Submitted:
30 August 2024
Posted:
30 August 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Antibiotic Mechanism of Action and Antibiotic Targets
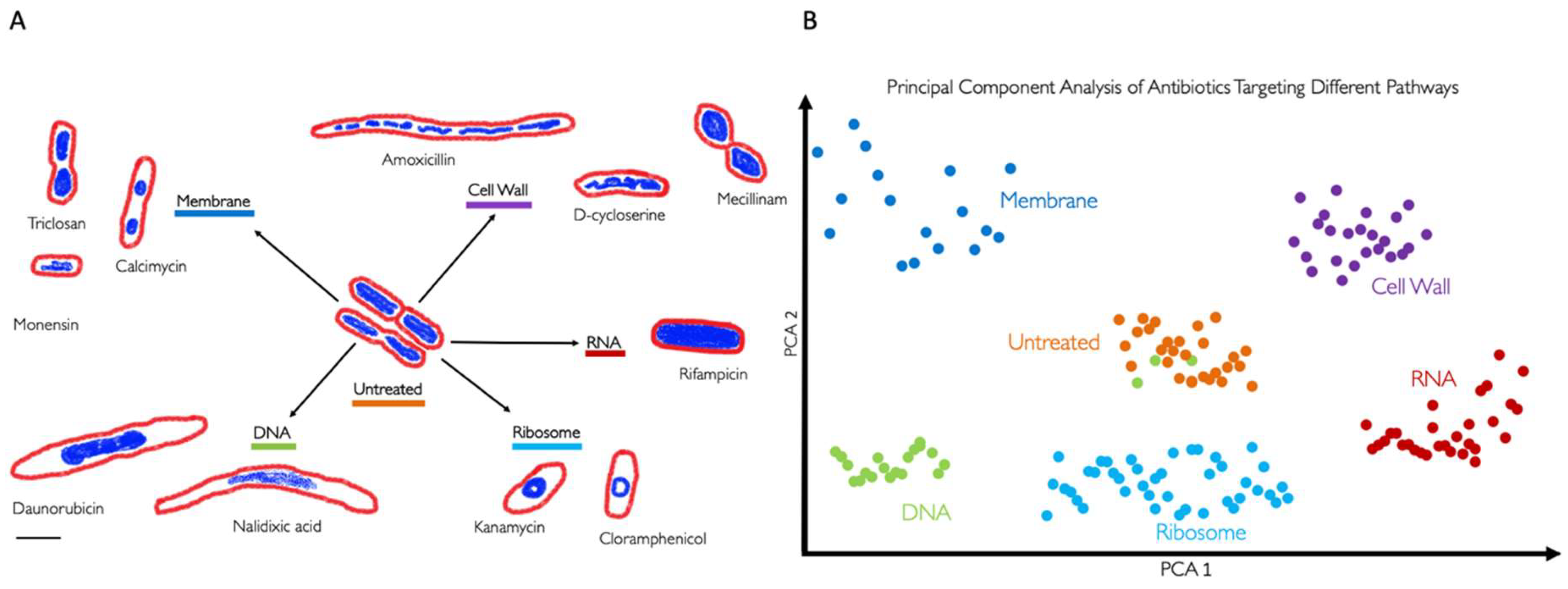
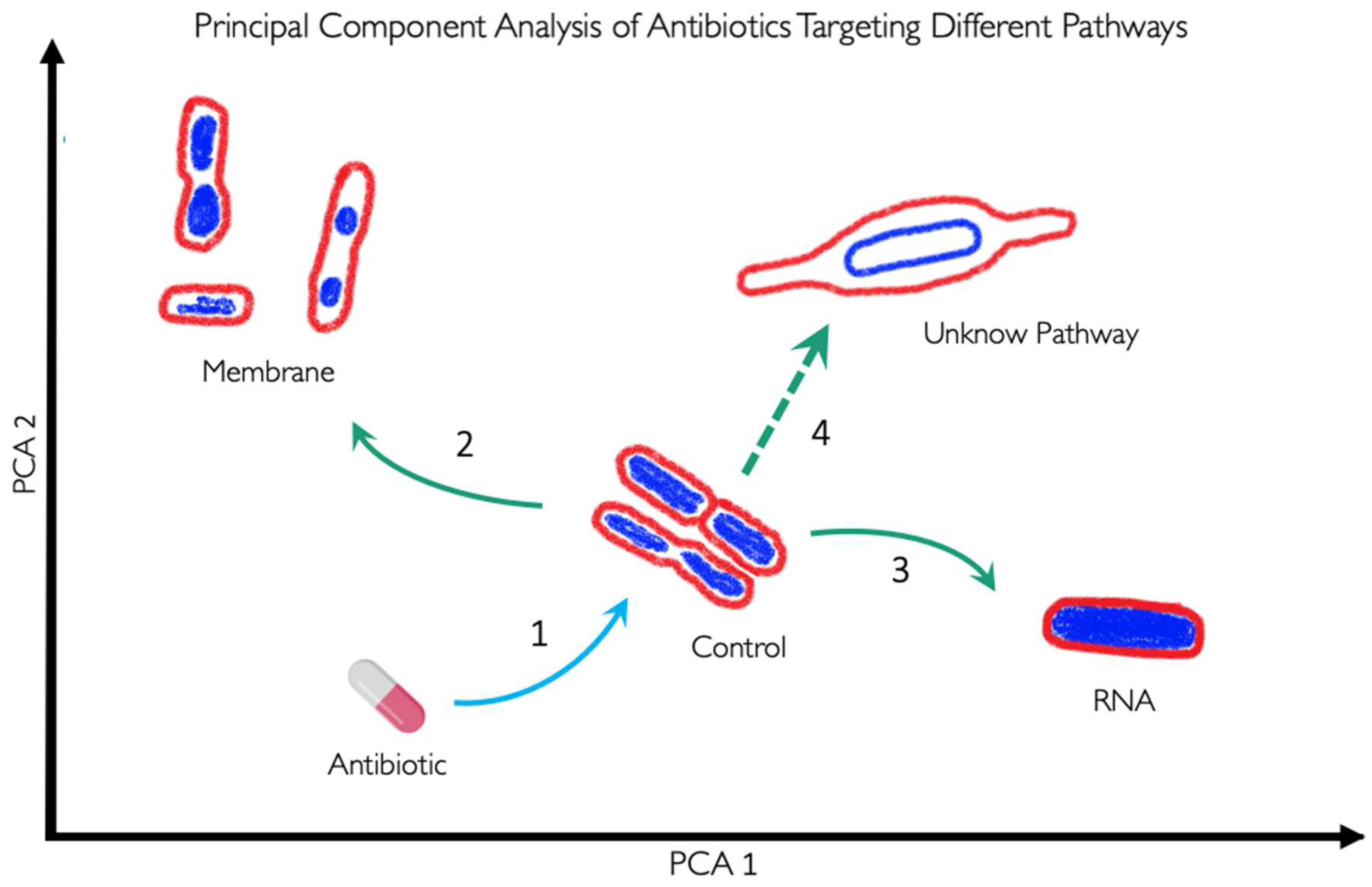
3. BCP to Identify the Mechanism of Action
4. BCP of Important Human Pathogens
5. BCP to Identify New Druggable Cell Pathways
6. BCP Limitations
7. BCP Potential Improvements
8. Image Analysis Tools for BCP and Data Availability
9. Conclusions
Acknowledgments
References
- Kim, C., Holm, M., Frost, I., Hasso-Agopsowicz, M. & Abbas, K. Global and regional burden of attributable and associated bacterial antimicrobial resistance avertable by vaccination: modelling study. BMJ Glob. Health 8, e011341 (2023). [CrossRef]
- Murray, C. J. L. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399, 629–655 (2022). [CrossRef]
- Centers for Disease Control and Prevention (U.S.). Antibiotic Resistance Threats in the United States, 2019. https://stacks.cdc.gov/view/cdc/82532 (2019) . [CrossRef]
- 160518_Final paper_with cover.pdf.
