1 µM of each primer, 100 ng of genomic DNA, and 0.625 units of Taq DNA Polymerase (5′Bio
®, Cuernavaca, México). The reaction begins with an initial denaturation at 95°C for 3 min, followed by 35 cycles: denaturation at 95°C; 30 sec, alignment at 55°C; 30 sec, extension at 72°C; 1 min, and a final 72°C extension for 10 min. The amplicons were visualized with 2% agarose gel, and the DNA purification was carried out using silica-gel membrane adsorption (Jena Bioscience, Jena, DE). The concentration of each DNA was determined with a NanoDrop
® 2000c spectrophotometer (Thermo Fischer Scientific, Waltham, MA, USA) using the A260/ A280 ratio and stored at -20 °C until subsequent use.
2.4. Setting the Conditions for the RPA Reaction
The RPA reaction of 20 µL was carried out in a 0.2 mL PCR tube under the conditions as in Juma et al. [
21] with some modifications: 50 mM Tris-HCl buffer (pH 8.6), 40 mM CH
3COOK, 6.0% PEG35000, 2 mM DTT, 650 µM dNTPs, 1 µM primer forward, 1 µM primer reverse, 20 mM phosphocreatine (Sigma Co., Burlington, MA, USA), 120 ng/µL creatine phosphokinase (Sigma Co., Burlington, MA, USA), 3.5 mM ATP (Thermo Fischer Scientific, Waltham, MA, USA), 40 ng/µL uvsY, 600 ng/µL gp32, 200 ng/µL Bsu DNA polymerase, 400 ng/µL uvsX and 14 mM Mg(OCOCH
3)
2 using ~20 ng of standard DNA. The common liquid solutions and salts for buffer and solutions preparations were purchased from Karal (León, GTO, MX). The most specialized reagents, when not specified, were purchased from Sigma Co. (Burlington, MA, USA).
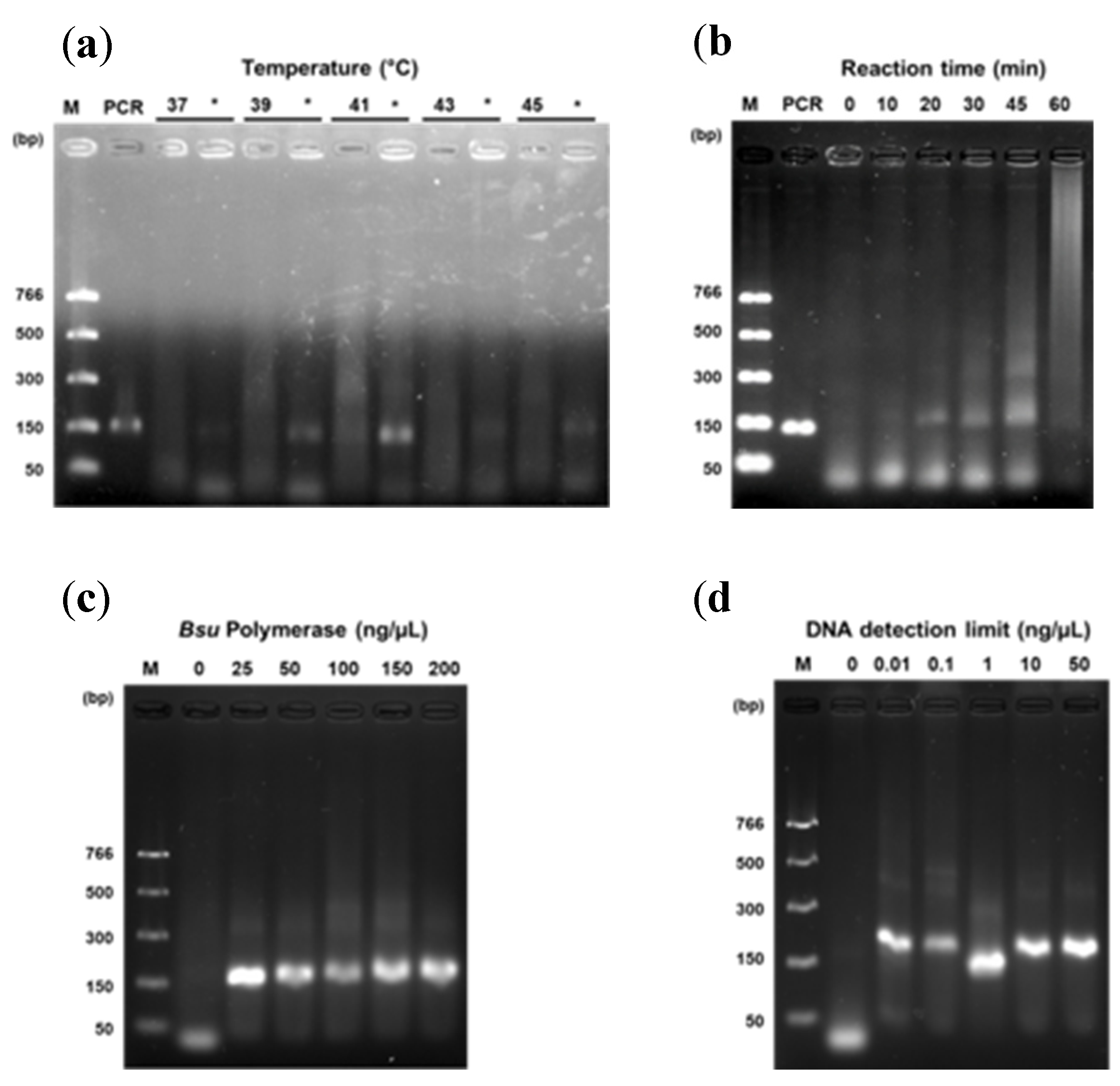
To standardize the reaction conditions, we evaluate the optimal temperature and time of amplification, the optimal concentration of Bsu DNA polymerase, and analytical sensitivity using the marker rpaCpTrnL to identify the female sex to set the optimal conditions to identify the other sexes: hermaphrodite and male. In this sense, RPA reactions were incubated, by duplicated, in a PCR MJ Mini thermal cycler (Bio-Rad, Hercules, CA, USA) to evaluate the optimal temperature conditions of amplification: 37, 39, 41, 43, and 45°C at 30 min. After that, the amplification time was tested at 0, 10, 20, 30, 45, and 60 min. Also, we evaluated optimal Bsu DNA polymerase concentration using 0, 25, 50, 100, 150, and 200 ng/µL. Using optimized conditions, we evaluate the detection limit using 0, 0.01, 0.1, 1, 10, and 50 ng of purified standard DNA. At the first seven minutes of incubation, reactions were mixed in a Vortex-Genie 2 (Scientific Industries, Inc., Bohemia, NY, USA) at 0.5 potency to improve amplification. Unless otherwise stated, the enzymes were inactivated at 85°C/ 5 min, according to Zou et al. [
14]. Subsequently, 5 µL was added to 2.0% (w/v) agarose gel (TAE 1X) and stained with GelRed (Biotium Inc., Fremont, CA, USA) to observe the amplification products.
