Submitted:
30 August 2024
Posted:
02 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
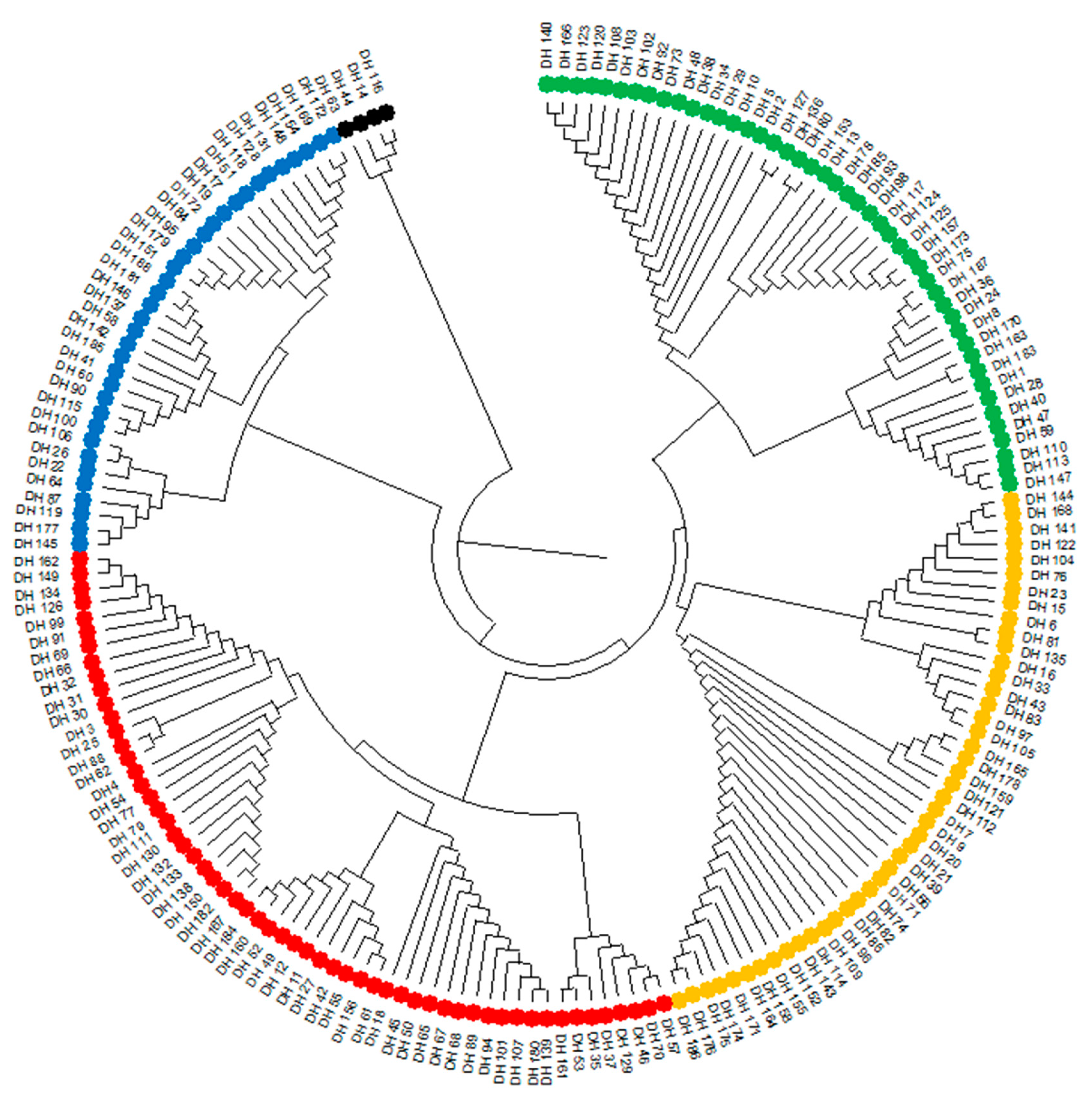
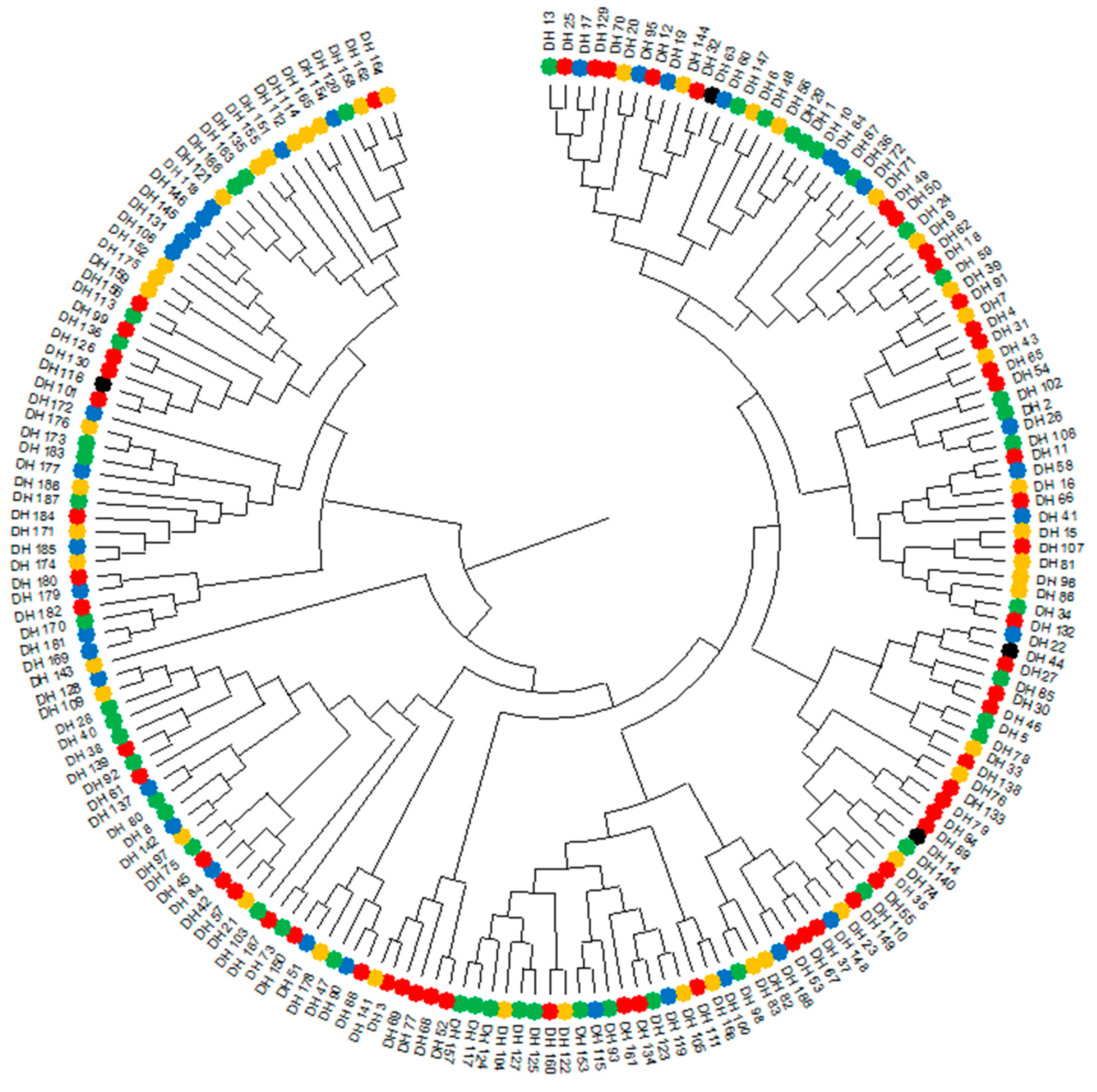
2.1. Phenotyping
2.2. Genotyping
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Field Assessment
4.3. DNA Extraction and Genotyping
4.4. Phenotypic and Genotypic Similarity
4.5. Genetic Parameters
4.5.1. Estimation Based on the Phenotype
4.5.2. Estimation Based on the Genotypic Observations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Qaim, M. Role of New Plant Breeding Technologies for Food Security and Sustainable Agricultural Development. Appl. Econ. Perspect. Policy 2020, 42, 129–150. [Google Scholar] [CrossRef]
- Benakanahalli, N.K.; Sridhara, S.; Ramesh, N.; Olivoto, T.; Sreekantappa, G.; Tamam, N.; Abdelbacki, A.M.M.; Elansary, H.O.; Abdelmohsen, S.A.M. A Framework for Identification of Stable Genotypes Basedon MTSI and MGDII Indexes: An Example in Guar (Cymopsis tetragonoloba L.). Agronomy 2021, 11, 1221. [Google Scholar] [CrossRef]
- Xu, P.; Wang, X.; Dai, S.; Cui, X.; Cao, X.; Liu, Z.; Shen, J. The multilocular trait of rapeseed is ideal for high-yield breeding. Plant Breed. 2021, 140, 65–73. [Google Scholar] [CrossRef]
- Ahmar, S.; Gill, R.A.; Jung, K.-H.; Faheem, A.; Qasim, M.U.; Mubeen, M.; Zhou, W. Conventional and Molecular Techniques from Simple Breeding to Speed Breeding in Crop Plants: Recent Advances and Future Outlook. Int. J. Mol. Sci. 2020, 21, 2590. [Google Scholar] [CrossRef] [PubMed]
- Salgotra, R.K.; Stewart, C.N.Jr. Functional Markers for Precision Plant Breeding. Int. J. Mol. Sci. 2020, 21, 4792. [Google Scholar] [CrossRef] [PubMed]
- Fehr, W.R. Principles of Cultivar Development, vol. 1, Theory and Technique. Macmillan Publishing Co., 866 Third Avenue, New York, N.Y. 536 pp, 1987.
- Cooper, M.; Powell, O.; Voss-Fels, K.P.; Messina, C.D.; Gho, C.; Podlich, D.E.; Technow, F.; Chapman, S.C.; Beveridge, C.A.; Ortiz-Barrientos, D.; Hammer, G.L. Modelling selection response in plant-breeding programs using crop models as mechanistic gene-to-phenotype (CGM-G2P) multi-trait link functions. In silico Plants 2021, 3, diaa016. [Google Scholar] [CrossRef]
- Bernardo, R. Reinventing quantitative genetics for plant breeding: something old, something new, something borrowed, something BLUE. Heredity 2020, 125, 375–385. [Google Scholar] [CrossRef]
- González Guzmán, M.; Cellini, F.; Fotopoulos, V.; Balestrini, R.; Arbona, V. New approaches to improve crop tolerance to biotic and abiotic stresses. Physiol. Plant. 2022, 174, e13547. [Google Scholar] [CrossRef]
