1. Introduction
As an important component of global aquatic ecosystems, freshwater lakes provide habitats for animals and plants and support global material cycling [
1,
2]. However, freshwater lakes, especially shallow lakes, are susceptible to climate change and human disturbances, resulting water scarcity, area loss, overexploitation, pollution, and ecological degradation [
3,
4]. Sediment, which is not only a reservoir of various substances but also an important interface for water-soil physical, chemical and biological reactions, is an essential component of shallow lake ecosystems [
5]. The highly heterogeneous spatial structure and complex chemical composition of resources in the sediment provide diverse microhabitats for biological groups [
6] so that sediment have high biomass and biodiversity. Fungi constitute a substantial proportion of microorganisms in the sediment and play a driving role in the process of material cycling and energy transfer [
7,
8]. However, owing to methodological limitations, the systematic analysis of fungal diversity and the ecological roles of fungi in shallow lake ecosystems had not progressed for years [
9].
The application and popularization of high-throughput sequencing technology and related methods provide powerful tools for studying microbiotic diversity [
10]. A large number of fungal species that are difficult to be observed by morphology or culture methods have been discovered, which greatly enriches the research on microbial diversity in ecological environments. However, little is known about the compositions of fungal communities and their ecological roles [
11], especially in lake sediment. Moreover, there are also gaps in the knowledge on the response of the compositions and dynamics of the fungal communities to the physical and chemical properties of sediments and the concentration of contaminants, which are influenced by human activities. Therefore, the study of fungal communities in sediments is helpful to reveal the mechanisms by which they are involved in ecological lake processes, as well as to exploring the overall impact of human activities on shallow lake ecosystems.
2. Materials and Methods
2.1. Study Area and Sampling
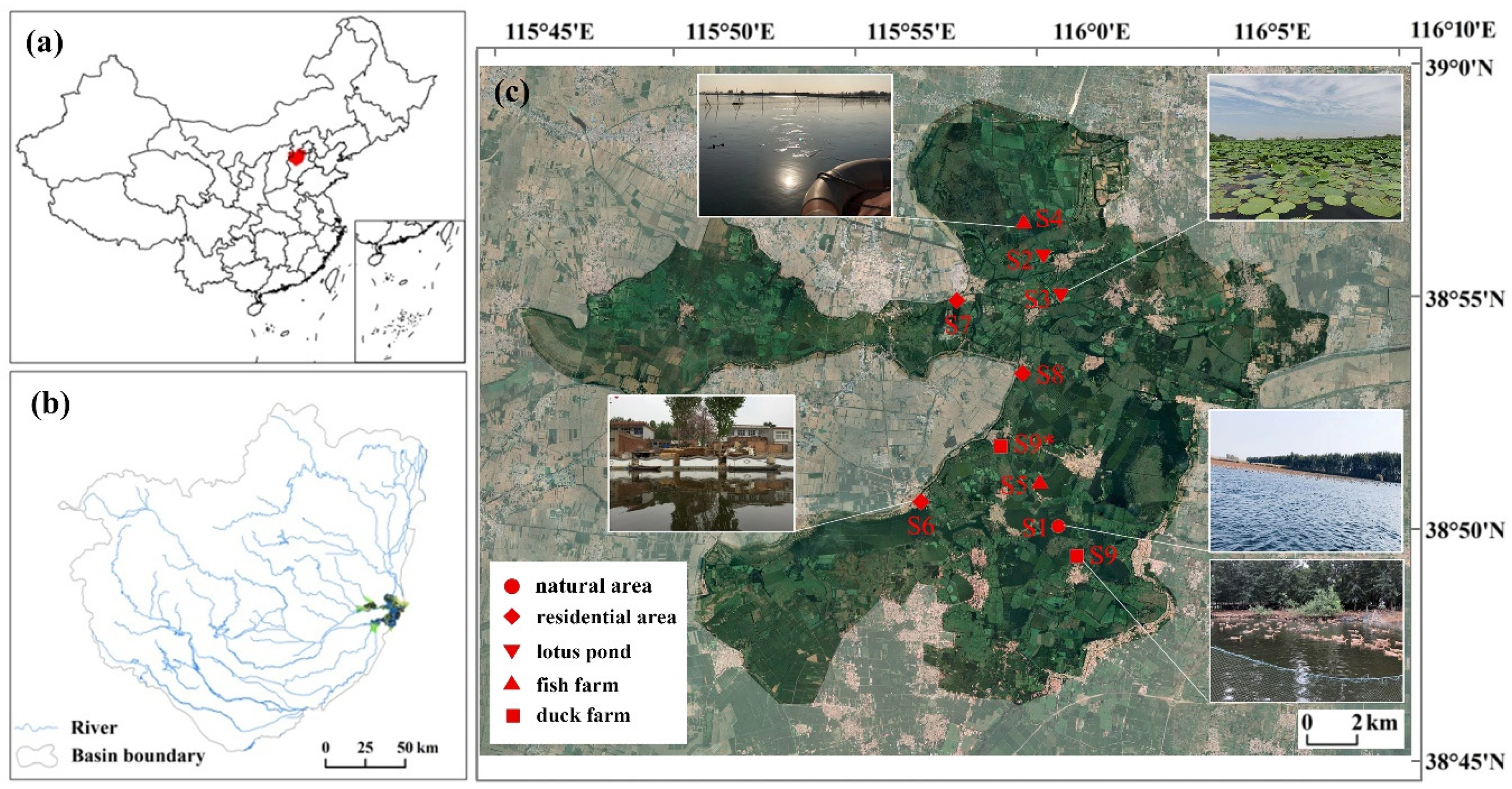
Baiyangdian Lake (BYD Lake) is the largest freshwater lake in the North China Plain and has important ecological functions [
12]. This macrophytic lake, which consists of 143 lakes and over 3,700 trenches, with a total area of 366 km2, locates 100 km southeast of Beijing and 40 km east of the city of Baoding (38°43′N ~ 39°02′N, 115°45′E ~ 116°07′E). According to the administrative divisions, 39 villages locate within or around the lake. The lake had been suffering from eutrophication during the 2000s as agricultural, aqua cultural, residential sewages used to directly discharge into the lake [
13].
9 sampling sites with different disturbance levels of human activities were selected in this study (
Figure 1). The sites included most typical habitats (natural areas, residential areas, lotus ponds, fish farms, duck farms) of BYD Lake. S1 located in the Baiyangdian National Aquatic Germplasm Reserve; S2 and S3 located in lotus ponds around villages; S4 and S5 located in fish farms with area of 10hm
2 and over 100 hm
2 respectively; S6, S7 and S8 located within or around villages with 10,000, 1000 and 4000 inhabitants respectively; S9 located in a duck farm which was removed in 2018 and was replaced by another duck farm S9*. Sediment samples were collected in December 2017, March 2018 and July 2018, which represented for winter, spring and summer respectively, with 3 parallel samples at each site. Sediments were collected using a Petersen mud grabber and then mixed, cooled and quickly sent to the lab to be stored at -20°C.