- GARDNER, A. D. Morphological Effects of Penicillin on Bacteria. Nature 146, 837–838 (1940). [CrossRef]
- Gardner, A. & others. Microscopical Effect of Penicillin on Spores and Vegetative Cells of Bacilli. Lancet 658–9 (1945). [CrossRef]
- Hutchings, M. I., Truman, A. W. & Wilkinson, B. Antibiotics: past, present and future. Curr. Opin. Microbiol. 51, 72–80 (2019).
- Walsh, C. Molecular mechanisms that confer antibacterial drug resistance. Nature 406, 775–781 (2000). [CrossRef]
- Luepke, K. H. et al. Past, Present, and Future of Antibacterial Economics: Increasing Bacterial Resistance, Limited Antibiotic Pipeline, and Societal Implications. Pharmacother. J. Hum. Pharmacol. Drug Ther. 37, 71–84 (2017). [CrossRef]
- Deep, A., Liang, Q., Enustun, E., Pogliano, J. & Corbett, K. D. Architecture and activation mechanism of the bacterial PARIS defence system. Nature 1–8 (2024) . [CrossRef]
- Tsunemoto, H., Sugie, J., Enustun, E., Pogliano, K. & Pogliano, J. Bacterial cytological profiling reveals interactions between jumbo phage φKZ infection and cell wall active antibiotics in Pseudomonas aeruginosa. PloS One 18, e0280070 (2023). [CrossRef]
- Birkholz, E. A. et al. An intron endonuclease facilitates interference competition between coinfecting viruses. Science 385, 105–112 (2024). [CrossRef]
- Corona, F. & Martinez, J. L. Phenotypic Resistance to Antibiotics. Antibiotics 2, 237–255 (2013). [CrossRef]
- Cotsonas King, A. & Wu, L. Macromolecular Synthesis and Membrane Perturbation Assays for Mechanisms of Action Studies of Antimicrobial Agents. Curr. Protoc. Pharmacol. 47, (2009). [CrossRef]
- Nonejuie, P., Burkart, M., Pogliano, K. & Pogliano, J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. 110, 16169–16174 (2013). [CrossRef]
- Lima, L. M., Silva, B. N. M. da, Barbosa, G. & Barreiro, E. J. β-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 208, 112829 (2020). [CrossRef]
- Baquero, F. & Levin, B. R. Proximate and ultimate causes of the bactericidal action of antibiotics. Nat. Rev. Microbiol. 19, 123–132 (2021). [CrossRef]
- Reynolds, P. E. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8, 943–950 (1989). [CrossRef]
- Jerala, R. Synthetic lipopeptides: a novel class of anti-infectives. Expert Opin. Investig. Drugs 16, 1159–1169 (2007). [CrossRef]
- O’Rourke, A. et al. Mechanism-of-Action Classification of Antibiotics by Global Transcriptome Profiling. Antimicrob. Agents Chemother. 64, e01207-19 (2020). [CrossRef]
- PubChem. Cerulenin. https://pubchem.ncbi.nlm.nih.gov/compound/5282054.
- Davis, B. D., Chen, L. L. & Tai, P. C. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc. Natl. Acad. Sci. U. S. A. 83, 6164–6168 (1986). [CrossRef]
- Chopra, I. & Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 65, 232–260 (2001). [CrossRef]
- Vázquez-Laslop, N. & Mankin, A. S. How Macrolide Antibiotics Work. Trends Biochem. Sci. 43, 668–684 (2018). [CrossRef]
- Tenson, T., Lovmar, M. & Ehrenberg, M. The Mechanism of Action of Macrolides, Lincosamides and Streptogramin B Reveals the Nascent Peptide Exit Path in the Ribosome. J. Mol. Biol. 330, 1005–1014 (2003). [CrossRef]
- Swaney, S. M., Aoki, H., Ganoza, M. C. & Shinabarger, D. L. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 42, 3251–3255 (1998). [CrossRef]
- Correia, S., Poeta, P., Hébraud, M., Capelo, J. L. & Igrejas, G. Mechanisms of quinolone action and resistance: where do we stand? J. Med. Microbiol. 66, 551–559 (2017).
