1. Introduction
The microorganisms to survive, can develop defense strategies against antibiotics called resistance mechanisms. There are different ways to antibiotic resistance by prevent entrance of antibiotic to the cells or DNA express the specific proteins, can inactivate antibiotics upon exposed which determine the mechanisms of resistance. Antimicrobial resistance is a naturally happen actions. However, increases in antimicrobial resistance are driven by a combination of bacteria exposed to antibiotics, and the spread of those bacteria and their resistance mechanisms by DNA mutation or horizontal gene transfer [
1,
2].
Considering its severe threat to public health and antibiotic treatment effectiveness, antibiotic resistance is a significant concern in microbiology. Increasing use and misuse of antimicrobials and other factor, such as pollution, create good conditions for Bacteria to develop resistance in humans and the environment. non-resistance Bacteria and normal flora in water, soil and air, can obtain resistance following touching with resistant bacteria. Human exposure to Antimicrobial Resistance in the environment can occur through contact with polluted waters, contaminated food, and other pathways that carry antimicrobial resistant microorganisms. Nevertheless, most studies on antibiotic resistance focused on infection-causing bacteria in humans, animals, and plants [
3]. Antibiotic resistance has also been reported in other organisms, including fungi and cyanobacteria [
4].
Cyanobacteria (also called Blue green algae: Cyanophyta) are photosynthetic Gram-negative prokaryotes, are unique among microbial world and grow in diverse habitats with a crucial role in ecosystems. It can be found in aquatic system (freshwater and sea water), moisture soil and other habitats such as epiphytes, epizoic and endozoic. cyanobacteria has wide range of diversity in morphology from Unicellular, filamentous, Aggregates and colonial form. Most of it has no complex DNA (single circular chromosome) so, they have ability to DNA transformation. their genome differ in size from 1.4 to 12 Mbp [
5,
6].
The microorganisms perform photosynthesis, fixed carbon and produce oxygen, biofuel and fix nitrogen gas, which increases soil fertility and treatment of industrial wastewaters by remove of heavy metals, phosphate and ammonia, synthesize a wide group of novel secondary metabolites compounds including antioxidants, Vitamins and other biologically active compounds has antibacterial, antiviral, antifungal, and anticancer activities [
7]. some cyanobacteria species can be engineered for biotechnological objectives in using DNA recombination to activation of the newly introduced genes of industrial interest [
8].
Nonetheless, cyanobacteria form harmful algal blooms (HABs) under certain conditions, producing toxins that could negatively impact humans, animals, aquatic health and ecosystems moreover, decrease water quality, alter the bacterial community structure and disrupt recreation and human health [
9]. Although cyanobacteria do not directly cause human infections, they could still develop antibiotic resistance. A reason for the phenomenon is the widespread utilization of antibiotics in medical and veterinary purposes, and other various industries, such as agriculture, aquaculture and promote the growth of livestock. The vast scale of antibiotic use and antibiotic misuse speed up the evolution of antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in the aquatic environment including cyanobacteria and other microorganism. [
10,
11].
The presence of antibiotic-resistant cyanobacteria in aquatic environments is concerning as the microorganisms could contribute to the dissemination of resistance genes to other bacteria, including those that are pathogenic to humans. Moreover, cyanotoxin production by particular cyanobacterial species might be increased under high antibiotic concentrations in water environments [
12]. Cyanotoxins are potent toxins that could harm humans and animals. so, the People exposed to the algal cyanotoxins by eating impure foods or dietary additive, or by swallowing contaminated water, may experience the following symptoms, such as Headache, nausea, Stomach pain, Neurological symptoms including: muscle feebleness, dizziness, depending on the type of cyanotoxins involved [
13].
The current study aimed to investigate and detection of antibiotic resistance genes in some locally isolated of freshwater Cyanobacteria (blue-green algae) species, then compare the results for same Antibiotic Resistance genes in local pathogenic bacteria isolated from disease cases to discover the presence and spread of these genes in some microorganisms (other than bacteria) in Iraqi freshwater.