2.2. Determination of Physicochemical Properties and Heavy Metals
The temperature of sediment was measured at the same time as sampling occurred. The sediment pH level was measured in a slurry of sediment and water (1:2.5) using a pH meter (ST3100/F, U.S.). The contents of carbon (C) and nitrogen (N) were detected using an elemental analyzer (VARIO EL, Germany). Before detection, the inorganic carbon in the sediment was removed by adding 5 mL of 1 mol/L hydrochloric acid (HCl) to a sample of 2 g of sediment, and then the sample, as well as 2 g of the raw sample, was oven-dried. Dried samples were weighed to obtain the ratio of raw sediment to acidified sediment, and then the acidified sample was ground and passed through a 100-mesh screen. The contents of total nitrogen (TN) and total organic carbon (TOC) of each sample were recalculated by multiplying the ratio of raw sediment to acidified sediment. Then, 100 ml of a 2 mol/L KCl solution was added to 10 g of sediment, and the mixture was shaken for 30 min to measure the concentrations of nitrate (NO
3-) by comparing the absorbance. Ammonium nitrogen (NH
4+) content was determined by continuous air-segmented flow analysis using a Skalar SAN Plus segmented flow analyzer (Skalar Inc., Breda, The Netherlands). Inductively coupled plasma atomic emission spectrometry (PECTRO ARCOS EOP, USA) was used to analyze the contents of total phosphorus (TP) and heavy metals (arsenic (As), cadmium (Cd), chromium (Cr), copper (Cu), lead (Pb), zinc (Zn), cobalt (Co), manganese (Mn), nickel (Ni) and iron (Fe)) of each sample, which had been digested with nitric acid-hydrofluoric acid-hydrochloric acid (HNO
3-HF-HCl) in an automatic digestion instrument (ST-60, China). The contents of different phosphorus forms derived from the summary of reference [
14].
2.3. DNA Extraction, PCR Amplification and Sequencing
Sediment DNA was extracted from 0.4 g samples using a Fast DNA Spin Kit (Mobio, USA) according to the instructions. The primers SSU 1196R and SSU 0817F [
15] were used to amplify the 18S rRNA gene of the fungi. The PCR products were purified by the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified by the QuantiFluorTM-ST (QuantiFluor, USA) instrument and then sequenced on the Illumina MiSeq platform (Illumina, USA). QIIME [
16] and MOTHUR [
17] were used to filter, stitch and control the original sequencing data. Sequences with 97% similarity were clustered into operational taxonomic units (OTUs) using UCLUST [
18]. The OTUs were compared with Unite 18S rRNA database [
19] to obtain the classifications of the OTUs. The China Biological Species List (2020 version) [
20] and Species 2000 (2019 version) [
21] were referred to for classifying uncertain species.
2.4. Statistical Analyses
Statistical analysis were implemented in R (version 3.6.2,
https://www.r-project.org/). The two-way analysis of variance test (Two-way ANOVA) was used to determine the differences in the contents of heavy metals, physicochemical properties and microbial alpha diversity indices among sampling sites and seasons with the
car package. The α diversity indexes (Sob (species observed), Chao1, Simpson and Shannon Index) were calculated with the
picante package. Principal co-ordinates analysis (PCoA) and analysis of similarities (ANOSIM) were used to analyze the β diversity of different sites and seasons with the
vegan package. Redundancy analysis (RDA) was used to investigate the influence of the environmental factors on fungal communities with the
vegan package, and the multicollinearity of different parameters were modified by checking the variance inflation factors (VIF). Pearson relationship was used to analyze the relationships of the environmental factors with the
Hmisc package. Figures were drawn with Origin (version 2017, OriginLab, USA) and R.
3. Results
3.1. Sediment Physical and Chemical Properties
The physical and chemical properties of the sediments are shown in
Table 1. The concentrations of total phosphorus (TP), calcium-bound phosphorus (HCl-P), As, Cd, Co, Zn and Mn showed no significant difference among seasons, while others varied significantly. Values of environmental factors of each site are shown in
Figure S1. The results also indicate significant differences among sites. For example, S2 and S3 had higher concentrations of total nitrogen (TN), total organic carbon (TOC), NH
4+, Cu and Ni. S9* had highest concentrations of TP, organic insoluble phosphorus (Res-P), metal-oxide-bound phosphorus (NaOH-P), HCl-P, while had lower NH
4+, As, Pb, Co and Mn. S6, S7 and S8 had higher weakly adsorbed phosphorus (NH
4Cl-P) and Fe.
The results of Pearson relationships were shown in
Table S1. The concentrations of some factors were highly correlated, for example TP and HCl-P (r=0.98, p < 0.01), TN and TOC (r=0.96, p < 0.01), Cu and Ni (r=0.83, p < 0.01), Co and Fe (r=0.88, p < 0.01), indicating that it’s difficult to distinguish these factors that affect fungal communities.
3.2. Compositions of Fungal Community in Sediments
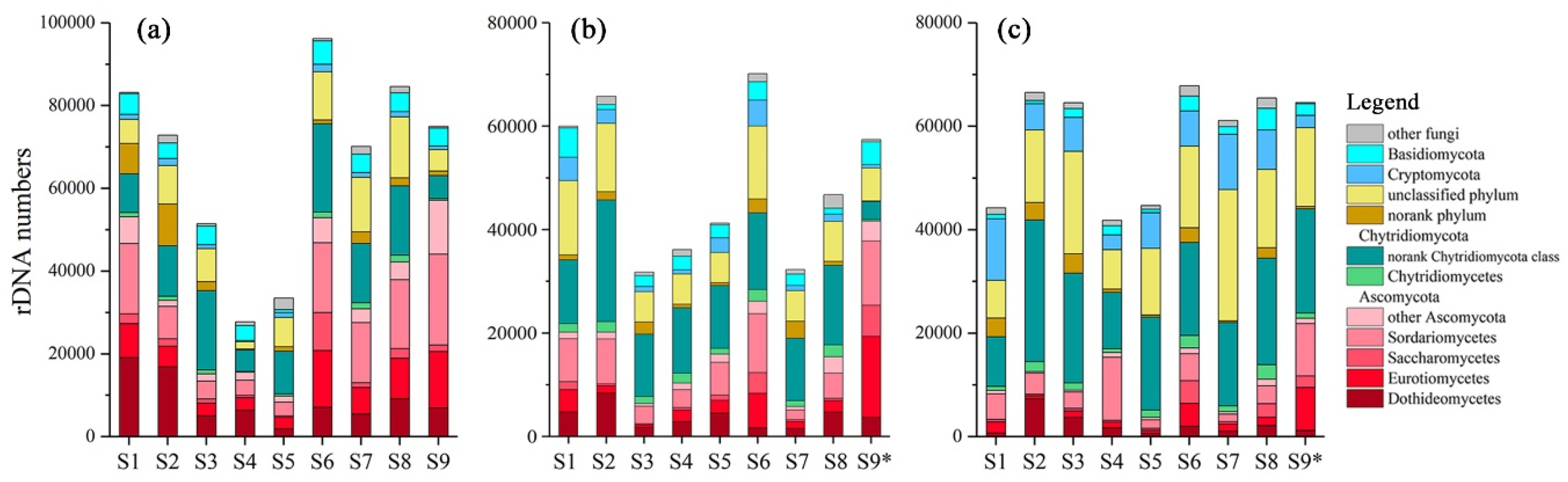
1,110,680, 851,215 and 989,346 sequences were obtained in winter, spring and summer respectively. These sequences were clustered into 772, 1031 and 1118 OTUs respectively, and the OTUs were identified into 537 species in total. 111, 99 and 89 species were filtered as fungi, accounting for 53.5%, 51.9% and 52.6% of sequences in winter, spring and summer respectively. The sequence numbers of fungi in different seasons and different sites were shown in
Figure 2. Sequence numbers and community compositions variated among seasons and sites. The fungi mainly belonged to Ascomycota and Chytridiomycota, accounting for 35.78% (7.92%~76.19%) and 28.79% (6.62%~44.01%) of all fungi sequences respectively. Dothideomycetes, Eurotiomycetes and Sordariomycetes were the most abundant classes of Ascomycota, while most Chytridiomycota belonged to a norank class. Basidiomycota, Blastocladiomycota and Cryptomycota accounted for 5.14% (0.92%~13.19%), 1.76% (0.11%%~7.54%) and 5.88 (1.00%~26.95%) respectively. Little Glomeromycota or Zygomycota were obtained, adding up to less than 1% of the fungi sequences. Besides, 3.74% (0.17%~13.86%) and 18.45% (6.37%~41.52%) sequences belonged to a norank phylum and an unclassified phylum respectively.