- Ojkic, N. et al. A Roadblock-and-Kill Mechanism of Action Model for the DNA-Targeting Antibiotic Ciprofloxacin. Antimicrob. Agents Chemother. 64, e02487-19 (2020). [CrossRef]
- Wong, W. R., Oliver, A. G. & Linington, R. G. Development of Antibiotic Activity Profile Screening for the Classification and Discovery of Natural Product Antibiotics. Chem. Biol. 19, 1483–1495 (2012). [CrossRef]
- Kohanski, M. A., Dwyer, D. J. & Collins, J. J. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8, 423–435 (2010). [CrossRef]
- WHO. AWaRe classification of antibiotics for evaluation and monitoring of use, 2023. (2023).
- Silver, L. L. Challenges of Antibacterial Discovery. Clin. Microbiol. Rev. 24, 71–109 (2011). [CrossRef]
- Hudson, M. A. & Lockless, S. W. Elucidating the Mechanisms of Action of Antimicrobial Agents. mBio 13, e02240-21. [CrossRef]
- Hage, D. S. et al. Pharmaceutical and biomedical applications of affinity chromatography: Recent trends and developments. J. Pharm. Biomed. Anal. 69, 93–105 (2012). [CrossRef]
- Franken, H. et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 10, 1567–1593 (2015). [CrossRef]
- Tacconelli, E. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018). [CrossRef]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 12, 371–387 (2013). [CrossRef]
- Lewis, K. et al. Sophisticated natural products as antibiotics. Nature 632, 39–49 (2024). [CrossRef]
- Gould, K. Antibiotics: from prehistory to the present day. J. Antimicrob. Chemother. 71, 572–575 (2016). [CrossRef]
- WHO. 2021 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis. (World Health Organization, 2022).
- Samernate, T. et al. High-Resolution Bacterial Cytological Profiling Reveals Intrapopulation Morphological Variations upon Antibiotic Exposure. Antimicrob. Agents Chemother. 67, e01307-22 (2023). [CrossRef]
- Balaban, N. Q. et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019). [CrossRef]
- Kussell, E., Kishony, R., Balaban, N. Q. & Leibler, S. Bacterial Persistence. Genetics 169, 1807–1814 (2005).
- Bailey, S. Principal Component Analysis with Noisy and/or Missing Data. Publ. Astron. Soc. Pac. 124, 1015–1023 (2012). [CrossRef]
- Van Den Broeck, T. et al. The Role of Single Nucleotide Polymorphisms in Predicting Prostate Cancer Risk and Therapeutic Decision Making. BioMed Res. Int. 2014, 1–16 (2014). [CrossRef]
- Regev, A. et al. The Human Cell Atlas. eLife 6, e27041 (2017).
- Liu, Z., Liu, J., Ghosh, S., Han, J. & Scarlett, J. Generative Principal Component Analysis. (2022).
- Ghadban, N., Honeine, P., Mourad-Chehade, F., Francis, C. & Farah, J. Diffusion strategies for in-network principal component analysis. in 2014 IEEE International Workshop on Machine Learning for Signal Processing (MLSP) 1–6 (IEEE, Reims, France, 2014). [CrossRef]
- Siirtola, H., Saily, T. & Nevalainen, T. Interactive Principal Component Analysis. in 2017 21st International Conference Information Visualisation (IV) 416–421 (IEEE, London, 2017). [CrossRef]
- Cushnie, T. P. T., O’Driscoll, N. H. & Lamb, A. J. Morphological and ultrastructural changes in bacterial cells as an indicator of antibacterial mechanism of action. Cell. Mol. Life Sci. 73, 4471–4492 (2016). [CrossRef]
- Gebicki, J. M. & James, A. M. The Preparation and Properties of Spheroplasts of Aerobacter aerogenes. J. Gen. Microbiol. 23, 9–18 (1960). [CrossRef]
- Errington, J. L-form bacteria, cell walls and the origins of life. Open Biol. 3, 120143 (2013). [CrossRef]
- Allan, E. J., Hoischen, C. & Gumpert, J. Bacterial L-forms. Adv. Appl. Microbiol. 68, 1–39 (2009).
- Mercier, R., Kawai, Y. & Errington, J. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife 3, e04629 (2014). [CrossRef]
- Errington, J. Cell wall-deficient, L-form bacteria in the 21st century: a personal perspective. Biochem. Soc. Trans. 45, 287–295 (2017). [CrossRef]
- Spratt, B. G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. U. S. A. 72, 2999–3003 (1975). [CrossRef]
- Spratt, B. G. & Pardee, A. B. Penicillin-binding proteins and cell shape in E. coli. Nature 254, 516–517 (1975). [CrossRef]
- Curtis, N. A., Orr, D., Ross, G. W. & Boulton, M. G. Affinities of penicillins and cephalosporins for the penicillin-binding proteins of Escherichia coli K-12 and their antibacterial activity. Antimicrob. Agents Chemother. 16, 533–539 (1979). [CrossRef]
- Di Modugno, E. et al. In vitro activity of the tribactam GV104326 against gram-positive, gram-negative, and anaerobic bacteria. Antimicrob. Agents Chemother. 38, 2362–2368 (1994). [CrossRef]
- Bernabeu-Wittel, M. et al. Morphological changes induced by imipenem and meropenem at sub-inhibitory concentrations in Acinetobacter baumannii. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 10, 931–934 (2004). [CrossRef]
- Differences in mode of action of (β-lactam antibiotics influence morphology, LPS release and in vivo antibiotic efficacy. CoLab https://colab.ws/articles/10.1177%2F096805199600300306.
- de Pedro, M. A., Donachie, W. D., Höltje, J.-V. & Schwarz, H. Constitutive Septal Murein Synthesis in Escherichia coli with Impaired Activity of the Morphogenetic Proteins RodA and Penicillin-Binding Protein 2. J. Bacteriol. 183, 4115–4126 (2001). [CrossRef]
- Sumita, Y., Fukasawa, M. & Okuda, T. Comparison of two carbapenems, SM-7338 and imipenem: affinities for penicillin-binding proteins and morphological changes. J. Antibiot. (Tokyo) 43, 314–320 (1990).