2. Materials and Methods
2.1. Algal Isolates
The blue-green algae species employed in this study:-
Spirulina laxa G.M.Smith,
Chroococcus minutes (Ktz.) Naegeli,
Oscillatoria princeps Vaucher,
Oscillatoria proteus Skuja,
Oscillatoria terebriformis Agardh, and
Lyngbya epiphytica Hieron. Algal samples were obtained from the postgraduate laboratory of the Department of Biology, College of Education for Pure Sciences, University of Anbar. The specimens were initially collected from Euphrates river in Ramadi city, Anbar province, west of Iraq, for genetic and physiological research [
14,
15]. By micropipette washing technique, the algal samples was isolate to get unialgal culture, subsequently, centrifuge washing technique for purification the algal samples towards axenic culture, and confirm it by streak plating technique [
16]. The study samples were cultured in BG11 medium (HIMEDIA, India) and prepared according to the guidelines provided by the manufacturer. by using 100 ml of BG11 medium in 500 ml conical flask, the cultures were incubated at 22 ±2 ℃ with a 14:10 hours of light : dark cycle to obtain the biomass required for algal DNA extraction. the biomass collected by centrifugation 5000 rpm for 5 minutes.
2.2. Bacterial Isolates
The current study utilized Escherichia coli and Klebsiella pneumoniae strains from the Microbiology Laboratory of the Department of Biology, College of Education for Pure Science, University of Anbar. The source of E. coli and K. pneumoniae was UTI and Sputum, respectively. The bacterial isolates were cultured in LB broth media, which consisted of 10% tryptone, 5% yeast extract, and 10% Sodium chloride (NaCl). The 5 ml of bacterial cultures in 15 ml test tube were incubated at 37 ℃ for 18 hours, then centrifuge it (8000 rpm for 10 minutes ) in 1.5 ml eppendrof tube to collect bacterial pellet for DNA Extraction.
2.3. Genomic DNA Extraction
The algal and bacterial DNA samples were extracted from the cultures utilizing a genomic DNA extraction kit supplied by Geneaid (Taiwan). The 100 mg of wet weight for algal culture and 1.5 mL overnight culture for bacteria. The extracted DNA confirmed using 0.8% agarose gel electrophoresis by dissolved 160mg agarose in 20ml TBE buffer. The extracted DNA stored in -20 ͦ C until use.
2.4. Polymerase Chain Reaction
Specific primers for six antibiotic-resistance genes were utilised during the polymerase chain reaction (PCR) performed in the present study. The antibiotics evaluated were gentamicin (Gm), spectinomycin (Sp), ampicillin (Ap), chloramphenicol (Cm), erythromycin (Em) and kanamycin (Km). The primer sequence, name, and PCR product for each gene are listed in
Table 1 [
17]. The reaction mixture was prepared using the Accupower
® GOLD Multiplex kit supplied by Bioneer (Republic of Korea) according to the manufacturer’s instructions. The PCR was performed using a thermo cycler (DLAB- T1000-G, USA). The PCR program involved initial denaturation at 95 C for 5 min. and 35 cycles (Denaturation at 95 ͦ C for 1 min. annealing at 56 ͦ C for 1 min. then Extension at 72 ͦ C for 1 min., and final Extension at 72 ͦ C for 5 min). the PCR products were run on 1.5% agarose gel (300 mg agarose in 20 ml TBE buffer.