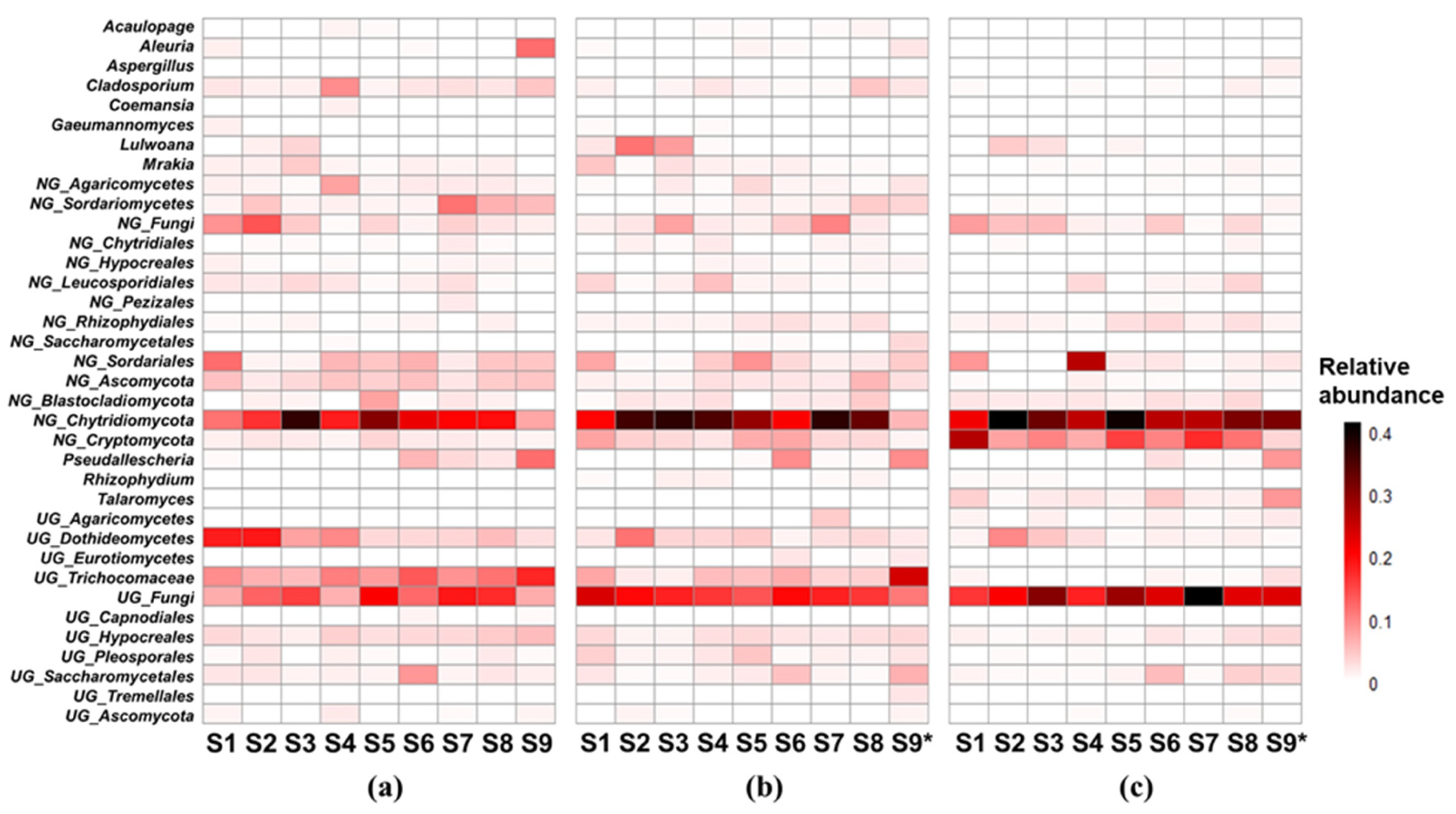
The patterns of dominant genera, which had relative abundances over 1% in at least one site, were shown in
Figure 3. There were 28, 29 and 25 dominant genera in winter, spring and summer respectively. 11, 4 and 3 genera dominated all sites in winter, spring and summer respectively. 3 genera, including an unclassified fungi genus (
UG_fungi), a norank Chytridiomycota genus (
NG_Chytridiomycota), and a norank Cryptomycota genus (
NG_Cryptomycota) dominated all sites and all seasons. The percentages of these 3 genera accounted for 35.40% (15.41%~55.49%), 36.14% (7.50%~50.80%) and 68.21% (55.12%~85.21%) in winter, spring and summer respectively.
3.3. Fungal Diversity in Sediment
The α diversity indexes were shown in
Figure 4. The ranges of the Sob, Chao1, Shannon and Simpson indices for all samples were 38~83, 38.0~90.5, 2.54~4.57 and 0.06~0.37, respectively. The Sob index and Chao1 index were higher in winter, indicating more species detected. Significantly lower Shannon index and higher Simpson index in summer indicated lower fungal diversity in this season. The fungal diversity was highest in winter and lowest in summer, and in spring the biodiversity was in between. The differences of indexes among sites were not as significant. Sob index of S1, S3 and S5 were significantly lower than S6, S7, S8 and S9(S9*), while the Chao1, Shannon and Simpson indexes among sites showed no significant differences.