- Dalhoff, A., Nasu, T. & Okamoto, K. Target affinities of faropenem to and its impact on the morphology of gram-positive and gram-negative bacteria. Chemotherapy 49, 172–183 (2003). [CrossRef]
- Horii, T., Kobayashi, M., Sato, K., Ichiyama, S. & Ohta, M. An in-vitro study of carbapenem-induced morphological changes and endotoxin release in clinical isolates of gram-negative bacilli. J. Antimicrob. Chemother. 41, 435–442 (1998). [CrossRef]
- Perumalsamy, H., Jung, M. Y., Hong, S. M. & Ahn, Y.-J. Growth-Inhibiting and morphostructural effects of constituents identified in Asarum heterotropoides root on human intestinal bacteria. BMC Complement. Altern. Med. 13, 245 (2013). [CrossRef]
- Nickerson, W. J. & Webb, M. Effect of folic acid analogues on growth and cell division of nonexacting microorganisms. J. Bacteriol. 71, 129–139 (1956). [CrossRef]
- Elliott, T. S. J., Shelton, A. & Greenwood, D. The response of Escherichia coli to ciprofloxacin and norfloxacin. J. Med. Microbiol. 23, 83–88 (1987). [CrossRef]
- Chen, K., Sun, G. W., Chua, K. L. & Gan, Y.-H. Modified Virulence of Antibiotic-Induced Burkholderia pseudomallei Filaments. Antimicrob. Agents Chemother. 49, 1002–1009 (2005). [CrossRef]
- Uphoff, S., Reyes-Lamothe, R., Garza de Leon, F., Sherratt, D. J. & Kapanidis, A. N. Single-molecule DNA repair in live bacteria. Proc. Natl. Acad. Sci. 110, 8063–8068 (2013). [CrossRef]
- Jones, E. C. & Uphoff, S. Single-molecule imaging of LexA degradation in Escherichia coli elucidates regulatory mechanisms and heterogeneity of the SOS response. Nat. Microbiol. 6, 981–990 (2021). [CrossRef]
- Jaramillo-Riveri, S. et al. Growth-dependent heterogeneity in the DNA damage response in Escherichia coli. Mol. Syst. Biol. 18, e10441 (2022).
- Banerjee, S. et al. Mechanical feedback promotes bacterial adaptation to antibiotics. Nat. Phys. 17, 403–409 (2021). [CrossRef]
- Ojkic, N., Serbanescu, D. & Banerjee, S. Antibiotic Resistance via Bacterial Cell Shape-Shifting. mBio 13, e00659-22 (2022). [CrossRef]
- Ojkic, N., Serbanescu, D. & Banerjee, S. Surface-to-volume scaling and aspect ratio preservation in rod-shaped bacteria. eLife 8, e47033 (2019). [CrossRef]
- Ojkic, N. & Banerjee, S. Bacterial cell shape control by nutrient-dependent synthesis of cell division inhibitors. Biophys. J. 120, 2079–2084 (2021). [CrossRef]
- Lin, L. et al. Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity Against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. EBioMedicine 2, 690–698 (2015). [CrossRef]
- Mohammad, H. et al. Bacteriological profiling of diphenylureas as a novel class of antibiotics against methicillin-resistant Staphylococcus aureus. PLOS ONE 12, e0182821 (2017). [CrossRef]
- Wu, Y. & Seyedsayamdost, M. R. The Polyene Natural Product Thailandamide A Inhibits Fatty Acid Biosynthesis in Gram-Positive and Gram-Negative Bacteria. Biochemistry 57, 4247–4251 (2018). [CrossRef]
- Sun, Y., Heidary, D. K., Zhang, Z., Richards, C. I. & Glazer, E. C. Bacterial Cytological Profiling Reveals the Mechanism of Action of Anticancer Metal Complexes. Mol. Pharm. 15, 3404–3416 (2018). [CrossRef]
- Dillon, N. A. et al. Characterizing the response of Acinetobacter baumannii ATCC 17978 to azithromycin in multiple in vitro growth conditions. Preprint a (2020). [CrossRef]
- Araújo-Bazán, L., Ruiz-Avila, L. B., Andreu, D., Huecas, S. & Andreu, J. M. Cytological Profile of Antibacterial FtsZ Inhibitors and Synthetic Peptide MciZ. Front. Microbiol. 7, (2016). [CrossRef]
- Andreu, J. M., Huecas, S., Araújo-Bazán, L., Vázquez-Villa, H. & Martín-Fontecha, M. The Search for Antibacterial Inhibitors Targeting Cell Division Protein FtsZ at Its Nucleotide and Allosteric Binding Sites. Biomedicines 10, 1825 (2022). [CrossRef]
- Soonthonsrima, T. et al. Phage-induced bacterial morphological changes reveal a phage-derived antimicrobial affecting cell wall integrity. Antimicrob. Agents Chemother. 67, e00764-23 (2023). [CrossRef]
- World Healt Organization. WHO Bacterial Priority Pathogens List, 2017. (2017).
- Di Bella, S. et al. Resistance to ceftazidime/avibactam in infections and colonisations by KPC-producing Enterobacterales: a systematic review of observational clinical studies. J. Glob. Antimicrob. Resist. 25, 268–281 (2021).
- Butler, M. S. et al. Analysis of the Clinical Pipeline of Treatments for Drug-Resistant Bacterial Infections: Despite Progress, More Action Is Needed. Antimicrob. Agents Chemother. 66, e01991-21. [CrossRef]
- Geneva: World Health Organization. WHO Bacterial Priority Pathogens List, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. (2024).
- Htoo, H. H. et al. Bacterial Cytological Profiling as a Tool To Study Mechanisms of Action of Antibiotics That Are Active against Acinetobacter baumannii. Antimicrob. Agents Chemother. 63, 10.1128/aac.02310-18 (2019). [CrossRef]
- Coram, M. A. et al. Morphological Characterization of Antibiotic Combinations. ACS Infect. Dis. 8, 66–77 (2022). [CrossRef]
- Montaño, E. T. et al. Bacterial Cytological Profiling Identifies Rhodanine-Containing PAINS Analogs as Specific Inhibitors of Escherichia coli Thymidylate Kinase In Vivo. J. Bacteriol. 203, 10.1128/jb.00105-21 (2021). [CrossRef]
- Nonejuie, P. et al. Application of bacterial cytological profiling to crude natural product extracts reveals the antibacterial arsenal of Bacillus subtilis. J. Antibiot. (Tokyo) 69, 353–361 (2016). [CrossRef]
- Sun, D. et al. Intrinsic Antibacterial Activity of Xeruborbactam In Vitro: Assessing Spectrum and Mode of Action. Antimicrob. Agents Chemother. 66, e00879-22. [CrossRef]
- Sridhar, S. et al. High-Content Imaging to Phenotype Antimicrobial Effects on Individual Bacteria at Scale. mSystems 6, e00028-21. [CrossRef]
- Smith, T. C. et al. Morphological profiling of tubercle bacilli identifies drug pathways of action. Proc. Natl. Acad. Sci. U. S. A. 117, 18744–18753 (2020). [CrossRef]
- Allen, R. et al. An arylsulphonamide that targets cell wall biosynthesis in Mycobacterium tuberculosis. 2024.07.22.604653 Preprint a (2024). [CrossRef]
- López-Jiménez, A. T. et al. High-content high-resolution microscopy and deep learning assisted analysis reveals host and bacterial heterogeneity during Shigella infection. eLife 13, (2024).