- Golebiowska-Paluch, G.; Dyda, M. The Genome Regions Associated with Abiotic and Biotic Stress Tolerance, as Well as Other Important Breeding Traits in Triticale. Plants 2023, 12, 619. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Phil. Trans. R. Soc. B 2008, 363, 557–572. [Google Scholar] [CrossRef]
- Song, L.; Wang, R.; Yang, X.; Zhang, A.; Liu, D. Molecular Markers and Their Applications in Marker-Assisted Selection (MAS) in Bread Wheat (Triticum aestivum L.). Agriculture 2023, 13, 642. [Google Scholar] [CrossRef]
- Arrones, A.; Vilanova, S.; Plazas, M.; Mangino, G.; Pascual, L.; Díez, M.J.; Prohens, J.; Gramazio, P. The Dawn of the Age of Multi-Parent MAGIC Populations in Plant Breeding: Novel Powerful Next-Generation Resources for Genetic Analysis and Selection of Recombinant Elite Material. Biology 2020, 9, 229. [Google Scholar] [CrossRef] [PubMed]
- Muranty, H.; Denancé, C.; Feugey, L.; Crépin, J.L.; Barbier, Y.; Tartarini, S.; Ordidge, M.; Troggio, M.; Lateur, M.; Nybom, H.; Paprstein, F.; Laurens, F.; Durel, C.E. Using whole-genome SNP data to reconstruct a large multi-generation pedigree in apple germplasm. BMC Plant Biol. 2020, 20, 2. [Google Scholar] [CrossRef]
- Sinha, D.; Maurya, A.K.; Abdi, G.; Majeed, M.; Agarwal, R.; Mukherjee, R.; Ganguly, S.; Aziz, R.; Bhatia, M.; Majgaonkar, A.; et al. Integrated Genomic Selection for Accelerating Breeding Programs of Climate-Smart Cereals. Genes 2023, 14, 1484. [Google Scholar] [CrossRef]
- Scott, M.F.; Fradgley, N.; Bentley, A.R.; Brabbs, T.; Corke, F.; Gardner, K.A.; Horsnell, R.; Howell, P.; Ladejobi, O.; Mackay, I.J.; Mott, R.; CockraM, J. Limited haplotype diversity underlies polygenic trait architecture across 70 years of wheat breeding. Genome Biol. 2021, 22, 137. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Arshad, K.; Abdullah, J.; Aslam, A.; Azam, A.; Bilal, M.; Asad, M.; Hamza, A.; Abdullah, M. A Comprehensive Review on Breeding Technologies and Selection Methods of Self-pollinated and Cross-Pollinated Crops. Asian J. Biotechnol. Genet. Eng. 2021, 4, 35–47. [Google Scholar]
- Muthoni, J.; Shimelis, H. Mating designs commonly used in plant breeding: A review. Aust. J. Crop Sci. 2020, 14, 1855–1869. [Google Scholar] [CrossRef]
- Tiwari, A.; Tikoo, S.K.; Angadi, S.P.; Kadaru, S.B.; Ajanahalli, S.R.; Vasudeva Rao, M.J. Inbred Line Development and Hybrid Breeding. In: Market-Driven Plant Breeding for Practicing Breeders. Springer, Singapore, 2022. [CrossRef]
- Boopathi, N.M. Marker-Assisted Selection (MAS). In: Genetic Mapping and Marker Assisted Selection. Springer, Singapore, 2020. [CrossRef]