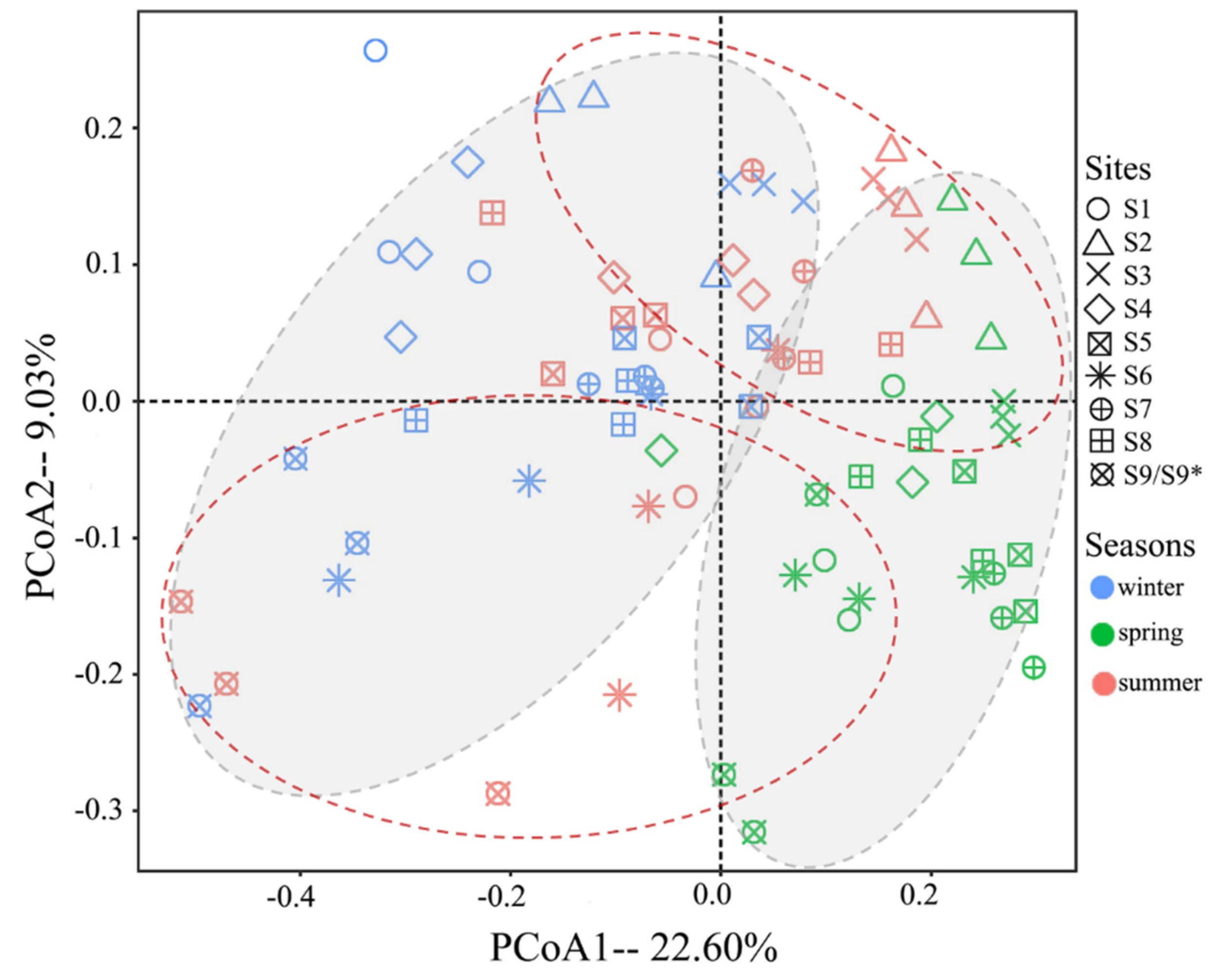
The results of PCoA were shown in
Figure 5. For seasonal analysis, winter and summer samples (in gray oval zones) were generally clustered into two groups, which distributed along the two sides of the first axis, and the spring samples were scattered among them. For sampling sites, most samples mixed, but lotus ponds (S2&S3) and duck farms (S9/S9*) were clustered into two groups (dashed red oval zones). ANOSIM results (
Figure S5) showed significant variations of fungal communities among both sites and seasons (p = 0.001), but when all species in the same season were composed and contrasted, the variations among seasons were low (R = 0.085).
3.4. RDA of Environmental Factors and the Fungal Community
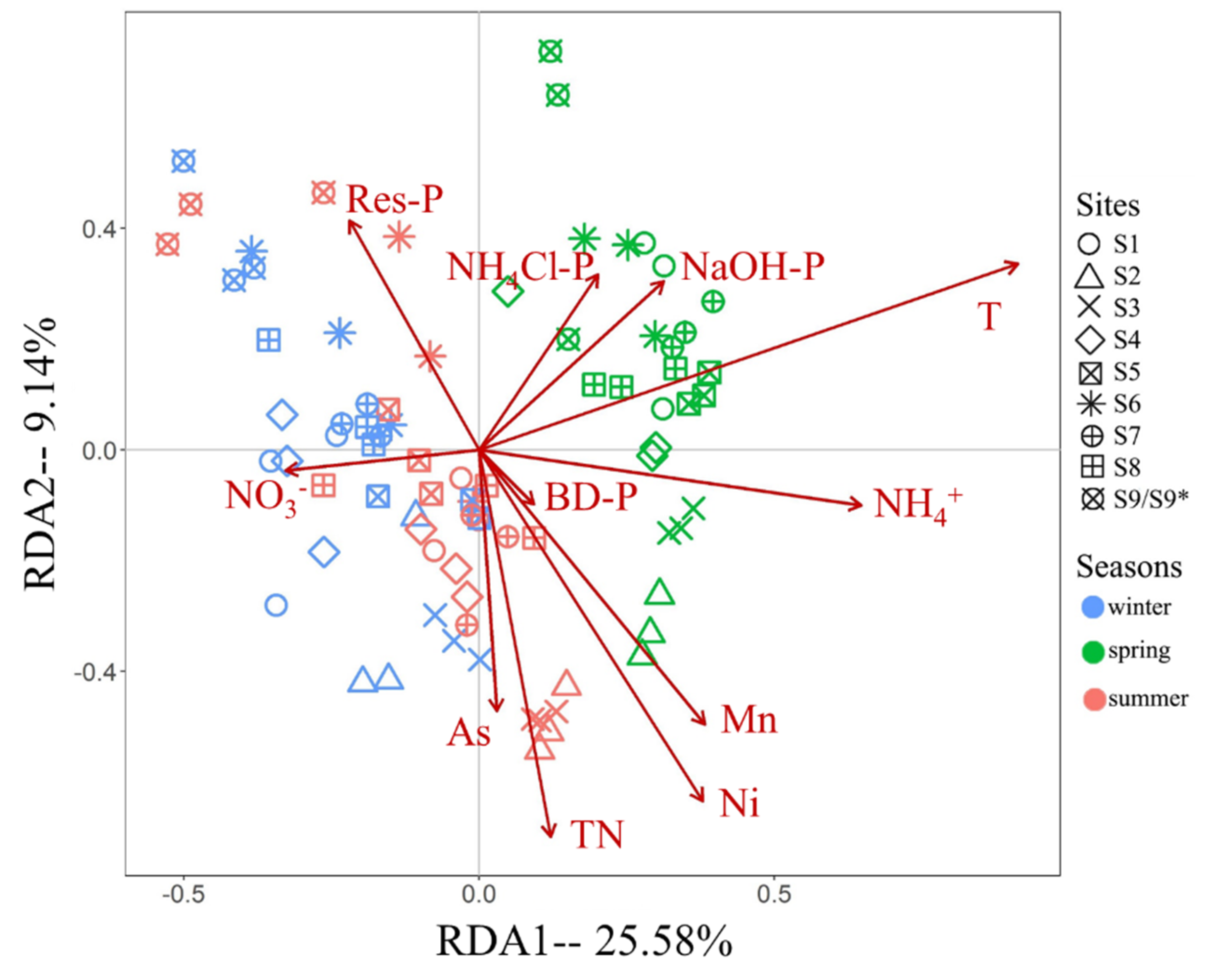
RDA result of the influence of environmental factors on the fungal communities of the BYD Lake sediment was shown in
Figure 6. Interpretation rates of significant environmental factors were shown in
Table 2. Parameters had significant effects explained 48.98% of the variations across three seasons, and the first and second axises accounted for 34.72%. The influence of temperature was the greatest. Phosphorus, including forms of Res-P, NaOH-P, BD-P and NH
4Cl-P, had the second greatest influence, accounting for 11.67% of the rates. TN, NO
3- and NH
4+ had almost entirely different effects, and their interpretation rates accounted for 8.08%. Significant heavy metals included As, Mn and Ni, and their influences accounted for 7.16%. Noted that high correlations (
Table S1) had been found between some factors. For example, Res-P and TP, TN and TOC, Cu and Ni, indicating the influences of some environmental factors may due to the related factors.
4. Discussion
4.1. The Compositions of Fungal Communities in the Sediment Environment of Shallow Lakes
The humus-rich anoxic environments of shallow lake sediments were suitable for fungi, which play important roles in the transformation of plant residues into fodder for invertebrates and other organisms [
22]. This study revealed highly diverse communities of fungi in the BYD Lake sediments, covering all known fungi phyla [
23]. Studies had found that Ascomycota accounts for a large percentage in the sediment [
24,
25,
26,
27], and this was consistent with that in the BYD Lake. However, the proportions of Chytridiomycota and other fungi such as Cryptomycota were much higher in the BYD Lake, especially in summer. Chytrids which produce spores with a motile cilium that helps to swim and/or crawl [
28], are adapted to aquatic habitats [
29,
30] and prefer saprophytic environments with animal and plant residues [
31]. Besides, a large number of chytrids parasitize algae [
32]. High proportion of Chytridiomycota in BYD Lake sediment may related to the abundance of plant residues, as well as algae that caused by eutrophication in summer [
33]. Cryptomycota species were commonly found as parasites in aquatic and soil ecosystems, and the abundance of this phylum often related to their host, which includes Chytridiomycota [
34].
Most Zygomycota species are terrestrial fungi [
35] and rare in aquatic habitats. However, Glomeromycota form symbionts of arbuscular mycorrhiza with plants [
36], and common reeds (
Phragmites australis) in BYD Lake were commonly colonized [