- Werth, B. J. et al. Defining Daptomycin Resistance Prevention Exposures in Vancomycin-Resistant Enterococcus faecium and E. faecalis. Antimicrob. Agents Chemother. 58, 5253–5261 (2014). [CrossRef]
- Quach, D. T., Sakoulas, G., Nizet, V., Pogliano, J. & Pogliano, K. Bacterial Cytological Profiling (BCP) as a Rapid and Accurate Antimicrobial Susceptibility Testing Method for Staphylococcus aureus. EBioMedicine 4, 95–103 (2016). [CrossRef]
- Kalla, G. Using Bacterial Cytological Profiling to Study the Interactions of Bacteria and the Defense Systems of Multicellular Eukaryotes. (University of California San Diego, USA, 2020).
- Blaskovich, M. A. T. et al. The antimicrobial potential of cannabidiol. Commun. Biol. 4, 1–18 (2021). [CrossRef]
- Sakoulas, G. et al. Examining the Use of Ceftaroline in the Treatment of Streptococcus pneumoniae Meningitis with Reference to Human Cathelicidin LL-37. Antimicrob. Agents Chemother. 59, 2428–2431 (2015). [CrossRef]
- Schäfer, A.-B. et al. Dissecting antibiotic effects on the cell envelope using bacterial cytological profiling: a phenotypic analysis starter kit. Microbiol. Spectr. 12, e03275-23 (2024). [CrossRef]
- Serbanescu, D., Ojkic, N. & Banerjee, S. Cellular resource allocation strategies for cell size and shape control in bacteria. FEBS J. 289, 7891–7906 (2022). [CrossRef]
- Htoo, H. H. et al. Bacterial Cytological Profiling as a Tool To Study Mechanisms of Action of Antibiotics That Are Active against Acinetobacter baumannii. Antimicrob. Agents Chemother. 63, 10.1128/aac.02310-18 (2019). [CrossRef]
- Samernate, T. et al. High-Resolution Bacterial Cytological Profiling Reveals Intrapopulation Morphological Variations upon Antibiotic Exposure. Antimicrob. Agents Chemother. 67, e01307-22 (2023). [CrossRef]
- Tsunemoto, H., Sugie, J., Enustun, E., Pogliano, K. & Pogliano, J. Bacterial cytological profiling reveals interactions between jumbo phage φKZ infection and cell wall active antibiotics in Pseudomonas aeruginosa. PLOS ONE 18, e0280070 (2023). [CrossRef]
- Lamsa, A. et al. Rapid Inhibition Profiling in Bacillus subtilis to Identify the Mechanism of Action of New Antimicrobials. ACS Chem. Biol. 11, 2222–2231 (2016). [CrossRef]
- Herschede, S. R., Salam, R., Gneid, H. & Busschaert, N. Bacterial cytological profiling identifies transmembrane anion transport as the mechanism of action for a urea-based antibiotic. Supramol. Chem. 34, 26–33 (2022). [CrossRef]
- Schäfer, A.-B. et al. Dissecting antibiotic effects on the cell envelope using bacterial cytological profiling: a phenotypic analysis starter kit. Microbiol. Spectr. 12, e03275-23 (2024). [CrossRef]
- Araújo-Bazán, L., Ruiz-Avila, L. B., Andreu, D., Huecas, S. & Andreu, J. M. Cytological Profile of Antibacterial FtsZ Inhibitors and Synthetic Peptide MciZ. Front. Microbiol. 7, (2016). [CrossRef]
- Santos, T. M. A. et al. Small Molecule Chelators Reveal That Iron Starvation Inhibits Late Stages of Bacterial Cytokinesis. ACS Chem. Biol. 13, 235–246 (2018). [CrossRef]
- Montaño, E. T. et al. Bacterial Cytological Profiling Identifies Rhodanine-Containing PAINS Analogs as Specific Inhibitors of Escherichia coli Thymidylate Kinase In Vivo. J. Bacteriol. 203, 10.1128/jb.00105-21 (2021). [CrossRef]
- Ulloa, E. R. et al. Azithromycin Exerts Bactericidal Activity and Enhances Innate Immune Mediated Killing of MDR Achromobacter xylosoxidans. Infect. Microbes Dis. 2, 10 (2020). [CrossRef]
- de Wet, T. J., Winkler, K. R., Mhlanga, M., Mizrahi, V. & Warner, D. F. Arrayed CRISPRi and quantitative imaging describe the morphotypic landscape of essential mycobacterial genes. eLife 9, e60083 (2020). [CrossRef]
- Smith, T. C. et al. Morphological profiling of tubercle bacilli identifies drug pathways of action. Proc. Natl. Acad. Sci. 117, 18744–18753 (2020). [CrossRef]
- Mayer, B., Schwan, M., Thormann, K. M. & Graumann, P. L. Antibiotic Drug screening and Image Characterization Toolbox (A.D.I.C.T.): a robust imaging workflow to monitor antibiotic stress response in bacterial cells in vivo. F1000Research 10, 277 (2022).
- Ouyang, X. et al. Classification of antimicrobial mechanism of action using dynamic bacterial morphology imaging. Sci. Rep. 12, 11162 (2022). [CrossRef]
- Mistretta, M. et al. Dynamic microfluidic single-cell screening identifies pheno-tuning compounds to potentiate tuberculosis therapy. Nat. Commun. 15, 4175 (2024). [CrossRef]
- El-sagheir, A. M. K. et al. Rational design, synthesis, molecular modeling, biological activity, and mechanism of action of polypharmacological norfloxacin hydroxamic acid derivatives. RSC Med. Chem. 14, 2593–2610 (2023).