- Merrick, L.F.; Carter, A. Comparison of genomic selection models for exploring predictive ability of complex traits in breeding programs. Plant Genome 2021, 14, e20158. [Google Scholar] [CrossRef]
- Gemenet, D.C.; Lindqvist-Kreuze, H.; De Boeck, B.; da Silva Pereira, G.; Mollinari, M.; Zeng, Z.B.; Yencho, G.C.; Campos, H. Sequencing depth and genotype quality: accuracy and breeding operation considerations for genomic selection applications in autopolyploid crops. Theor. Appl. Genet. 2020, 133, 3345–3363. [Google Scholar] [CrossRef]
- Merrick, L.F.; Herr, A.W.; Sandhu, K.S.; Lozada, D.N.; Carter, A.H. Optimizing Plant Breeding Programs for Genomic Selection. Agronomy 2022, 12, 714. [Google Scholar] [CrossRef]
- Obšteter, J.; Jenko, J.; Gorjanc, G. Genomic Selection for Any Dairy Breeding Program via Optimized Investment in Phenotyping and Genotyping. Front. Genet. 2021, 12, 637017. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Lin, Z.; Li, X.; Zhao, Y,; Zhao, B. ; Wu, G.; Ma, X.; Wang, H.; Xie, Y.; Li, Q.; Song, G.; Kong, D.; Zheng, Z.; Wei, H.; Shen, R.; Wu, H.; Chen, C.; Meng, Z.; Wang, T.; Li, Y.; Li, X.; Chen, Y.; Lai, J.; Hufford, M.B.; Ross-Ibarra, J.; He, H.; Wang, H. Genome-wide selection and genetic improvement during modern maize breeding. Nat. Genet. 2020, 52, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Sarkar, A.; Carbonetto, P.; Stephens, M. A Simple New Approach to Variable Selection in Regression, with Application to Genetic Fine Mapping. J. R. Stat. Soc. B 2020, 82, 1273–1300. [Google Scholar] [CrossRef] [PubMed]
- Swarup, S.; Cargill, E.J.; Crosby, K.; Flagel, L.; Kniskern, J.; Glenn, K.C. Genetic diversity is indispensable for plant breeding to improve crops. Crop Sci. 2021, 61, 839–852. [Google Scholar] [CrossRef]
- Gibson, A.K. Genetic diversity and disease: The past, present, and future of an old idea. Evolution 2022, 76(s1), 20–36. [Google Scholar] [CrossRef]
- Tuvesson, S.D.; Larsson, CT.; Ordon, F. Use of Molecular Markers for Doubled Haploid Technology: From Academia to Plant Breeding Companies. In: Segui-Simarro, J.M. (eds) Doubled Haploid Technology. Methods in Molecular Biology, 2021, 2288. Humana, New York, NY. [CrossRef]
- Hooghvorst, I.; Nogués, S. Chromosome doubling methods in doubled haploid and haploid inducer-mediated genome-editing systems in major crops. Plant Cell Rep. 2021, 40, 255–270. [Google Scholar] [CrossRef]
- Yali, W. Haploids and Doubled Haploid Technology Application in Modern Plant Breeding. J. Plant Sci. 2022, 10, 71–75. [Google Scholar] [CrossRef]
- Maqbool, M.A.; Beshir, A.; Khokhar, E.S. Doubled haploids in maize: Development, deployment, and challenges. Crop Sci. 2020, 60, 2815–2840. [Google Scholar] [CrossRef]