Once optimal conditions were assessed, we carried out RPA reactions to evaluate primers designed to determine the sex of papaya using the DNA standard mentioned before in duplicate for each sex. In addition, we evaluated the RPA performance with the addition of betaine 0.8 M to enhance specificity, according to Luo et al. [
29]. Finally, we evaluated these conditions with crude extracts of hermaphrodite leaves isolated by the NaOH method, according to Satya et al. [
30], in duplicate.
3. Results
3.1. Expression and Purification of Recombinant Enzymes uvsX, uvsY, gp32, and Bsu DNA Polymerase.
A set of enzymes used to amplify DNA related to papaya sex by an RPA assay was produced using E. coli (DE3) cells. From four liters of culture, the stock concentration was as follows: 1.29 mg/mL uvsY, 2.04 mg/mL uvsX, 1.15 mg/mL gp32, and 1.64 mg/mL Bsu-Pol.
3.2. RPA Assay Optimization for Papaya Sex Determination
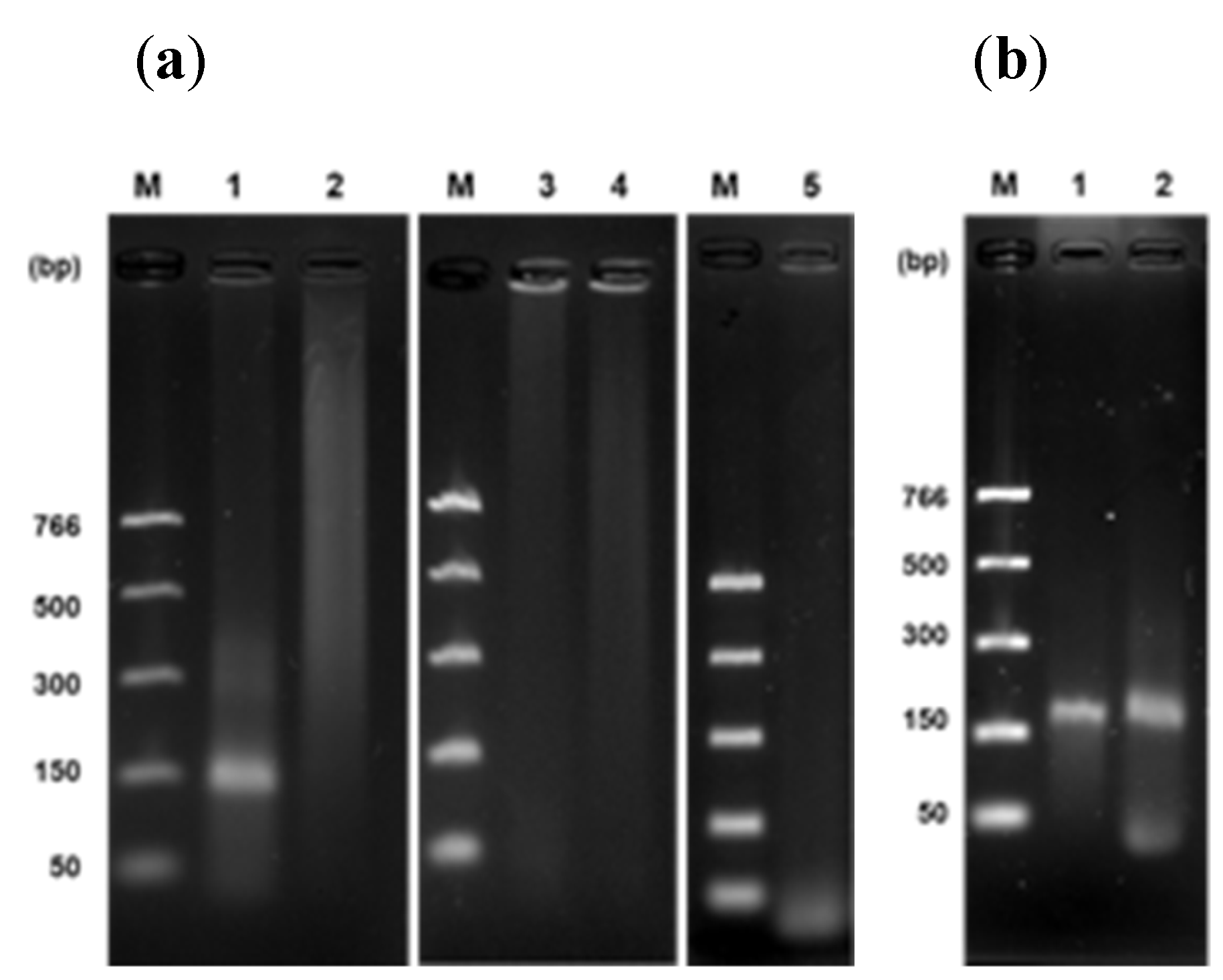
A detection system of papaya DNA related to sex was established according to several conditions by an isothermal approach using uvsY, uvsX, gp32, and Bsu-DNAP preparations. We used a fragment of a DNA sequence for each sex as a standard. In addition, primers for rpaCptrnL and female standard DNA were evaluated for temperature, reaction time, Bsu-DNAP concentration, and