37]. It was unreasonable that little Glomeromycota species was identified in this study. We guess the primers may not be suitable for PCR amplification of this phylum. Otherwise, those norank fungi (
NG_Fungi) or unclassified fungi (
UG_Fungi) that account for large proportions may be belong to Glomeromycota, as most genome of arbuscular mycorrhizal fungi were not clear and the genome of only a few species were published in recent years [
38,
39,
40,
41].
Besides fungi, a great number of eukaryotes species were identified from the OTUs (accounted for almost half sequence numbers,
Table S2). Most of them were flagellates or ciliates, belonging to Choanozoa or Ciliophora. Unexpectedly, only a handful algae species were identified, and most of them belonged to Cryptophyta, which consist of single-celled algae with two flagella. These microorganisms have similar physiological structures and closer phylogeny with fungi, as cilium or flagellum also present in fungi [
42]. This may be the reason why these species presented in the PCR products.
4.2. The Temporal and Spatial Variations of Fungal Communities
Temporal and spatial dynamics of fungal communities had been revealed in freshwater ecosystems, but researchers have not reached a consensus whether seasons or habitats have a greater influence [
43]. In this study, the results of ANOSIM (
Figure S2) indicated high specificity of fungal communities in different habitats. When samples from the same season were composed and analyzed, there was almost no difference between intra group and inter group differences. This indicated that habitat has higher influence on the compositions of fungal communities. It was contrary to the α diversity indexes that showed highest biodiversity in winter and no significant differences among most sites. However, the β diversity verified that the compositions of fungal communities differed among sites, especially those in lotus ponds and duck farms. Dominant genera showed in
Figure 3 indicated that the most dominant fungi maintained their dominance across seasons. Meanwhile, other taxa, which took a relatively small part in the relative abundance but composed a considerable part of the species numbers, resulted in higher α diversity index [
44].
The effects of habitats on fungal communities were also manifest in their compositions and biomass. The genera contributing to similarity of fungal composition between lotus ponds and duck farms (
Table S4) revealed significantly higher percentages of
NG_Chytridiomycota,
NG_Cryptomycota, and
UC_Dothideomycetes in lotus ponds, as well as higher percentages of
UG_Trichocomaceae,
Pseudallescheria and
Talaromyces in duck farms. Some fungi only thrive in habitats with intense human activities. For instance, Pseudallescheria boydii is a common human pathogen [
44], and in this study we found this species only enriched in S6, S7, S8 and S9/S9*, where there were more human activities. There may be more species related to human activities present, but it is hard to judge due to the poor understanding of their taxonomy and ecological functions. Moreover, although the fungal diversity was not significantly lower in fish farms than other habitats, the sequence numbers were the least in S4 and S5. This may relate to fish cultivation measures that inhibit microorganisms, and the abundance of Ichthyosporea (
Table S2), which include several common fish parasites [
45], supported this option.