- Hu, H.; Meng, Y.; Liu, W.; Chen, S.; Runcie, D.E. Multi-Trait Genomic Prediction Improves Accuracy of Selection among Doubled Haploid Lines in Maize. Int. J. Mol. Sci. 2022, 23, 14558. [Google Scholar] [CrossRef]
- Hale, B.; Ferrie, A.M.R.; Chellamma, S.; Samuel, J.P.; Phillips, G.C. Androgenesis-Based Doubled Haploidy: Past, Present, and Future Perspectives. Front. Plant Sci. 2022, 12, 751230. [Google Scholar] [CrossRef]
- Hooghvorst, I.; Nogués, S. Chromosome doubling methods in doubled haploid and haploid inducer-mediated genome-editing systems in major crops. Plant Cell Rep. 2021, 40, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Z.; Chen, Q.; Tang, J.; Lübberstedt, T.; Li, H. Mapping of QTL for Grain Yield Components Based on a DH Population in Maize. Sci. Rep. 2020, 10, 7086. [Google Scholar] [CrossRef]
- Zhang, J.; She, M.; Yang, R.; Jiang, Y.; Qin, Y.; Zhai, S.; Balotf, S.; Zhao, Y.; Anwar, M.; Alhabbar, Z.; et al. Yield-Related QTL Clusters and the Potential Candidate Genes in Two Wheat DH Populations. Int. J. Mol. Sci. 2021, 22, 11934. [Google Scholar] [CrossRef]
- Cazzola, F.; Bermejo, C.J.; Gatti, I.; Cointry, E. Speed breeding in pulses: an opportunity to improve the efficiency of breeding programs. Crop Pasture Sci. 2021, 72, 165–172. [Google Scholar] [CrossRef]
- Bocianowski, J.; Kozak, M.; Liersch, A.; Bartkowiak-Broda, I. A heuristic method of searching for interesting markers in terms of quantitative traits. Euphytica 2011, 181, 89–100. [Google Scholar] [CrossRef]
- Robert, P.; Auzanneau, J.; Goudemand, E.; Oury, F.X.; Rolland, B.; Heumez, E.; Bouchet, S.; Le Gouis, J.; Rincent, R. Phenomic selection in wheat breeding: identification and optimisation of factors influencing prediction accuracy and comparison to genomic selection. Theor. Appl. Genet. 2022, 135, 895–914. [Google Scholar] [CrossRef] [PubMed]
- Pélabon, C.; Hilde, C.H.; Einum, S.; Gamelon, M. On the use of the coefficient of variation to quantify and compare trait variation. Evol. Lett. 2020, 4, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Labroo, M.R.; Studer, A.J.; Rutkoski, J.E. Heterosis and Hybrid Crop Breeding: A Multidisciplinary Review. Front. Genet. 2021, 12, 643761. [Google Scholar] [CrossRef]
- Lanzl, T.; Melchinger, A.E.; Schön, C.C. Influence of the mating design on the additive genetic variance in plant breeding populations. Theor. Appl. Genet. 2023, 136, 236. [Google Scholar] [CrossRef]