sensitivity in the RPA optimization. The optimal conditions for rpaW11 and rpaPMSM2 primers were also verified. Several temperatures were analyzed, and all amplified a 150 bp product at 30 min, being 41°C with the more intense band (
Figure 2a). The DNA amplification was observed at 20, 30, 45, and 60 min; however, at 60 min, the band was less intense than others. We decided the RPA reaction’s optimum time was 30 min (
Figure 2b). We used 200 ng/µL of Bsu-DNAP for the above experiments, yielding clear bands. Nevertheless, we screened for lower concentrations. Our minimal quantity tested was 25 ng/µL with good performance, compared to 50, 75, and 100 ng/µL (
Figure 2c). Finally, the RPA assay showed an intense band at the lower concentration of 0.01 ng/µL of standard DNA (
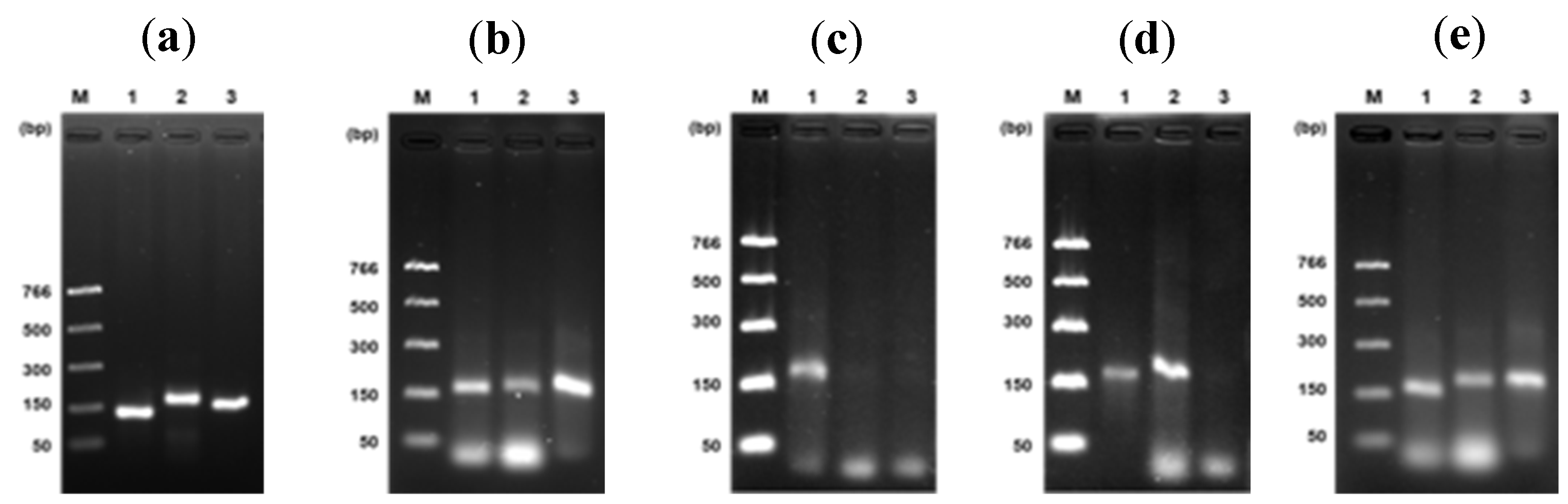
Figure 2d). Once optimal reaction conditions were established, we used them with the two primers rpaW11 and rpaPMSM2.
Figure 3b shows clear bands from each primer pair compared to PCR bands (
Figure 3a).
Figure 3c-e represents an RPA sexing of papaya under optimal temperature, time, and polymerase concentration conditions. Thus, only one band should be observed in the female, two in the hermaphrodite, and three in the male sexes, according to primer specificity described previously [
28].