- Kuru, E. et al. In Situ Probing of Newly Synthesized Peptidoglycan in Live Bacteria with Fluorescent D -Amino Acids. Angew. Chem. Int. Ed. 51, 12519–12523 (2012). [CrossRef]
- Vollmer, W., Blanot, D. & de Pedro, M. A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 (2008). [CrossRef]
- Young, K. D. The Selective Value of Bacterial Shape. Microbiol. Mol. Biol. Rev. 70, 660–703 (2006). [CrossRef]
- Stratford, J. P. et al. Electrically induced bacterial membrane-potential dynamics correspond to cellular proliferation capacity. Proc. Natl. Acad. Sci. 116, 9552–9557 (2019). [CrossRef]
- Prindle, A. et al. Ion channels enable electrical communication in bacterial communities. Nature 527, 59–63 (2015). [CrossRef]
- De Souza-Guerreiro, T. C. et al. Membrane potential dynamics unveil the promise of bioelectrical antimicrobial susceptibility Testing (BeAST) for anti-fungal screening. mBio 15, e01302-24 (2024).
- Wong, F. et al. Cytoplasmic condensation induced by membrane damage is associated with antibiotic lethality. Nat. Commun. 12, 2321 (2021). [CrossRef]
- Ma, J. et al. The multimodality cell segmentation challenge: toward universal solutions. Nat. Methods 21, 1103–1113 (2024). [CrossRef]
- Cutler, K. J., Stringer, C., Wiggins, P. A. & Mougous, J. D. Omnipose: a high-precision morphology-independent solution for bacterial cell segmentation. [CrossRef]
- Bali, A. & Singh, S. N. A Review on the Strategies and Techniques of Image Segmentation. in 2015 Fifth International Conference on Advanced Computing & Communication Technologies 113–120 (2015). [CrossRef]
- Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 2020 181 18, 100–106 (2020). [CrossRef]
- StarDist: Application of the deep-learning tool for phase-contrast cell images - 2020 - Wiley Analytical Science. Analytical Science Article DO Series https://analyticalscience.wiley.com/do/10.1002/was.000400090/.
- Lou, W. et al. Multi-stream Cell Segmentation with Low-level Cues for Multi-modality Images. Preprint at http://arxiv.org/abs/2310.14226 (2023).
- Paintdakhi, A. et al. Oufti: An integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol. Microbiol. 99, 767–777 (2016).
- Danielsen, J. & Nordenfelt, P. Computer Vision-Based Image Analysis of Bacteria. in Bacterial Pathogenesis: Methods and Protocols (eds. Nordenfelt, P. & Collin, M.) 161–172 (Springer, New York, NY, 2017). [CrossRef]
- THE ANALYSIS OF CELL IMAGES* - Prewitt - 1966 - Annals of the New York Academy of Sciences - Wiley Online Library. https://nyaspubs.onlinelibrary.wiley.com/doi/abs/10.1111/j.1749-6632.1965.tb11715.x.
- Mendelsohn, M. L., Kolman, W. A., Perry, B. & Prewitt, J. M. S. Computer Analysis of Cell Images. Postgrad. Med. 38, 567–573 (1965). [CrossRef]
- Stylianidou, S., Brennan, C., Nissen, S. B., Kuwada, N. J. & Wiggins, P. A. SuperSegger: robust image segmentation, analysis and lineage tracking of bacterial cells. Mol. Microbiol. 102, 690–700 (2016). [CrossRef]
- Chai, B., Efstathiou, C., Yue, H. & Draviam, V. M. Opportunities and challenges for deep learning in cell dynamics research. Trends Cell Biol. 0, (2023). [CrossRef]
- Jan, M., Spangaro, A., Lenartowicz, M. & Mattiazzi Usaj, M. From pixels to insights: Machine learning and deep learning for bioimage analysis. BioEssays 46, 2300114 (2024). [CrossRef]
- O’Mahony, N. et al. Deep Learning vs. Traditional Computer Vision. in Advances in Computer Vision (eds. Arai, K. & Kapoor, S.) 128–144 (Springer International Publishing, Cham, 2020). [CrossRef]
- Falk, T. et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019). [CrossRef]
- Panigrahi, S. et al. Misic, a general deep learning-based method for the high-throughput cell segmentation of complex bacterial communities. eLife https://elifesciences.org/articles/65151 (2021). [CrossRef]
- Osokin, A., Chessel, A., Salas, R. E. C. & Vaggi, F. GANs for Biological Image Synthesis. in 2017 IEEE International Conference on Computer Vision (ICCV) 2252–2261 (2017). doi:10.1109/ICCV.2017.245.