- Aakanksha; Yadava, S. K.; Yadav, B.G.; Gupta, V.; Mukhopadhyay, A.; Pental, D.; Pradhan, A.K. Genetic Analysis of Heterosis for Yield Influencing Traits in Brassica juncea Using a Doubled Haploid Population and Its Backcross Progenies. Front. Plant Sci. 2021, 12, 721631. [Google Scholar] [CrossRef]
- Mackay, T. Epistasis and quantitative traits: using model organisms to study gene–gene interactions. Nat. Rev. Genet. 2014, 15, 22–33. [Google Scholar] [CrossRef]
- Gjuvsland, A.B.; Hayes, B.J.; Omholt, S.W.; Carlborg, Ö. Statistical Epistasis Is a Generic Feature of Gene Regulatory Networks. Genetics 2007, 175, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Bürger, R.; Arnold, S. Epistasis and natural selection shape the mutational architecture of complex traits. Nat. Commun. 2014, 5, 3709. [Google Scholar] [CrossRef]
- Bocianowski, J. Epistasis interaction of QTL effects as a genetic parameter influencing estimation of the genetic additive effect. Genet. Mol. Biol. 2013, 36, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Bocianowski, J.; Nowosad, K. Mixed linear model approaches in mapping QTLs with epistatic effects by a simulation study. Euphytica 2015, 202, 459–467. [Google Scholar] [CrossRef]
- Cyplik, A.; Bocianowski, J. Analytical and numerical comparisons of two methods of estimation of additive × additive × additive interaction of QTL effects. J. Appl. Genet. 2022, 63, 213–221. [Google Scholar] [CrossRef]
- Cyplik, A.; Sobiech, A.; Tomkowiak, A.; Bocianowski, J. Genetic Parameters for Selected Traits of Inbred Lines of Maize (Zea mays L.). Appl. Sci. 2022, 12, 6961. [Google Scholar] [CrossRef]
- Cyplik, A.; Bocianowski, J. A Comparison of Methods to Estimate Additive–by–Additive–by–Additive of QTL×QTL×QTL Interaction Effects by Monte Carlo Simulation Studies. Int. J. Mol. Sci. 2023, 24, 10043. [Google Scholar] [CrossRef] [PubMed]
- Cyplik, A.; Czyczyło-Mysza, I.M.; Jankowicz-Cieslak, J.; Bocianowski, J. QTL×QTL×QTL Interaction Effects for Total Phenolic Content of Wheat Mapping Population of CSDH Lines under Drought Stress by Weighted Multiple Linear Regression. Agriculture 2023, 13, 850. [Google Scholar] [CrossRef]
- Cyplik, A.; Piaskowska, D.; Czembor, P.; Bocianowski, J. The use of weighted multiple linear regression to estimate QTL × QTL × QTL interaction effects of winter wheat (Triticum aestivum L.) doubled-haploid lines. J. Appl. Genet. 2023, 64, 679–693. [Google Scholar] [CrossRef]
- Hill, W.G.; Goddard, M.E.; Visscher, P.M. Data and Theory Point to Mainly Additive Genetic Variance for Complex Traits. PLoS Genet. 2008, 4, e1000008. [Google Scholar] [CrossRef]