3.3. Performance of Enzymes in RPA Reaction
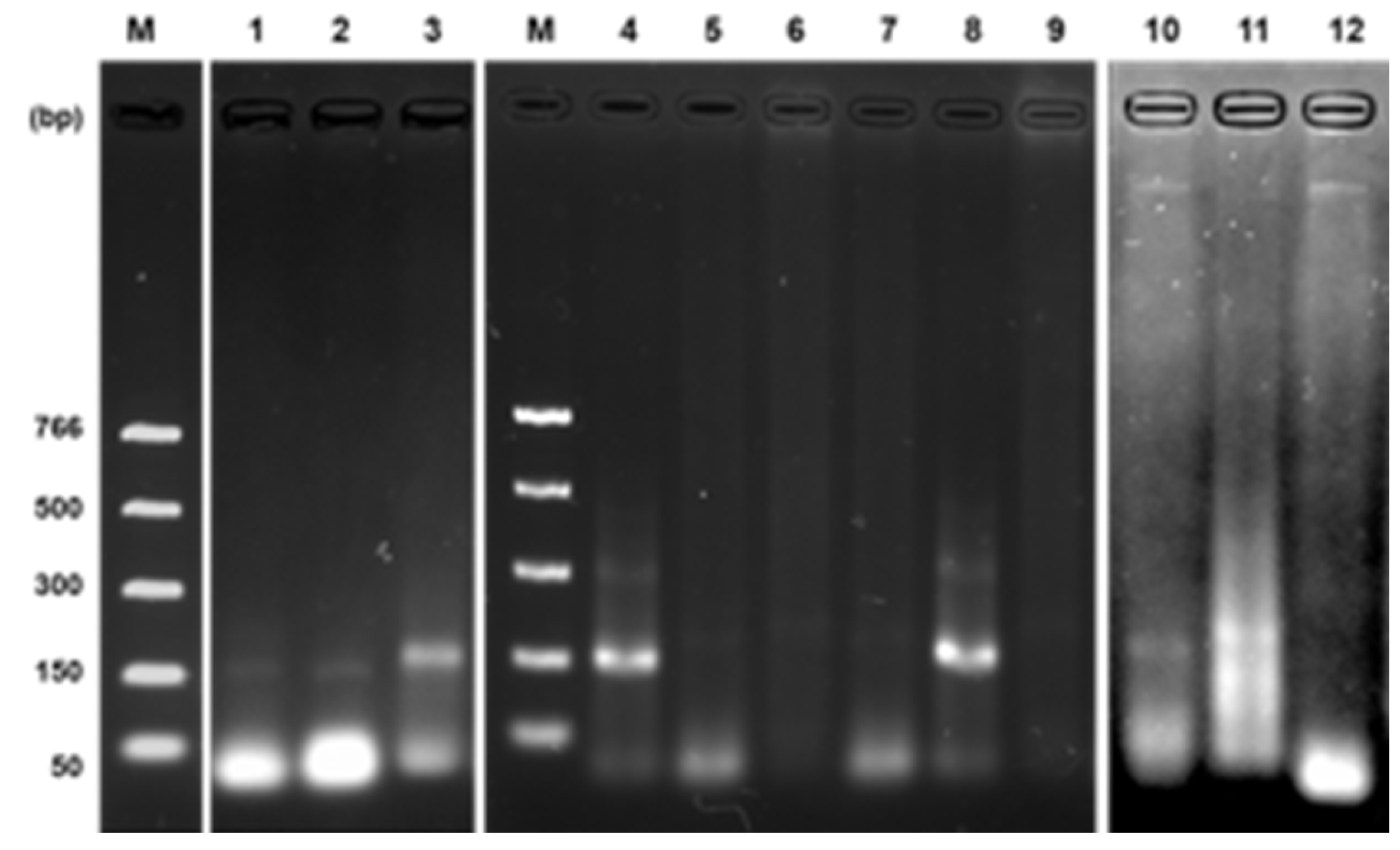
The effect of some components concerning DNA amplification was observed during the optimization of the RPA assay. In the presence of all components, a band was clearly observed. Conversely, no band was observed in the absence of DNA standard, polymerase, uvsY, uvsX, gp32, or ATP. On the other hand, we observed stability of uvsX, uvsY, and gp32 for almost six months at -20°C; after time, no amplification was obtained (
Figure 4a). In the case of Bsu-DNAP, we observed activity for more than one year at -20°C.
Likewise, we noted that sample mixing at seven minutes and denaturation at 85°C/5min after the time of incubation is dependent on the concentration of enzymes, so a high concentration of some of them (e.g., ~5 mg/mL uvsY, ~13 mg/mL gp32) the reaction does not require mixing or heat denaturation strategies, notwithstanding we recommend their application for better results (
Figure 4b).
3.4. Evaluation of Betaine Addition on the RPA Efficiency and Primer Specificity Validation
The in silico analysis of each pair of primers denoted specificity for the DNA sequence to be amplified. Nevertheless, in the RPA reactions, a weak amplification was observed using non-specific primers to DNA targets, e.g., female or hermaphrodite. However, a bright band was detected in the male target DNA (
Figure 5, lanes 1-3). Then, to evaluate whether the specificity of the primers could be improved, betaine 0.8 M was added to the RPA reaction cocktail using standard DNA.
Figure 5 (lanes 4-9) shows the performance of betaine in eliminating unspecific amplification by the primers. Duplicates evaluated such conditions. With these conditions, we could use crude extracts to identify the hermaphrodite sex; only two bands were clearly observed corresponding to such sex (
Figure 5, lanes 10-12).
4. Discussion
In this study, we developed an isothermal DNA detection assay for papaya sex determination by the recombinase polymerase amplification (RPA) method using the four enzymes necessary for the reaction: a recombinase uvsX, a single-stranded DNA binding protein gp32, a recombinase loading factor uvsY and Bsu DNA polymerase. The RPA optimization produced an expected band of the specific DNA target used as a template at the optimal temperature of 41°C; likewise, the other temperatures tested also produced an amplification band, but a faint one that was observed once sample denaturation was applied. Amplification of DNA fragments was detected as early as twenty minutes. However, after one hour, the band was less visible, probably due to the strand displacement activity of the polymerase not being fully functional or the loss of recombinase activity.
The sensitivity of the RPA method is similar or even higher than that of PCR. Thus, minimal DNA concentration at 9.6 or 10 pg has been reported to detect a specific target [
31,
32,
33,
34]. In our study, 0.01 ng/µL was our detection limit, similar to the authors reported above. The RPA method represents an advantage over PCR in amplification time and eliminates the need for a thermocycler for constant heating and cooler cycles. However, one of the challenges we faced at the end of the RPA reaction was the visualization of the results on the agarose gel. In the beginning, we only observed a slight band on a swept background (
Figure 2a), and to eliminate this noise and increase the amplification signal, we applied a heating step of the sample at 85°C for 5 minutes after the end of the reaction for enzyme denaturation according to Zou et al. [
14]. This heating allowed us to have more precise bands and avoid false negatives. Other methods have been reported apart from applying heat at different temperatures (65°C for 10 min and 95°C for 10 min), such as adding SDS, formamide, and even purification with columns. However, this represents a high cost [
33]. On the other hand, we also realized that denaturation was unnecessary when having a high concentration of the enzymes in stock since a smaller volume was added to the reaction tube. This observation was particularly for uvsY and gp32 (
Figure 4b).