4.3. The Factors Affect Sediment Fungal Community Compositions
Generally, temperature is negative to fungal diversity [
46]. In this study, we found there were more dominant genera in winter and spring, including some psychrophilic fungi genera, for example Cladosporium and Mrakia [
47]. The psychrophilic fungi were common in winter but rare in summer. However, there were also some fungi adapted to high temperature. For example, Talaromyces which is a mold in the environment and a yeast in the tissues at high temperature [
48], was rare in winter or spring but common in summer. The percentages of the parasitic fungi Cryptomycota were much higher in summer, and it may relate to their higher biological activities at higher temperatures. Meanwhile, the influence of temperature on Chytridiomycota is not significant since the percentages of Chytridiomycota did not vary greatly among seasons.
Researchers have different views about the effects of nutrient on microorganisms, suggesting phosphorus and nitrogen may have positive or negative effects [
49,
50], and the types of nutrient exert different degrees of effects on the fungal structure [
51]. According to the results of this study, NO
3- and NH
4+ had opposite effects on the fungal communities, and their effects were almost completely different from that of TN, which mainly composed of organic nitrogen [
52]. These may due to the difference of mycorrhizal fungi in utilization different nitrogen sources [
53,
54,
55].However, there were no evidence that certain fungus species were capable to take up nitrogen in a certain form, partly because most fungus species are poorly identified. Studies have verified that soil microorganism communities were affected by active phosphorus [
49], as P is one of the most important macronutrient for organisms. Unexpectedly, inert Res-P also showed significant influence and had the highest interpretation rate among all P forms. This indicates that some fungi in the sediment may be effective phosphate-solubilizing [
56].
In most cases, the accumulation of heavy metals causes adverse effects on microorganisms [
57]. However, studies have also illustrated that heavy metals have no influence on microbial community compositions [
58]. In this study, Except for Cr, there was no significant difference of heavy metal concentrations between the sediments and the regional background values (
Table S3, [
59]), and the influence of Cr on fungal communities was not significant. Heavy metals contributed to the interpretation of the influence of environmental factors on fungal community composition; however, the influence and the mechanisms were not clear, so more research needs to be done.
5. Conclusions
High-throughput sequencing technology provides a convenient means for detecting microbial diversity in the environment, but there are still many shortcomings. More than 100 fungal species had been identified in the sediment of BYD Lake, but most of their information was not clear, including some important fungi, such as Glomeromycota. More studies are required to further explore their ecological functions.
In lake sediment, more fungal species prefer cold environment, and the number of rare species in winter is significantly higher than those in other seasons, resulting to higher biodiversity. There was no significant difference of fungal diversity among different habitats, while the composition of the communities was quite different. The impacts of human activities on fungal communities were not as strong as expected. Some human-related fungi only concentrated in habitats with intensive human activities, and the fungal biomass was lower in fish farms due to fish cultivation measures.
Environmental factors have various effects on fungal communities. In general, the most influential factor temperature is negative to fungal diversity, but some fungi such as Cryptomycota, dominant at high temperatures. Different forms of N (NO3-, NH4+ and organic N) have almost completely different roles in shaping the fungal community. Different forms of P also showed different effects. The effects of heavy metals are not clear and more research is needed.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org, Figure S1. Contents of sediment physical and chemical properties among all habitats in three seasons. Error bars represented the standard deviations. Figure S2. ANOSIM results of fungal communities in different sites and seasons: (a) sites, (b) seasons. Table S1. Pearson relationships of the environmental factors of Baiyangdian Lake sediments. Table S2. Secequance numbers of fungi at phylum level in BYD Lake sediments. Table S3. Background value of the concentrations of heavy metals in soil of Hebei Province. Table S4. Key genera contributing to similarity of fungal composition between lotus ponds and duck farms.
Author Contributions
Conceptualization, Yujun Yi; methodology, Yujun Yi and Senlu Yin; software, Senlu Yin; validation, Yujun Yi and Senlu Yin; formal analysis, Senlu Yin; investigation, Yujun Yi and Senlu Yin; resources, Yujun Yi; data curation, Senlu Yin; writing—original draft preparation, Senlu Yin; writing—review and editing, Yujun Yi and Senlu Yin; visualization, Senlu Yin; supervision, Yujun Yi; project administration, Yujun Yi; funding acquisition, Yujun Yi. All authors have read and agreed to the published version of the manuscript.
Funding
Please add: This work was supported by the National Science Fund for Distinguished Young Scholars (52025092), the Innovation Research Group Project of the National Science Foundation of China (52221003), and the Joint Funds of the National Natural Science Foundation of China (U2243236).
Data Availability Statement
Acknowledgments
The authors would like to thank Wenjun Wang and Chuqiao Lin for their assistance with the field campaign.
Conflicts of Interest
The authors declared that they have no conflicts of interest to this manuscript. The authors declared that they do not have any commercial or associative interest that represents a conflict of interest in connection with the manuscript submitted.
References
- Postel, S., Carpenter, S., 1997. Freshwater ecosystem services. Daily, G.C.(Ed.) Nature’s Services: Societal Dependence on Natural Ecosystems, Island Press, pp. 195–214.