- Chan, C. K., Hadjitheodorou, A., Tsai, T. Y.-C. & Theriot, J. A. Quantitative comparison of principal component analysis and unsupervised deep learning using variational autoencoders for shape analysis of motile cells. 2020.06.26.174474 Preprint a (2020). [CrossRef]
- Goldsborough, P., Pawlowski, N., Caicedo, J. C., Singh, S. & Carpenter, A. E. CytoGAN: Generative Modeling of Cell Images. 227645 Preprint a (2017). [CrossRef]
- Cylke, C., Si, F. & Banerjee, S. Effects of antibiotics on bacterial cell morphology and their physiological origins. Biochem. Soc. Trans. 50, 1269–1279 (2022). [CrossRef]
- Chessel, A. & Carazo Salas, R. E. From observing to predicting single-cell structure and function with high-throughput/high-content microscopy. Essays Biochem. 63, 197–208 (2019). [CrossRef]
- Chong, Y. T. et al. Yeast Proteome Dynamics from Single Cell Imaging and Automated Analysis. Cell 161, 1413–1424 (2015). [CrossRef]
- McMahon, C. L. et al. Development of an Imaging Flow Cytometry Method for Fungal Cytological Profiling and Its Potential Application in Antifungal Drug Development. J. Fungi 9, 722 (2023). [CrossRef]
- McDiarmid, A. H. et al. Morphological profiling in human neural progenitor cells classifies hits in a pilot drug screen for Alzheimer’s disease. Brain Commun. 6, fcae101 (2024). [CrossRef]
- Ren, E. et al. Deep Learning-Enhanced Morphological Profiling Predicts Cell Fate Dynamics in Real-Time in hPSCs. (2021). [CrossRef]
- Perlman, Z. E. et al. Multidimensional Drug Profiling By Automated Microscopy. Science 306, 1194–1198 (2004). [CrossRef]
| TARGET | CHEMICAL STRUCTURE | MECHANISM OF ACTION (MOA) | GENERIC NAME EXAMPLES | USE |
|---|---|---|---|---|
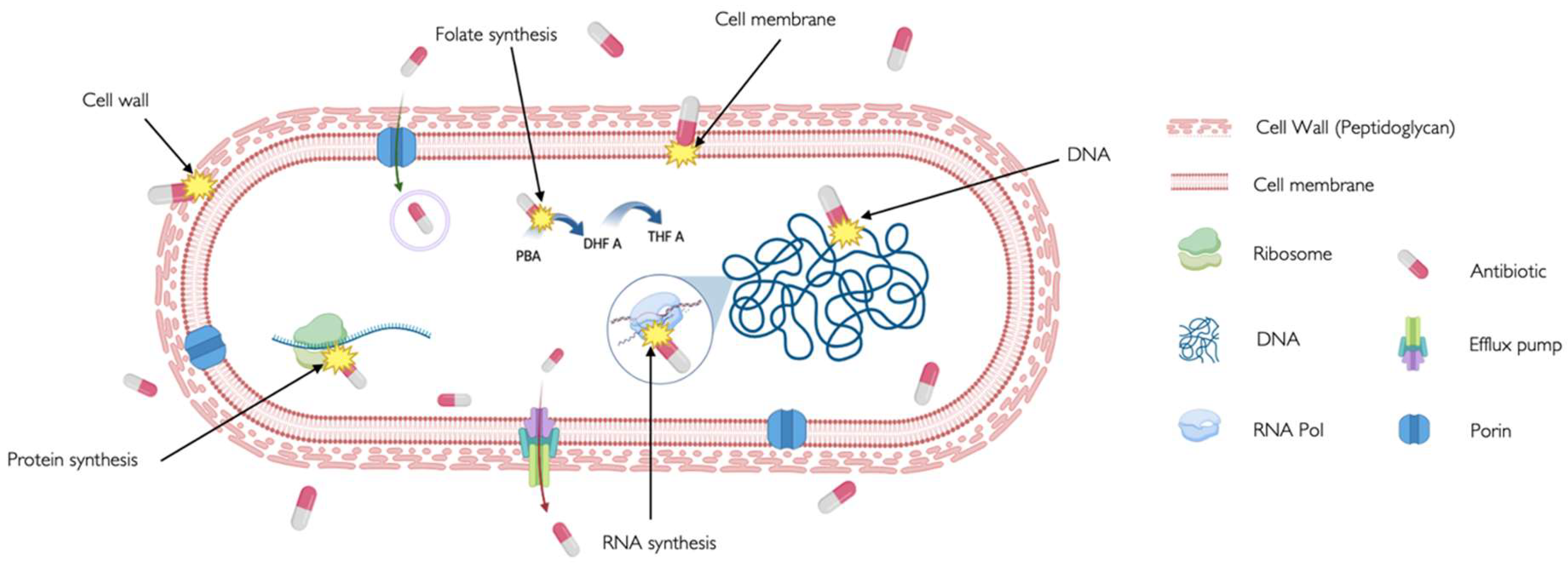
| CELL WALL | β-Lactams | Inhibit penicillin-binding proteins (PBPs) that crosslink peptidoglycan chains in the bacterial cell wall [16], disrupting cell wall integrity and causing cell lysis [17]. | Penicillins, cephalosporins, cephamycins, carbapenems, and others. | To treat a variety of infections, including skin infections, chest infections, urinary tract infections sepsis and meningitis. |
| Glycopeptides | Target gram-positive bacteria by binding to the acyl-D-Ala-D-Ala terminus to the growing peptidoglycan and then cross-linking peptides within and between peptidoglycan [18]. | Vancomicyn | Last resort medication for the treatment of sepsis and lower respiratory tract, skin, and bone infections caused by Gram-positive bacteria. | |
| MEBRANE | Lipopeptides | Insert in the cell membrane and cause depolarization, reducing the ability to create ATP and cell death [19]. | Daptomycin, Colistin | For treatment of complicated skin and skin-structure infections associated to Gram-positive bacteria. |
| FATTY ACID SYNTHESIS | Chlorophenol | Block the reduction step of the fatty acid synthesis pathway by inhibiting an enoyl-ACP reductase (fabI) [20]. | Triclosan | Added to many consumer products as soaps, body washes and toothpastes, intended to reduce or prevent bacterial contamination. |
| Oxirane carboxylic acids | Irreversibly binds to fatty acid synthase, specifically b-ketoacyl-acyl carrier protein synthase. In sterol synthesis, inhibits HMG-CoA synthetase activity [21]. |
Cerulenin | Antifungal agent whose activity interferes with or otherwise acts to prevent the formation of fatty acids and sterols. With selective cytotoxicity to cancer cells | |
| PROTEIN SYNTHESIS | Aminoglycosides | Interact with the 30s ribosomal subunit of 16S RNA causing misreading and/or truncated proteins and cell death [17], [22]. | Gentamicin, tobramicin, kanamycin | To treat mainly very serious illnesses and infections such as sepsis, as they can cause very serious side effects. |
| Tetracyclines | Inhibit translation by binding to 16S rRNA of the 30S ribosomal subunit, preventing tRNA binding to 30S [23]. | Tetracycline, doxycycline and lymecycline | To treat a wide range of infections, acne and skin conditions as rosacea. | |
| Macrolides | It binds to the 23S rRNA of the 50S ribosomal subunit, leading to the production of incomplete peptide chains [24]. | Azithromicin, erythromycin and clarithromicyn | Particularly useful to treat lung and chest infections, as an alternative for people with a penicillin allergy or penicillin-resistant strains. | |
| Lincosamide | It binds to the 50S ribosome subunit to stimulate dissociation of the peptidyl- tRNA molecule from the ribosomes during elongation [25] | Clindamicyn | Primarily used to treat gram-positive bacterial infections in which there is resistance or intolerance to penicillin. | |