The efficiency of the RPA reaction to amplify a target DNA fragment for papaya sex determination was achieved at the appropriate time and constant temperature, along with all enzymes and components. No amplification was observed without standard DNA, polymerase, uvsY, uvsX, gp32, or ATP (
Figure 4a). These data confirm the results obtained by Kojima and co-workers [
19], who also reported that in the absence of uvsY, creatine kinase, or DTT, a weak amplification band was produced, being these components not indispensable but necessary to improve the reaction. In our work, the uvsY protein was indispensable for increasing the reaction’s efficiency, the ATP, and the ATP regeneration system. In the RPA system, the recombinase is the essential component of the recombination system, and their high efficiency depends on ATP and the ability to regenerate it to maintain optimal concentration [
10,
20].
Advances for in situ sex determination of papaya seedlings by isothermal methods have been proposed via loop-mediated isothermal amplification (LAMP) [
35,
36]. Notwithstanding, the amplification temperatures are higher (65°C) and need up to six primers to work than the RPA method. However, the recombinase polymerase amplification has not been explored for papaya sex identification. For that reason, in this study, we aimed to develop an RPA assay using a specific DNA sequence for each sex, as previously reported for PCR [
28]. Although by the in silico analyses, the specificity and oligonucleotide sequence requirements were according to the literature [
27], the RPA primers displayed an amplification band in non-specific DNA at the experimental level. One reason could be that the recombinase can incorrectly bind the primers to off-targets due to traces of DNA from
Escherichia coli during the purification process of the enzymes used in this study, as previously reported [
37]. In our attempts to eliminate that, betaine 0.8 M was added to the RPA reaction, and the results revealed a specific amplification of primers and more product. This high specificity was also observed when crude leaf extracts from hermaphrodite papaya plants were used in the assay. We decided to use the extract of a hermaphrodite plant, as this is the most important sex in terms of papaya fruit productivity worldwide [
38].
On the other hand, it has been reported that the addition of betaine in isothermal reactions, such as LAMP and RPA, can decrease the melting temperature of DNA and serve as a molecular barrier for intermolecular hybridization between the two strands of DNA (ssDNA), thus hindering the hybridization between the template and primers and increasing the specificity of amplification in the isothermal reactions [
29,
39]. Likewise, the RPA inventors recommend a probe-based detection method that maintains a blockage of 3’-end to prevent a primer amplification by itself [
10]. This approach should be pursued in further analysis for our primer sets to apply it to points of need, such as papaya seedling greenhouses, and thus improve the sexing time. Compared to PCR, RPA could amplify in minutes with lateral flow strips. Likewise, a new set of primers for each sex should be considered for testing and to enhance the specificity of the method. Meanwhile, freeze-drying of reagents and optimization of enzyme production should also be addressed in further studies.
5. Conclusions
The reaction conditions of RPA allowed us to reliably identify the papaya’s sex. The isothermal-based DNA amplification method demonstrated sensitivity and specificity to perform constantly in thirty minutes and at 41°C. Our results might contribute to generating a standardized and practical method of RPA for applying it to papaya seedlings and enhance the development of point-of-need strategies such as detection in lateral flow strips.
Author Contributions
Conceptualization, J.G.A.-H., A.C.-R. and A.M.-A.; methodology, J.G.A.-H., A.C.-A., M.A.P.-A., F.J. CA.; validation, J.G.A.-H., A.C.-A.; project administration, A.M.-A.; formal analysis, J.G.A.-H., A.M.-A.; investigation, J.G.A.-H.; resources, A.M.-A.; writing—original draft preparation, J.G.A.-H.; funding acquisition, A.M.-A.; writing—review and editing, J.G.A.-H., A.C.-A., A.M.-A.; visualization, J.G.A.-H.; supervision, A.M.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by IDEA Guanajuato grant number MA-CFINN0926 to A.M-A for developing the RPA assay and CONAHCYT infrastructure grant number 317147 for equipment and the APC to A.M-A.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.
Acknowledgments
We thank Dr. Luis G. Brieba for giving us the plasmids and methodologies for purifying uvsX, gp32, uvsY, and Bsu DNA polymerase. José Guadalupe Ávila-Hernández thanks to Consejo Nacional de Humanidades, Ciencias y Tecnologías (CONAHCYT, México) and Cinvestav-Irapuato for the financial support for his master studies. The authors thank Claudia Geraldine León-Ramírez and Diego de Jesús Pantoja Gutiérrez for their technical assistance in determining protein concentrations.
Conflicts of Interest
The authors declare they do not have a conflict of interest.