- Starosta, E.; Jamruszka, T.; Szwarc, J.; Bocianowski, J.; Jędryczka, M.; Grynia, M.; Niemann, J. DArTseq-Based, High-Throughput Identification of Novel Molecular Markers for the Detection of Blackleg (Leptosphaeria spp.) Resistance in Rapeseed. Int. J. Mol. Sci. 2024, 25, 8415. [Google Scholar] [CrossRef]
- Tian, Z.; Wang, J.-W.; Li, J.; Han, B. Designing future crops: challenges and strategies for sustainable agriculture. Plant J. 2021, 105, 1165–1178. [Google Scholar] [CrossRef]
- Habib-ur-Rahman, M.; Ahmad, A.; Raza, A.; Hasnain, M.U.; Alharby, H.F.; Alzahrani, Y.M.; Bamagoos, A.A.; Hakeem, K.R.; Ahmad, S.; Nasim, W.; Ali, S.; Mansour, F.; EL Sabagh, A. Impact of climate change on agricultural production; Issues, challenges, and opportunities in Asia. Front. Plant Sci. 2022, 13, 925548. [Google Scholar] [CrossRef]
- Kozak, M.; Bocianowski, J.; Liersch, A.; Tartanus, M.; Bartkowiak-Broda, I.; Piotto, F.A.; Azevedo, R.A. Genetic divergence is not the same as phenotypic divergence. Mol. Breed. 2011, 28, 277–280. [Google Scholar] [CrossRef]
- Seyis, F.; Snowdon, R.J.; Luhs, W.; Friedt, W. Molecular characterization of novel resynthesized rapeseed (Brassica napus) lines and analysis of their genetic diversity in comparison with spring rapeseed cultivars. Plant Breed. 2003, 122, 473–478. [Google Scholar] [CrossRef]
- Hannan, A.; Islam, M.M.; Rahman, M.S.; Hoque, M.N.; Sagor, G.H.M. Morpho-genetic Evaluation of Rice Genotypes (Oryza sativa L.) Including Some Varieties and Advanced Lines Based on Yield and Its Attributes. Journal of the Bangladesh Agricultural University 2024, 18, 923–933. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, Y.; Liu, X.; Chen, B.; Tu, J.; Tingdong, F. Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor. Appl. Genet. 2007, 115, 849–858. [Google Scholar] [CrossRef]
- Carlborg, Ö.; Haley, C.S. Epistasis: too often neglected in complex trait studies? Nat. Rev. Genet. 2004, 5, 618–625. [Google Scholar] [CrossRef]
- Miko, I. Epistasis: gene interaction and phenotype effects. Nature Education 2008, 1, 197. [Google Scholar]
- Phillips, P.C. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008, 9, 855–867. [Google Scholar] [CrossRef]
- Germain, D.P.; Jurca-Simina, I.E. Principles of Human Genetics and Mendelian Inheritance. In: Burlina, A. (eds) Neurometabolic Hereditary Diseases of Adults. Springer, Cham, 2018. [CrossRef]
- Schrodi, S.J.; Mukherjee, S.; Shan, Y.; Tromp, G.; Sninsky, J.J.; Callear, A.P.; Carter, T.C.; Ye, Z.; Haines, J.L.; Brilliant, M.H.; Crane, P.K.; Smelser, D.T.; Elston, R.C.; Weeks, D.E. Genetic-based prediction of disease traits: prediction is very difficult, especially about the future. Front. Genet. 2014, 5, 162. [Google Scholar] [CrossRef]