3. Results
The studied isolated
Spirulina laxa G.M.Smith (
S. laxa),
Chroococcus minutes (Ktz.) Naegeli (
C. minutes),
Oscillatoria princeps Vaucher (
O. princeps),
Oscillatoria proteus Skuja (
O. proteus),
Oscillatoria terebriformis Agardh (
O. terebriformis), and
Lyngbya epiphytica Hieron (
L. epiphytica) was identified depended on Bellinger and Sigee 2010 [
18]. Then, the identification for studies samples confirmed via amplified of 16srRNA gene post-chromosomal DNA extraction. The sequences were submitted to NCBI for alignment and obtain the accession numbers [
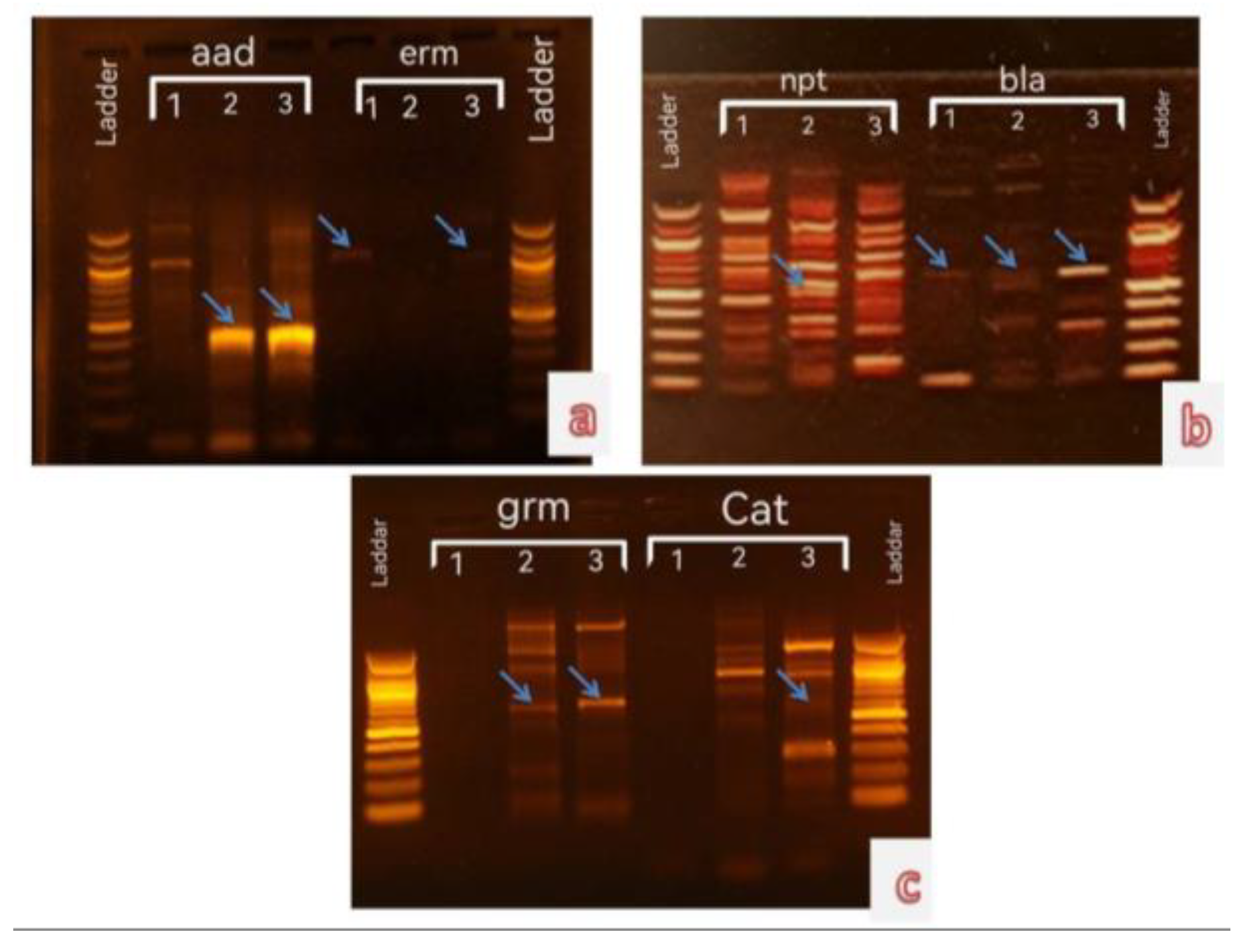
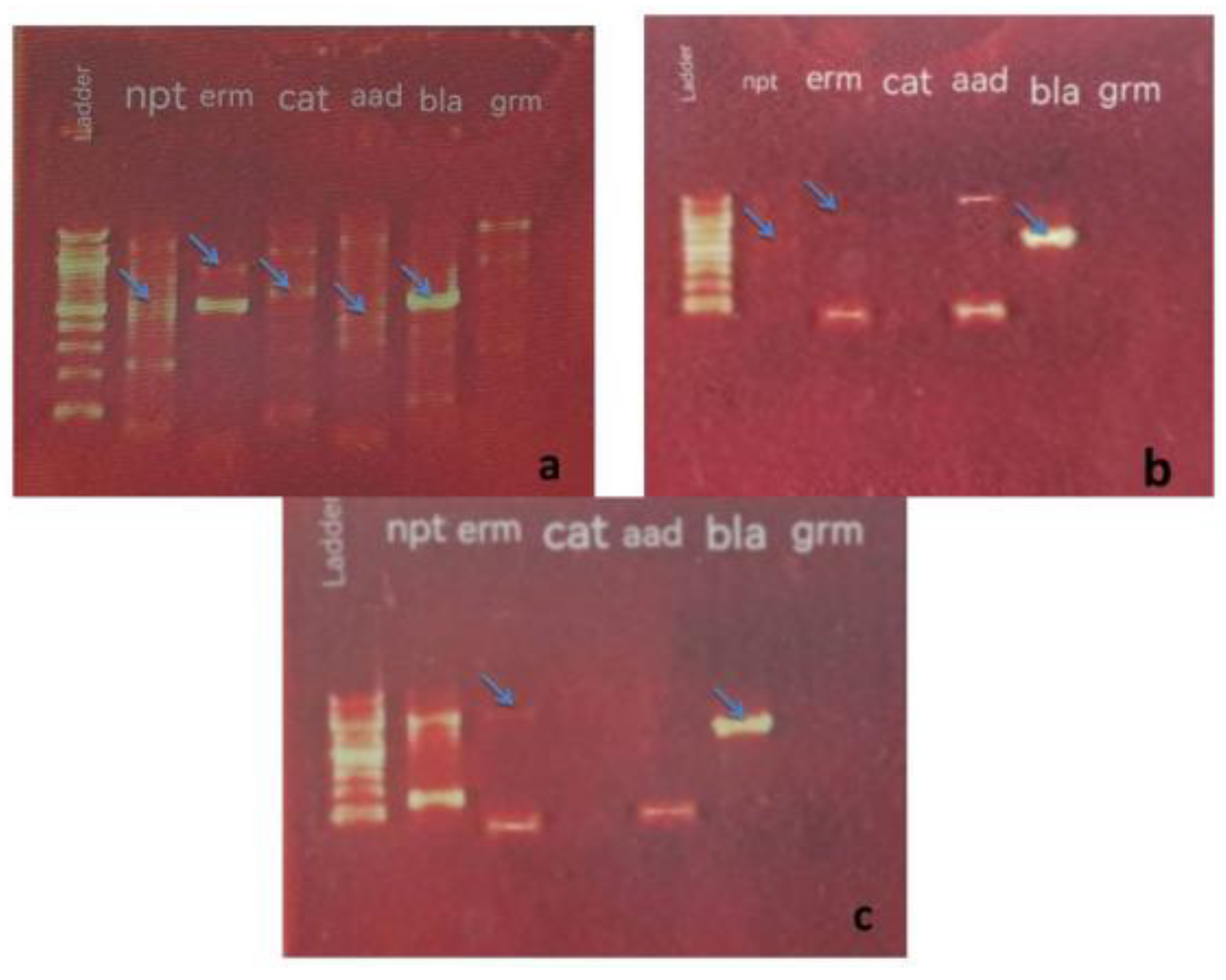
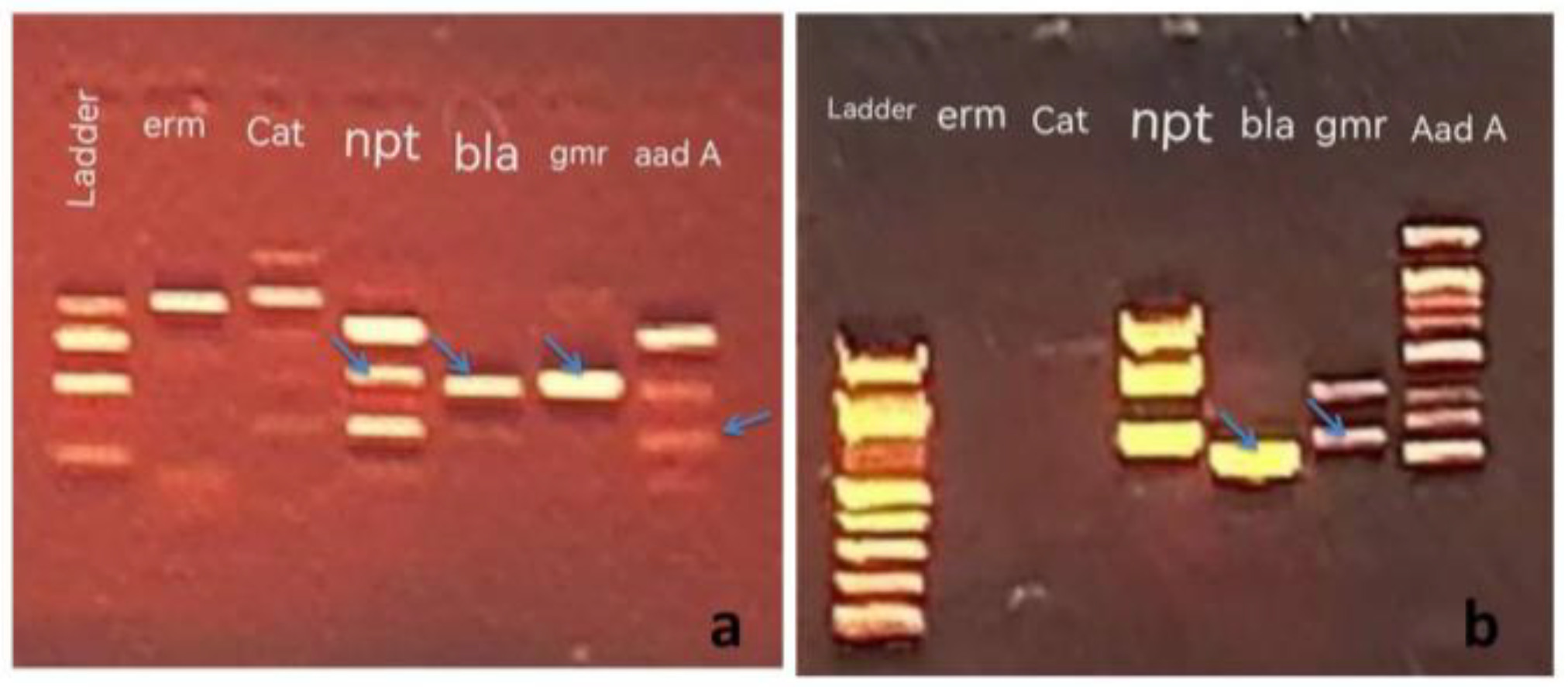
19]. The algal and bacterial DNA extract yields from the specimens in the present study were assessed in 0.8% agarose gel electrophoresis. Subsequently, the DNA was employed as a template to detect the antibiotic resistance genes of different antibiotics utilizing specific primers.
A previous study reported the ability of several blue-green algae species to grow in a BG11 medium that consisted of different concentrations of numerous antibiotics [
20]. The findings suggested that the studied algae possessed ability for antibiotic resistance. The PCR Resultantly, the algal samples and pathogenic bacteria contained similar DNA bands for different antibiotic resistance genes , which represent antibiotic resistance genes in both types of Microorganisms (see
Figure 1,
Figure 2 and
Figure 3).
The ARGs were detected in the algae and pathogenic bacteria samples evaluated in this study. The bla gene ( Ap
R) was present in all algal samples ( Blue-Green Algae and Bacteria) but the
ermC gene (Em
R) was not observed in
O. proteus and
K. pneumonia. While, the
aacC1 (Gm
R) and
cat (Cm
R) genes were the least apparent in all algal samples, only two algal species for each gene. (
Table 2). On the other hand, the alga
S. laxa and
O.
terebriformis has five out of six studied genes, whereas
O.princeps has only two antibiotic resistance genes was study.