References
- Deputy, J.; Ming, R.; Ma, H.; Liu, Z.; Fitch, M.; Wang, M.; Manshardt, R.; Stiles, J. Molecular markers for sex determination in papaya (Carica papaya L.). Theor Appl Genet 2002, 106, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Saalau-Rojas, E.; Barrantes-Santamaría, W.; Loría-Quirós, C.L.; Brenes-Angulo, A.; Gómez-Alpízar, L. Identificación mediante PCR del sexo de la papaya (Carica papaya L. ), híbrido “Pococí.” Agron. Mesoam 2009, 20, 311–317. [Google Scholar] [CrossRef]
- Aspeitia-Echegaray, V.; Torres-Tapia, Ma.A.; Mendoza-Rodríguez, D.V; Reyes-Valdés, M.H. Evaluación de marcadores genéticos para discriminación entre hembras y hermafroditas de papaya (Carica papaya L.) variedad “Maradol”. Rev. Fitotec. Mex 2014, 37, 193–197. [Google Scholar] [CrossRef]
- Ruiz Ruiz, F.G.; Anaya López, J.L.; Rodríguez Vera, A.P. Evaluación de marcadores moleculares en la predicción temprana de plantas de papaya (Carica papaya L.) de tipo hermafrodita var. Maradol en la zona Costa de Oaxaca. Temas de Ciencia y Tecnología 2017, 21, 22–32. [Google Scholar]
- Araya-Valverde, E.; Bogantes, A.; Holst, A.; Vargas-Mora, C.; Gómez-Alpízar, L.; Brenes, A.; Sánchez-Barrantes, E.; Chavarría, M.; Barboza-Barquero, L. Field performance of hermaphrodite papaya plants obtained through molecular selection and micropropagation. Crop Breed Appl Biotechnol 2019, 19, 420–427. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 2000, 28, e63–e63. [Google Scholar] [CrossRef] [PubMed]
- Compton, J. Nucleic acid sequence-based amplification. Nature 1991, 350, 91–92. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.T.; Fraiser, M.S.; Schram, J.L.; Little, M.C.; Nadeau, J.G.; Malinowski, D.P. Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res 1992, 20, 1691–1696. [Google Scholar] [CrossRef]
- Banér, J.; Nilsson, M.; Mendel-Hartvig, M.; Landegren, U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res 1998, 26, 5073–5078. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol 2006, 4, e204. [Google Scholar] [CrossRef]
- Tapia-Sidas, D.A.; Vargas-Hernández, B.Y.; Ramírez-Pool, J.A.; Núñez-Muñoz, L.A.; Calderón-Pérez, B.; González-González, R.; Brieba, L.G.; Lira-Carmona, R.; Ferat-Osorio, E.; López-Macías, C.; Ruiz-Medrano, R.; Xoconostle-Cázares, B. Starting from scratch: Step-by-step development of diagnostic tests for SARS-CoV-2 detection by RT-LAMP. PloS One 2023, 18, e0279681. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, X.; Wu, R.; Xiao, X.; Xing, F. Development of an on-spot and rapid recombinase polymerase amplification assay for Aspergillus flavus detection in grains. Food Control 2021, 125, 107957. [Google Scholar] [CrossRef]
- Wand, N.I.V.; Bonney, L.C.; Watson, R.J.; Graham, V.; Hewson, R. Point-of-care diagnostic assay for the detection of Zika virus using the recombinase polymerase amplification method. J. Gen. Virol 2018, 99(8), 1012–1026. [Google Scholar] [CrossRef]
- Zou, Y.; Mason, M.G.; Botella, J.R. Evaluation and improvement of isothermal amplification methods for point-of-need plant disease diagnostics. PloS One 2020, 15, e0235216. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Yang, X.; Hu, T.; Jiao, B.; Xu, Y.; Zheng, X.; Shen, D. Comparative evaluation of a novel recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay, LAMP, conventional PCR, and leaf-disc baiting methods for detection of Phytophthora sojae. Front. Microbiol 2019, 10, 1884. [Google Scholar] [CrossRef]
- El Wahed, A.A.; Patel, P.; Maier, M.; Pietsch, C.; Rüster, D.; Böhlken-Fascher, S.; Kissenkötter, J.; Behrmann, O.; Frimpong, M.; Diagne, M.M. Suitcase lab for rapid detection of SARS-CoV-2 based on recombinase polymerase amplification assay. Anal. Chem 2021, 93, 2627–2634. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Kapoor, R.; Kumar, R.; Kumar, S.; Saritha, R.K.; Kumar, S.; Baranwal, V.K. Rapid diagnosis of Cucumber mosaic virus in banana plants using a fluorescence-based real-time isothermal reverse transcription-recombinase polymerase amplification assay. J. Virol. Methods 2019, 270, 52–58. [Google Scholar] [CrossRef]
- Lobato, I. M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trac-Trend Anal Chem 2018, 98, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.; Juma, K.M.; Akagi, S.; Hayashi, K.; Takita, T.; O’Sullivan, C.K.; Fujiwara. S.; Nakura. Y.; Yanagihara. I.; Yasukawa, K. Solvent engineering studies on recombinase polymerase amplification. JBB 2021, 131, 219–224. [Google Scholar] [CrossRef]
- Kojima, K.; Morimoto, K.; Juma, K.M.; Takita, T.; Saito, K.; Yanagihara, I.; Fujiwara. S.; Yasukawa, K. Application of recombinant human pyruvate kinase in recombinase polymerase amplification. JBB 2023, 136, 341–346. [Google Scholar] [CrossRef]
- Juma, K.M.; Inoue, E.; Asada, K.; Fukuda, W.; Morimoto, K.; Yamagata, M.; Takita, T.; Kojima, K.; Suzuki, K.; Nakura, Y.; Yanagihara, I.; Fujiwara, S.; Yasukawa, K. Recombinase polymerase amplification using novel thermostable strand-displacing DNA polymerases from Aeribacillus pallidus and Geobacillus zalihae. JBB 2023, 135, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Juma, K.M.; Kojima, K.; Takita, T.; Natsuaki, K.T.; Yasukawa, K. Comparison of sensitivity and rapidness of PCR, recombinase polymerase amplification, and RNA-specific amplification for detection of Rice yellow mottle virus. J. Biol. Macromol 2021, 21, 27. [Google Scholar] [CrossRef]
- Juma, K.M.; Takita, T.; Ito, K.; Yamagata, M.; Akagi, S.; Arikawa, E.; Kojima, K.; Biyani, M.; Fujiwara, S.; Nakura, Y.; Yanagihara, I.; Yasukawa, K. Optimization of reaction condition of recombinase polymerase amplification to detect SARS-CoV-2 DNA and RNA using a statistical method. Biochem. Biophys. Res. Commun 2021, 567, 195–200. [Google Scholar] [CrossRef]
- Juma, K.M.; Takita, T.; Yamagata, M.; Ishitani, M.; Hayashi, K.; Kojima, K. , Suzuki, K.; Ando, Y.; Fukuda, W.; Fujiwara, S.; Nakura, Y.; Yanagihara, I.; Yasukawa, K. Modified uvsY by N-terminal hexahistidine tag addition enhances efficiency of recombinase polymerase amplification to detect SARS-CoV-2 DNA. Mol. Biol. Rep 2022, 49, 2847–2856. [Google Scholar] [CrossRef] [PubMed]
- Seidman, C.E.; Struhl, K.; Sheen, J.; Jessen, T. Introduction of plasmid DNA into cells. Curr. Protoc. Mol. Biol 1997, 37, 1.8.1–1.8.10. [Google Scholar] [CrossRef]
- Cordoba-Andrade, F.; Peralta-Castro, A.; García-Medel, P.; Castro-Torres, E.; González-González, R.; Castro-Lara, A.Y.; Mora Garduño, J.D.; Raygoza, C.D.; Baruch-Torres, N.; Díaz-Quezada, C. An open-source bacteriophage T4 recombinase polymerase amplification (RPA) system for pathogen detection: A proof of concept on SARS-CoV-2 detection. In Preparation.