- Sun, X.; Ma, P.; Mumm, R.H. Nonparametric Method for Genomics-Based Prediction of Performance of Quantitative Traits Involving Epistasis in Plant Breeding. PLoS ONE 2012, 7, e50604. [Google Scholar] [CrossRef]
- Li, M.; Lou, X.Y.; Lu, Q. On Epistasis: A Methodological Review for Detecting Gene-Gene Interactions Underlying Various Types of Phenotypic Traits. Recent Pat. Biotechnol. 2012, 6, 230–236. [Google Scholar] [CrossRef]
- Raffo, M.A.; Sarup, P.; Guo, X.; Liu, H.; Andersen, J.R.; Orabi, J.; Jahoor, A.; Jensen, J. Improvement of genomic prediction in advanced wheat breeding lines by including additive-by-additive epistasis. Theor. Appl. Genet. 2022, 135, 965–978. [Google Scholar] [CrossRef]
- Das, A.K.; Choudhary, M.; Kumar, P.; Karjagi, C.G.; KR, Y.; Kumar, R.; Singh, A.; Kumar, S.; Rakshit, S. Heterosis in Genomic Era: Advances in the Molecular Understanding and Techniques for Rapid Exploitation. Crit. Rev. Plant Sci. 2021, 40, 218–242. [Google Scholar] [CrossRef]
- Jeon, D.; Kang, Y.; Lee, S.; Choi, S.; Sung, Y.; Lee, T.-H.; Kim, C. Digitalizing breeding in plants: A new trend of next-generation breeding based on genomic prediction. Front. Plant Sci. 2023, 14, 1092584. [Google Scholar] [CrossRef]
- Pang, E.C.K.; Halloran, G.M. The genetics of blackleg [Leptosphaeria maculans (Desm.) Ces, et De Not.] resistance in rapeseed (Brassica napus L.). Theor. Appl. Genet. 1996, 93, 932–940. [Google Scholar] [CrossRef]
- Kumar, V.; Paillard, S.; Fopa-Fomeju, B.; Falentin, C.; Deniot, G.; Baron, C.; Vallée, P.; Manzanares-Dauleux, M.J.; Delourme, R. Multi-year linkage and association mapping confirm the high number of genomic regions involved in oilseed rape quantitative resistance to blackleg. Theor. Appl. Genet. 2018, 131, 1627–1643. [Google Scholar] [CrossRef]
- Pilet, M.; Delourme, R.; Foisset, N.; Renard, M. Identification of loci contributing to quantitative field resistance to blackleg disease, causal agent Leptosphaeria maculans (Desm.) Ces. et de Not., in Winter rapeseed (Brassica napus L.). Theor. Appl. Genet. 1998, 96, 23–30. [Google Scholar] [CrossRef]
- Zhao, J.; Meng, J. Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2003, 106, 759–764. [Google Scholar] [CrossRef]
- Larkan, N.J.; Raman, H.; Lydiate, D.J.; Robinson, S.J.; Yu, F.; Barbulescu, D.M.; Raman, R.; Luckett, D.J.; Burton, W.; Wratten, N.; Salisbury, P.A.; Rimmer, S.R.; Borhan, M.H. Multi-environment QTL studies suggest a role for cysteine-rich protein kinase genes in quantitative resistance to blackleg disease in Brassica napus. BMC Plant Biol. 2016, 16, 183. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.N.; DeClerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef] [PubMed]