The studied pathogenic bacteria
E. coli and
K. pneumonia has 4 Antibiotic resistance genes (
bla,
ermC,
aacC and
npt) and 3 Antibiotic resistance genes (
bla,
aacC and
npt), respectively (
Table 2).
4. Discussion
Freshwater Ecosystems play the important role in succour life by helping as agricultural environments and providing a source of drinking freshwater. So, the pollution or changes in physical and chemical factors for these ecosystem, including cyanoHAB causes a change in the interactions between living microorganisms, which causes variations in the communities of the aquatic environment and spread of AR [
21].
The results showed the presence of antibiotic resistance genes in some studied species of blue-green algae isolated from the local environment. This indicates the possibility of the spread of antibiotic resistance in aquatic microorganisms in the local environment. The global distribution of cyanobacteria indicates their ability to cope with a wide range of global environmental stresses, such as high and low temperatures, nitrogen starvation, anaerobic stress and osmotic tension, photooxidation, salinity and drought. They have developed a several of mechanisms by which cyanobacteria defend themselves against environmental stressors [
22].
Antibiotic contamination is a serious environmental and health challenge. Residual antibiotics from municipal, industrial, and agricultural wastewater, and sewage discharge, are continuously released into freshwater environments, where they contribute to the evolution and spread of antibiotic resistance. Therefore, it has a serious impact on aquatic organisms, especially microalgae and cyanobacteria, which play an important role as primary producers in the water ecosystem [
23]. Antibiotics, also have significant negative effects on the growth of cyanophyta and their chlorophyll content, and production of algal toxin like microcystin. Cyanophyta being more susceptible to the effect as they are prokaryotic [
24].
Two isolates of pathogenic bacteria resistant to antibiotics were used as a positive control to identify the extent of similarity in the feature of possessing antibiotic resistance genes between bacteria and blue-green algae. The results refers to the similarity in presence of antibiotic resistance genes in both types of microorganisms ( Bacteria and Cyanobacteria). This might be indicate the high level of contamination by antibiotics for local freshwater by different reasons, which led to the transfer of the antibiotic resistance feature to microorganisms that did not possess this feature.
Antibiotics are occasionally employed in the industries to prevent bacterial infections. Nevertheless, when antibiotics are released into the environment, they might exert selective pressure on bacteria, including cyanobacteria, leading to resistance development [
25]. Numerous studies have reported antibiotic resistance genes (ARGs) in cyanobacteria isolated from different environments, including freshwater and marine ecosystems. The genes reportedly confer resistance to various antibiotics, including tetracyclines, beta (β)-lactams, and macrolides [
26,
27,
28].
Antibiotic resistance is a critical issue on a global scale, the ARGs are deemed environmental contaminants that could be transmitted between antibiotic-resistant and non-antibiotic-resistant bacteria via multiple mobile genetic elements (MGEs). Furthermore, the microorganisms are being spread in various bodies of water, including surface, drinking, sewage, and natural water [
29,
30,
31]. Antibiotic resistance in cyanobacteria could also impact the control of cyanobacterial harmful blooms leads to damage the water ecology by consume oxygen in the water, and produce high concentration of toxins which can kill fish and other living organisms [
32].
Although cyanobacteria are ubiquitous in aquatic ecosystems and are exposed to pollution or antibiotic resistance, their role in AR expansion in natural ecosystems remains unknown [
33]. Some studies hypothesised cyanobacteria might contain AR genes, considering their MGEs, such as plasmids. The MGEs are the primary AR gene transfer mechanism between microorganisms. Some reports suggested that plasmids might determine cyanobacterial resistance to antibiotics [
34,
35,
36].
The plasmids play critical roles in transfer the ARGs among microorganisms by Horizontal gene transfer. Some studies refers to presence the plasmids in Cyanobacteria, comprising a total of 256 plasmids distributed across 145 cyanobacterial species belonging to the Oscillatoriophycideae, Synechococcales, Nostocales, Pleurocapsales, and Pseudanabaenales. More than 69 of which have one or more extrachromosomal elements and 43 of these harbours large plasmids over 100 kbp, and contribute to the distribution of antimicrobial resistance genes [
37].