| Oxazolidinones | Limit translation by binding to 23S rRNA of the 50S subunit and preventing the formation of a functional 70S subunit [26]. | Linezolid | Active against multidrug-resistant staphylococci, streptococci, and enterococci. | |
| DNA SYNTHESIS | Fluoroquinoles | Inhibit DNA replication by targeting DNA gyrase and topoisomerase IV[27,28]. | Ciprofloxacin and levofloxacin | Broad-spectrum antibiotics that are used to treat a wide range of infections, especially respiratory and urinary tract infections. Not commonly used due to their risk of serious side effects. |
| Sulfonamides | Competitive inhibitor of Dihydropteroate synthase (DHPS) involved in folate synthesis [29] | Sulfamethazine, sulfapyridine | Utilized in the treatment of tonsillitis, septicemia, meningococcal meningitis, bacillary dysentery, and number of infections of urinary tract | |
| RNA SYNTHESIS | Rifamycins | It binds to the RNA polymerase and blocks the RNA synthesis [30] | Rifapentine, Rifampin | Effective against mycobacteria, and are therefore used to treat tuberculosis, leprosy, and mycobacterium avium complex (MAC) infections. |
| Bacteria | Resistant to | Bacterial Cytological Profilling (BCP) |
|---|---|---|
| Priority 1. Critical group | ||
| Acinetobacter baumannii | Carbapenems | Yes[41,77,81,89] |
| Enterobacteriaceae* | Third generation cephalosporine | Yes[15,80,90,91,92] |
| Enterobacteriaceae** | Carbapenems, ESBL-producing | Yes[77,93,94] |
| Rifampicin-Resistant Tuberculosis (RR-TB)*** | Rifampicin | Yes[95,96] |
| Priority 2. High group | ||
| Salmonella Thypi | Fluoroquinolones | Yes [94] |
| Shigella spp. | Fluoroquinolones | Yes [97] |
| Enterococcus faecium | Vancomycin | Yes [98] |
| Pseudomonas aeruginosa | Carbapenems | Yes[11,77,93,94] |
| Non-typhoidal Salmonella | Fluoroquinoles | No |
| Neisseria gonorrhoeae | Cephalosporin, Fluoroquinolonas | No |
| Staphylococcus aureus | Methicillin and vancomycin | Yes[78,99,100,101] |
| Priority 3. Medium group | ||
| Group A Streptococci | Macrolide | No |
| Streptococcus pneumoniae | Macrolide/No sensitivity to penicillin | Yes [102] |
| Haemophilus influenzae | Ampicillin | No |
| Group B Streptococci | Penicillin | No |
| Organism | Dyes/Fluorophores | Processed Data available | Segmentation | Feature extraction | Source |
|---|---|---|---|---|---|
| Acinetobacter baumannii and E. coli | FM4-64 DAPI SYTOX-Green |
Yes | CellProfiler | CellProfiler | [105] |
| Acinetobacter baumannii | FM4-64 DAPI SYTOX-Green |
No | Ilastic | CellProfiler | [106]* |
| Pseudomonas aeruginosa | FM4-64 DAPI |
Yes | Manually (FIJI/ImageJ ) |
Manually (FIJI/ImageJ) |
[107]* |
| S. aureus | FM4-64 DAPI SYTOX-Green |
Yes | Semi-Manual (FIJI/ImageJ) | Semi-Manual (FIJI/ImageJ) | [15]* |
| S. aureus | FM4-64 DAPI SYTOX-Green WGA-647 |
Yes | CellProfiler | CellProfiler | [99]* |
|
S. aureus, S. Typhimurium, and K. pneumoniae |
FM4-64 DAPI SYTOX-Green |
Yes | Harmony | Harmony | [94] |
| B. subtilis | FM 4-64 DAPI SYTOX Green |
Yes | CellProfiler | CellProfiler FIJI |
[108]* |
| B. subtilis | FM4-64 DAPI SYTOX-Green |
No | CellProfiler | CellProfiler | [109]* |
| B. subtilis | Nile red DAPI |
No | MicrobeJ | MicrobeJ | [110]* |
|
Bacillus subtilis, E. coli |
FM4-64 DAPI GFP |
No | Wasabi software (Hamamatsu) | Wasabi software (Hamamatsu) | [111] |
| E. coli | FM4-64 Hoechst-33342 Dendra2 protein |
No | FIJI/ImageJ | FIJI/ImageJ | [80] |
|
E. coli Caulobacter crescentus |
FM4-64 DAPI |
No | Oufti | Oufti | [112]* |
| E. coli | FM4-64 DAPI SYTOX-Green |
No | Semi-Manual (FIJI/ImageJ) | Semi-Manual (FIJI/ImageJ) | [113]* |
| Achromobacter xylosoxidans | FM4-64 DAPI SYTOX-Green NBD Azithromycin |
No | FIJI/ImageJ and CellProfiler | FIJI/ImageJ and CellProfiler | [114] |
| M. smegmatis | ParB-mCherry | Yes | MicrobeJ | MicrobeJ | [115]* |
| M. tuberculosis Erdman | FM4-64FX SYTO 24 |
Yes | MorphEUS | MorphEUS | [116]* |
| Shewanella putrefaciens | Ffh-mVenus FtsY-mVenus uL1-mVenus |
Yes | FIJI/ImageJ | FIJI/ImageJ | [117]* |
| V. parahaemolyticus | FM4-64 DAPI |
No | FIJI/ImageJ | FIJI/ImageJ | [84] |
| Bacillus subtilis | FM4-64 DAPI SYTOX-Green SYTO-9 |
No | - | Manual w/ FIJI | [118]* |
|
M. smegmatis M. tuberculosis |
FM4-64 GFP CellROX |
Yes | Omnipose | Manual w/ FIJI or with Cell Counter installation in FIJI Custom python script |
[119] |
|
E. coli B. subtilis |
FM4-64 DAPI GFP DiSC |
No | FIJI/ImageJ and MicrobeJ | FIJI/ImageJ and MicrobeJ | [120] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