- Zedler, J.B. Progress in wetland restoration ecology. Trends in Ecology & Evolution 2000, 15, 402–407. [Google Scholar]
- Brönmark, C.; Hansson, L.A. Environmental issues in lakes and ponds: Current state and perspectives. Environmental Conservation. 2002, 29, 290–307. [Google Scholar] [CrossRef]
- Jeppesen, E.; Brucet, S.; Naselli-Flores, L.; et al. Ecological impacts of global warming and water abstraction on lakes and reservoirs due to changes in water level and related changes in salinity. Hydrobiologia 2015, 750, 201–227. [Google Scholar] [CrossRef]
- Webster, J.; Ridgway, I. The application of the equilibrium partitioning approach for establishing sediment quality criteria at two UK sea disposal and outfall sites. Mar Pollut Bull. 1994, 28, 653–661. [Google Scholar] [CrossRef]
- Ferris, H.; Tuomisto, H. Unearthing the role of biological diversity in soil health. Soil Biology and Biochemistry 2015, 85, 101–109. [Google Scholar] [CrossRef]
- Azam, F.; Fenchel, T.; Field, J.; Gray, J.S.; Meyer, L.; Thingstad, T. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 1983, 10, 257–263. [Google Scholar] [CrossRef]
- Wurzbacher, C.; Bärlocher, F.; Grossart, H.P. Fungi in lake ecosystems. Aquatic Microbial Ecology 2010, 59, 125–149. [Google Scholar] [CrossRef]
- Grossart, H.; Rojas-Jimenez, K. Aquatic fungi: targeting the forgotten in microbial ecology. Curr. Opin. Microbiol. 2016, 31, 140–145. [Google Scholar] [CrossRef]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; Dodsworth, J.A.; Hedlund, B.P.; Tsiamis, G.; Sievert, S.M.; Liu, W.T.; Eisen, J.A.; Hallam, S.J.; Kyrpides, N.C.; Stepanauskas, R.; Rubin, E.M.; Hugenholtz, P.; Woyke, T. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013, 499, 431–7. [Google Scholar] [CrossRef]
- Grossart, H.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat Rev Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Luo, X.; Li, F.; Dai, J.; Guo, J.; Chen, S.; Hong, C.; Mai, B.; Xu, M. Organochlorine compounds and polycyclic aromatic hydrocarbons in surface sediment from BYD Lake, North China: concentrations, sources profiles and potential risk. J Environ SCI-China. 2010, 22, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yi, Y.; Yang, Y.; Liu, H.; Yang, Z. Modelling phosphorus loading to the largest shallow lake in northern china in different shared socioeconomic pathways J. Clean. Prod. 2021, 297, 126537. [Google Scholar] [CrossRef]
- Liu, X.; Dong, L.; Cheng; et al. Typical Pollutant Migration Mechanism and Risk Assessment in Baiyangdian Typical Pollutant Migration Mechanism and Risk Assessment in Baiyangdian; Beijing Science Press, 2017. [Google Scholar]
- Borneman, J.; Hartin, R.J. PCR primers that amplify fungal rRNA genes from environmental samples. Applied and Environmental Microbiology 2000, 66, 4356–4360. [Google Scholar]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, G.; Knight, R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Current protocols in bioinformatics. 2011, 36, 10–7. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.; Westcott, S.; Ryabin, T.; Hall, J.; Hartmann, M.; Hollister, E.; Lesniewski, R.; Oakley, B.; Parks, D.; Robinson, C.; Sahl, J.; Stres, B.; Thallinger, G.; Van Horn, D.; Weber, C. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied & Environmental Microbiology. 2009, 75, 7537–7541. [Google Scholar]
- Edgar, R. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013, 10, 996–8. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; Douglas, B.; Drenkhan, T.; Eberhardt, U.; Dueñas, M.; Grebenc, T.; Griffith, G.W.; Hartmann, M.; Kirk, P.M.; Kohout, P.; Larsson, E.; Lindahl, B.D.; Lücking, R.; Martín, M.P.; Matheny, P.B.; Nguyen, N.H.; Niskanen, T.; Oja, J.; Peay, K.G.; Peintner, U.; Peterson, M.; Põldmaa, K.; Saag, L.; Saar, I.; Schüßler, A.; Scott, J.A.; Senés, C.; Smith, M.E.; Suija, A.; Taylor, D.L.; Telleria, M.T.; Weiss, M.; Larsson, K.-H. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- The Biodiversity Committee of Chinese Academy of Sciences, 2020. Catalogue of Life China: 2020 Annual Checklist, Beijing, China.
- Roskov, Y., Ower, G., Orrell, T., Nicolson, D., Bailly, N., Kirk, P.M., Bourgoin, T., DeWalt, R.E., Decock, W., Nieukerken, E., van Zarucchi, J., Penev, L., eds. (2019). Species 2000 & ITIS Catalogue of Life, 2019 Annual Checklist. Digital resource at www.catalogueoflife.org/annual-checklist/2019. Species 2000: Naturalis, Leiden, the Netherlands. ISSN 2405-884X.
- Dudka, I.A. Fungi as Components of Freshwater Biocenoses, Mikol. Fitopatol. 1974, 8, 444–449. [Google Scholar]
- Kirk, P.M., Cannon, P.F., Minter, D.W. and Stalpers, J.A., 2008. Dictionary of the Fungi. 10th Edition, CAB International, Wallingford.
- Lepère, C.; Boucher, D.; Jardillier, L.; Domaizon, I.; Debroas, D. Succession and regulation factors of small eukaryote community composition in a lacustrine ecosystem (Lake Pavin). Appl Environ Microbiol 2006, 72, 2971–2981. [Google Scholar] [CrossRef]
- Tian, J.; Zhu, D.; Wang, J.; et al. Environmental factors driving fungal distribution in freshwater lake sediments across the Headwater Region of the Yellow River, China. Sci Rep. 2018, 8, 3768. [Google Scholar] [CrossRef]
- Li, F.; Zhang, X.; Xie, Y.; Wang, J. Sedimentary DNA reveals over 150 years of ecosystem change by human activities in Lake Chao, China. Environment International. 2019, 133, 105214. [Google Scholar] [CrossRef] [PubMed]
- de Souza, L.M.D.; Lirio, J.M.; Coria, S.H.; et al. Diversity, distribution and ecology of fungal communities present in Antarctic lake sediments uncovered by DNA metabarcoding. Sci Rep. 2022, 12, 8407. [Google Scholar] [CrossRef] [PubMed]
- Medina, E.M.; Buchler, N.E. Chytrid fungi. Current Biology 2020, 30, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Morin, E.; Miyauchi, S.; San Clemente, H.; Chen, E.C.H.; Pelin, A.; de la Providencia, I.; Ndikumana, S.; Beaudet, D.; Hainaut, M.; Drula, E.; Kuo, A.; Tang, N.; Roy, S.; Viala, J.; Henrissat, B.; Grigoriev, I.V.; Corradi, N.; Roux, C.; Martin, F.M. Comparative genomics of Rhizophagus irregularis, R. cerebriforme, R. diaphanus and Gigaspora rosea highlights specific genetic features in Glomeromycotina. New Phytol. 2019, 222, 1584–1598. [Google Scholar]
- Shi, X.; Li, S.; Zhang, M.; Liu, C.; Wu, Q. Temperature mainly determines the temporal succession of the photosynthetic picoeukaryote community in Lake Chaohu, a highly eutrophic shallow lake. Science of the Total Environment. 2020, 702, 134803. [Google Scholar] [CrossRef] [PubMed]
- Barr, D.J.S. Chytridiomycota. Systematics and Evolution; Springer: Berlin, Heidelberg, 2001; pp. 93–112. [Google Scholar]
- Voronin, L.V. Zoosporic fungi in freshwater ecosystems. Inland Water Biol 2008, 1, 341–346. [Google Scholar] [CrossRef]
- Tang, C.; Yi, Y.; Zhou, Y. Planktonic indicators of trophic states for a shallow lake (BYD Lake, China). Limnologica. 2019, 78, 125712. [Google Scholar] [CrossRef]
- Gleason, F.H.; Carney, L.T.; Lilje, O.; Glockling, S.L. Ecological potentials of species of Rozella cryptomycota Fung. Ecol. 2012, 5, 651–656. [Google Scholar]
- Volk T. J., 2001. Fungi. Editor(s): Levin S. A., Encyclopedia of Biodiversity. Elsevier. pp. 141–163.