- Jędryczka, M. Epidemiology and Damage Caused by Stem Canker of Oilseed Rape in Poland. Ph.D. Thesis, Dissertations and Monographs, Abstract of Habilitation Thesis Institute of Plant Genetics, Polish Academy of Sciences, Poznań, Poland, 2006; pp. 42, 150. (In Polish, with English abstract).
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Bocianowski, J.; Niemann, J.; Jagieniak, A.; Szwarc, J. Comparison of Six Measures of Genetic Similarity of Interspecific Brassicaceae Hybrids F2 Generation and Their Parental Forms Estimated on the Basis of ISSR Markers. Genes 2024, 15, 1114. [Google Scholar] [CrossRef]
- Bocianowski, J.; Krajewski, P. Comparison of the genetic additive effect estimators based on phenotypic observations and on molecular marker data. Euphytica 2009, 165, 113–122. [Google Scholar] [CrossRef]
- Choo, T.M.; Reinbergs, E. Analyses of Skewness and Kurtosis for Detecting Gene Interaction in a Doubled Haploid Population. Crop Sci. 1982, 22, 231–235. [Google Scholar] [CrossRef]
- Bocianowski, J. A comparison of two methods to estimate additive-by-additive interaction of QTL effects by a simulation study. J. Theor. Biol. 2012, 308, 20–24. [Google Scholar] [CrossRef]
- Bocianowski, J. Analytical and numerical comparisons of two methods of estimation of additive additive interaction of QTL effects. Sci. Agric. 2012, 69, 240–246. [Google Scholar] [CrossRef]
- VSN International Genstat for Windows 23nd Edition. VSN International, Hemel Hempstead, UK, 2023.
| Genetic parameter | Phenotypic method | Genotypic method |
|---|---|---|
| additive, a | 2.450 *** | 1.794 *** |
| additive-additive, aa | 0.435 * | -0.532 ** |
| additive-additive-additive, aaa | 0.952 ** | -0.478 * |
| Marker Number | Marker Type | Chromosome | Marker Position on Chromosome (bp) | Marker Sequence |
|---|---|---|---|---|
| 7853 | SilicoDArT | A06 | 5,968,268–5,968,337 | TGCAGAAAATGGAATGTTCTTGAGAGATCCTAGTGGAGAATGGGTGACAAATATGCCTCAAGACATGAA |
| 11720 | SNP (18:A>T) | C06 | 34,959,583–34,959,652 | TGCAGAAGCAGCCATGAGACAGTATTGCTGTTGAGATATATTGTTGCTGTACCTTGGGGAGGAAGCAAC |
| 12134 | SNP (29:G>A) | C06 random | 2,170,846–2,170,777 | TGCAGCTTCTACTTTTAGTTGGACAGAGCGCTCAAAGTCAACAATTACAGATCGGAAGAGCGGTTCAGC |
| 12232 | SNP (46:T>A) | C03 | 1,024,177–1,024,246 | TGCAGAAAAAGATTCAGGTTCCCGGGACCTGAAGATCACTGGATTGTCTGATGCTGTGTTAGGATGCAT |
| 12456 | SNP (45:A>C) | A08 | 13,650,920–13,650,989 | TGCAGTTTCTACACGTACATATCCAATATTTTAGTTTACTTAGGAAGAAATTTGAAATTTGATTTTATT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