- Strayer-Scherer, A.; Jones, J.B.; Paret, M.L. Recombinase polymerase amplification assay for field detection of tomato bacterial spot pathogens. Phytopathology 2019, 109, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Hernández, J.G.; Camas-Reyes, A.; Martínez-Antonio, A. Sex determination of papaya var. ‘Maradol’ reveals hermaphrodite-to-male sex reversal under greenhouse conditions. Crop Breed Appl Biotechnol 2023, 23, e457923312. [Google Scholar] [CrossRef]
- Luo, G.C.; Yi, T.T.; Jiang, B.; Guo, X.L.; Zhang, G.Y. Betaine-assisted recombinase polymerase assay with enhanced specificity. Anal. Biochem 2019, 575, 36–39. [Google Scholar] [CrossRef]
- Satya, P.; Mitra, S.; Ray, D.P.; Mahapatra, B.S.; Karan, M.; Jana, S.; Sharma, D.A. Rapid and inexpensive NaOH based direct PCR for amplification of nuclear and organelle DNA from ramie (Boehmeria nivea), a bast fibre crop containing complex polysaccharides. Ind Crop Prod 2013, 50, 532–536. [Google Scholar] [CrossRef]
- Babujee, L.; Witherell, R.A.; Mikami, K.; Aiuchi, D.; Charkowski, A.O.; Rakotondrafara, A.M. Optimization of an isothermal recombinase polymerase amplification method for real-time detection of Potato virus YO and N types in potato. J. Virol. Methods 2019, 267, 16–21. [Google Scholar] [CrossRef]
- Larrea-Sarmiento, A.; Stack, J.P.; Alvarez, A.M.; Arif, M. Multiplex recombinase polymerase amplification assay developed using unique genomic regions for rapid on-site detection of genus Clavibacter and C. nebraskensis. Sci. Rep 2021, 11, 12017. [Google Scholar] [CrossRef] [PubMed]
- Londoño, M.A.; Harmon, C.L.; Polston, J.E. Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics. Virol. J 2016, 13, 1–9. [Google Scholar] [CrossRef]
- Munawar, M.; Toljamo, A.; Martin, F.; Kokko, H. Recombinase polymerase amplification assay for fast, sensitive and on-site detection of Phytophthora cactorum without DNA extraction. Eur. J. Hortic. Sci 2019, 84, 14–19. [Google Scholar] [CrossRef]
- Hsu, T.H.; Gwo, J.C.; Lin, K.H. Rapid sex identification of papaya (Carica papaya) using multiplex loop-mediated isothermal amplification (mLAMP). Planta 2012, 236, 1239–1246. [Google Scholar] [CrossRef]
- Tsai, C.C.; Shih, H.C.; Ko, Y.Z.; Wang, R.H. , Li, S.J.; Chiang, Y.C. Direct LAMP assay without prior DNA purification for sex determination of papaya. Int. J. Mol. Sci 2016, 17, 1630. [Google Scholar] [CrossRef] [PubMed]
- Lupan, I.; Ianc, M.B.; Ochis, C.; Popescu, O. The evidence of contaminant bacterial DNA in several commercial Taq polymerases. Rom. Biotechnol. Lett 2013, 18, 8007–12. [Google Scholar]
- Ávila-Hernández, J.G.; Cárdenas-Aquino, M.d.R.; Camas-Reyes, A.; Martínez-Antonio, A. Sex determination in papaya: Current status and perspectives. Plant Sci. J 2023, 335, 111814. [Google Scholar] [CrossRef]
- Ma, C.; Wang, Y.; Zhang, P.; Shi, C. Accelerated isothermal nucleic acid amplification in betaine-free reaction. Anal. Biochem 2017, 530, 1–4. [Google Scholar] [CrossRef]
Figure 1.
Mode of action of recombinase polymerase amplification method.
Figure 1.
Mode of action of recombinase polymerase amplification method.
Figure 2.