- Redecker, D.; Raab, P. Phylogeny of the glomeromycota (arbuscular mycorrhizal fungi): recent developments and new gene markers. Mycologia. 2006, 98, 885–95. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhao, J.; He, X. Symbiotic characteristics of arbuscular mycorrhizal fungi and dark septate endophytes in roots of Baiyang Lake. China Sciencepaper. 2016, 11, 1762–1768, (in Chinese with English abstract). [Google Scholar]
- Tisserant, E.; Malbreil, M.; Kuo, A.; Kohler, A.; Symeonidi, A.; Balestrini, R.; Charron, P.; Duensing, N.; Frei Dit Frey, N.; Gianinazzi-Pearson, V.; Gilbert, L.B.; Handa, Y.; Herr, J.R.; Hijri, M.; Koul, R.; Kawaguchi, M.; Krajinski, F.; Lammers, P.J.; Masclaux, F.G.; Murat, C.; Morin, E.; Ndikumana, S.; Pagni, M.; Petitpierre, D.; Requena, N.; Rosikiewicz, P.; Riley, R.; Saito, K.; San Clemente, H.; Shapiro, H.; Van Tuinen, D.; Becard, G.; Bonfante, P.; Paszkowski, U.; Shachar-Hill, Y.Y.; Tuskan, G.A.; Young, J.P.W.; Sanders, I.R.; Henrissat, B.; Rensing, S.A.; Grigoriev, I.V.; Corradi, N.; Roux, C.; Martin, F. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci 2013, 110, 20117–20122. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Kobayashi, Y.; Kameoka, H.; et al. Evidence of non-tandemly repeated rDNAs and their intragenomic heterogeneity in Rhizophagus irregularis. Commun Biol. 2018, 1, 87. [Google Scholar] [CrossRef] [PubMed]
- Monchy, S.; Sanciu, G.; Jobard, M.; Rasconi, S.; Gerphagnon, M.; Chabé, M.; Cian, A.; Meloni, D.; Niquil, N.; Christaki, U.; Viscogliosi, E.; Sime-Ngando, T. Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing. Environ. Microbiol. 2011, 13, 1433–1453. [Google Scholar] [CrossRef] [PubMed]
- Malar, C.M.; Krüger, M.; Krüger, C.; Wang, Y.; Stajich, J.E.; Keller, J.; Chen, E.C.H.; Yildirir, G.; Villeneuve-Laroche, M.; Roux, C.; et al. The genome of Geosiphon pyriformis reveals ancestral traits linked to the emergence of the arbuscular mycorrhizal symbiosis Curr. Biol. 2021, 31, 1570–1577. [Google Scholar]
- Dick, M.W. Fungi, flagella and phylogeny. Mycological Research. 1997, 101, 385–394. [Google Scholar] [CrossRef]
- Simon, M.; López-García, P.; Deschamps, P.; et al. Marked seasonality and high spatial variability of protist communities in shallow freshwater systems. ISME J 2015, 9, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Guarro, J.; Kantarcioglu, A.S.; Horré, R.; Rodriguez-Tudela, J.L.; Cuenca Estrella, M.; et al. Scedosporium apiospermum: changing clinical spectrum of a therapy-refractory opportunist. Med. Mycol 2006, 44, 295–327. [Google Scholar] [CrossRef]
- Glockling, S.L.; Marshall, W.L.; Gleason, F.H. Phylogenetic interpretations and ecological potentials of the Mesomycetozoea (Ichthyosporea). Fungal Ecology. 2013, 6, 237–247. [Google Scholar] [CrossRef]
- Větrovský, T.; Kohout, P.; Kopecký, M.; et al. A meta-analysis of global fungal distribution reveals climate-driven patterns. Nat Commun 2019, 10, 5142. [Google Scholar] [CrossRef]
- Hassan, N.; Rafiq, M.; Hayat, M.; et al. Psychrophilic and psychrotrophic fungi: a comprehensive review. Rev Environ Sci Biotechnol 2016, 15, 147–172. [Google Scholar] [CrossRef]
- Kauffman, C.A. 2017. 33—Fungal Pneumonias, Editor(s): Jonathan Cohen, William G. Powderly, Steven M. Opal, Infectious Diseases (Fourth Edition). Elsevier. pp.292-299.
- Huang, J.; Hu, B.; Qi, K.; Chen, W.; Pang, X.; Bao, W.; Tian, G. Effects of phosphorus addition on soil microbial biomass and community composition in a subalpine spruce plantation. European Journal of Soil Biology 2016, 72, 35–41. [Google Scholar] [CrossRef]
- Tian, J.; Zhu, D.; Wang, J.; Wu, B.; Hussain, M.; Liu, X. Environmental factors driving fungal distribution in freshwater lake sediments across the Headwater Region of the Yellow River, China. Scientific reports 2018, 8, 3768. [Google Scholar] [CrossRef]
- Liu, J.; Wang, J.; Gao, G.; Bartlam, M.G.; Wang, Y. Distribution and diversity of fungi in freshwater sediments on a river catchment scale. Frontiers in microbiology 2015, 6, 329. [Google Scholar]
- Su, J.; Zhang, L.; Yu, M.; Sun, T.; Wang, B.; Zhao, J.; Wang, Z.; Sun, Y.; Zhou, S. Characteristics and distribution of nitrogen forms insediments of Baiyangdian lake in summer and autumn. Environmental Engineering. 2022, 40, 53–58, (in Chinese with English abstract). [Google Scholar]
- Talbot, J.M.; Treseder, K.K. Controls over mycorrhizal uptake of organic nitrogen. Pedobiologia 2010, 53, 169–179. [Google Scholar] [CrossRef]
- Kranabetter, J.M.; Hawkins, B.J.; Jones, M.D.; Robbins, S.; Dyer, T.; Li, T. Species turnover (β-diversity) in ectomycorrhizal fungi linked to NH4+ uptake capacity. Mol Ecol. 2015, 24, 5992–6005. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Lu, F.; Huang, P.; Tang, M.; Xu, F.; Zhang, W.; Zhou, J.; Zhao, P.; Jia, Y.; Dai, C. Root endophyte differentially regulates plant response to NO3− and NH4+ nutrition by modulating N fluxes at the plant–fungal interface. Plant, Cell & Environment 2022, 45, 1813–1828. [Google Scholar]
- Jones, D.L.; Oburger, E. Solubilization of phosphorus by soil microorganisms. In Phosphorus in Action; Springer, 2011; pp. 169–198. [Google Scholar]
- Wang, F.; Yao, J.; Si, Y.; Chen, H.; Russel, M.; Chen, K.; Qian, Y.; Zaray, G.; Bramanti, E. Short-time effect of heavy metals upon microbial community activity. Journal of Hazardous Materials. 2010, 173, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Stefanowicz, A.M.; Kapusta, P.; Szarek-Łukaszewska, G.; Grodzińska, K.; Niklińska, M.; Vogt, R.D. Soil fertility and plant diversity enhance microbial performance in metal-polluted soils. Science of the total environment 2012, 439, 211–219. [Google Scholar] [CrossRef]
- China Environmental Monitoring Center. Background value of soil elements in China; China Environmental Science Press: Beijing, China, 1990. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).