Effect of temperature, time, Bsu-DNAP concentration, and sensitivity on the reaction efficiency of RPA. RPA reaction (20 µL) was carried out with the following conditions: 50 mM Tris-HCl buffer (pH 8.6), 40 mM CH3COOK, 6.0% PEG35000, 2 mM DTT, 650 µM dNTPs, 1 µM primer forward, 1 µM primer reverse, 20 mM phosphocreatine, 120 ng/µL creatine phosphokinase, 3.5 mM ATP, 40 ng/µL uvsY, 600 ng/µL gp32, 200 ng/µL (a,b) and 25 ng/µL (d) Bsu DNA polymerase, 400 ng/µL uvsX and 14 mM Mg(OCOCH3)2. Time of reaction, 30 min (a,c,d); temperature, 41°C (b,c,d); standard DNA ~20 ng/µL (a–c). In (a), * means the amplification before heat denaturation of the sample.
Figure 2.
Effect of temperature, time, Bsu-DNAP concentration, and sensitivity on the reaction efficiency of RPA. RPA reaction (20 µL) was carried out with the following conditions: 50 mM Tris-HCl buffer (pH 8.6), 40 mM CH3COOK, 6.0% PEG35000, 2 mM DTT, 650 µM dNTPs, 1 µM primer forward, 1 µM primer reverse, 20 mM phosphocreatine, 120 ng/µL creatine phosphokinase, 3.5 mM ATP, 40 ng/µL uvsY, 600 ng/µL gp32, 200 ng/µL (a,b) and 25 ng/µL (d) Bsu DNA polymerase, 400 ng/µL uvsX and 14 mM Mg(OCOCH3)2. Time of reaction, 30 min (a,c,d); temperature, 41°C (b,c,d); standard DNA ~20 ng/µL (a–c). In (a), * means the amplification before heat denaturation of the sample.
Figure 3.
Evaluation of set of primers related to sex of papaya. PCR (a) and RPA (b) test of each primer individually; rpaCpTrnL (lane 1), rpaW11 (lane2), and rpaPMSM2 (lane 3), using genomic DNA (a) or standard DNA (b). (c–e) Sex determination by RPA using standard DNA and specific primers according to their sex, female (c), hermaphrodite (d), and male (e). Female, 150 bp; hermaphrodite, 165 bp; and male, 150 bp. M: molecular weight marker. Temperature, 41°C; time, 30 min; and Bsu-Pol concentration, 25 ng/µL.
Figure 3.
Evaluation of set of primers related to sex of papaya. PCR (a) and RPA (b) test of each primer individually; rpaCpTrnL (lane 1), rpaW11 (lane2), and rpaPMSM2 (lane 3), using genomic DNA (a) or standard DNA (b). (c–e) Sex determination by RPA using standard DNA and specific primers according to their sex, female (c), hermaphrodite (d), and male (e). Female, 150 bp; hermaphrodite, 165 bp; and male, 150 bp. M: molecular weight marker. Temperature, 41°C; time, 30 min; and Bsu-Pol concentration, 25 ng/µL.
Figure 4.
Effect of RPA components on the reaction efficiency. (a) Lane 1, optimized RPA reaction (20 µL) with all components; effect of absence of enzymes: lane 2, uvsY; lane 3, uvsX; lane 4, gp32; lane 5, absence of ATP. (b) Effect of mixing and heat denaturation: applied (lane 1) or not applied (lane 2). M: molecular weight marker.
Figure 4.
Effect of RPA components on the reaction efficiency. (a) Lane 1, optimized RPA reaction (20 µL) with all components; effect of absence of enzymes: lane 2, uvsY; lane 3, uvsX; lane 4, gp32; lane 5, absence of ATP. (b) Effect of mixing and heat denaturation: applied (lane 1) or not applied (lane 2). M: molecular weight marker.
Figure 5.
Non-specific amplification of primers on RPA reaction. RPA evaluation of primers using standard DNA of male (lanes 1-3), female (lanes 4-6), and hermaphrodite (lanes 7-9). Lanes 1, 4, and 7, RPA reaction with rpaCpTrnL primers; lanes 2, 5, and 8, RPA reaction with rpaW11 primers; lanes 3, 6, and 9, RPA reaction with rpaPMSM2 primers. The effect of the addition of betaine 0.8 M to the RPA reaction is observed in lanes 4-9. In crude extract evaluation from the hermaphrodite plant, two bands were observed confirming sex identification (lanes 10-12).
Figure 5.
Non-specific amplification of primers on RPA reaction. RPA evaluation of primers using standard DNA of male (lanes 1-3), female (lanes 4-6), and hermaphrodite (lanes 7-9). Lanes 1, 4, and 7, RPA reaction with rpaCpTrnL primers; lanes 2, 5, and 8, RPA reaction with rpaW11 primers; lanes 3, 6, and 9, RPA reaction with rpaPMSM2 primers. The effect of the addition of betaine 0.8 M to the RPA reaction is observed in lanes 4-9. In crude extract evaluation from the hermaphrodite plant, two bands were observed confirming sex identification (lanes 10-12).
Table 1.
RPA primers used in this study.
Table 1.
RPA primers used in this study.
| Sex to identify |
Primer name |
Sequence (5´-3´) |
Size of RPA product |
%GC content |
| Female |
rpaCpTrnL-F
rpaCpTrnL-R |
GGGGATATGGCGAAATCGGTAGACGCTACGGA
TGTTTGTTCTCGTAAAACAGGATTTGGCTCAG |
150 bp |
56
41 |
| Hermaphrodite |
rpaW11-F
rpaW11-R |
TGGATCGTGCTCCTAGTGCTCATGGTGACACC
CTGATGCGTGTGTGGCTCTATCTATATGTGTG |
165 bp |
56
47 |
| Male |
rpaPMSM2-F
rpaPMSM2-R |
GCGATGCTTCAAGTGTTGACATAAAGGCAGTT
AATATCCCTCTAATACTCTCACCAAGGCATAC |
150 bp |
44
41 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).