Several cyanobacterial species reportedly resist several antibiotics, including penicillin and ampicillin [
25]. For instance, a gene encoding penicillin-binding protein was recorded in a
Thermosynechococcus elongatus cyanobacterium [
24]. Although the finding explained the weak β-lactamase activity in the bacterium, its physiological role has not been determined. Nonetheless, a recent study reported that cyanobacteria are hosts of ARGs. Cyanobacteria are also associated with CyanoHABs when the microbial community structures in freshwater are altered. as Accordingly, other biotic (such as interactions with other microorganisms) and abiotic factors necessitate investigation [
38,
39,
40].
Promoting responsible antibiotic utilization in all sectors, including agriculture, aquaculture, and human medicine, is essential in addressing the cyanobacteria antibiotic resistance issue. Proper waste management and treatment could prevent antibiotic release into the environment. Monitoring antibiotic resistance in cyanobacteria and other bacteria is crucial in comprehending the extent of the issue and developing appropriate strategies to mitigate the impact. Nonetheless, no local studies have addressed antibiotic-resistance genes in blue-green algae. The importance of the blue-green algae in producing toxins might be linked to their resistance to antibiotics. Consequently, future studies could focus on toxin-producing and antibiotic resistance genes in similar species.
5. Conclusions
In conclusion The prevalence of Antibiotic Resistance Genes (ARGs) in the locally isolated blue-green algae species evaluated in this study resembled the pathogenic bacteria specimens. The results highlighted the importance of this study. Significant similarities between antibiotic resistance genes in cyanobacteria and pathogenic bacteria were observed in the present study. Nevertheless, future studies on Antibiotic Resistance Genes of cyanobacteria in other localities are necessary and correlation it with ability of toxins production and pollution of freshwater and the change in DNA materials by natural selection towards appear a new generation of harmful cyanobacteria.
Author Contributions
Conceptualization, H.K.; methodology, H.K., N.A.; software, D.A.; formal analysis, N.A.; data curation, H.K., D.A.; writing—original draft preparation, H.K.; writing—review and editing, N.A and D.A.; supervision, H.K.; project administration, H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
For their assistance in completing this work, the authors are grateful to all of the employees at the Department of Biology, College of Education for Pure Science, University Of Anbar, as well as their colleagues at the Molecular Biology Lab.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J basic Microbiol 2021, 61, 1049–1070. [Google Scholar] [CrossRef]
- Nwobodo, D.C.; Ugwu, M.C.; Anie, C.O.; Al-Ouqaili, M.T.S.; Ikem, J.C.; Chigozie, U.V.; et al. Antibiotic resistance: The challenges and some emerging strategies for tackling a global menace. J Clin Lab Anal 2022, 36, e24655. [Google Scholar] [CrossRef] [PubMed]
- Timms, V.J.; Hassan, K.A.; Pearson, L.A.; Neilan, B.A. Cyanobacteria as a critical reservoir of the environmental antimicrobial resistome. Environ Microbiol 2023, 25, 2266–2276. [Google Scholar] [CrossRef] [PubMed]
- Cassier-Chauvat, C.; Veaudor, T.; Chauvat, F. Comparative Genomics of DNA Recombination and Repair in Cyanobacteria: Biotechnological Implications. Front Microbiol 2016, 7, 1809. [Google Scholar] [CrossRef]
- Labella, J.I.; Llop, A.; Contreras, A. The default cyanobacterial linked genome: an interactive platform based on cyanobacterial linkage networks to assist functional genomics. FEBS Lett 2020, 594, 1661–1674. [Google Scholar] [CrossRef]
- Mehdizadeh Allaf, M.; Peerhossaini, H. Cyanobacteria: Model Microorganisms and Beyond. Microorganisms 2022, 10, 696. [Google Scholar] [CrossRef]
- Cassier-Chauvat, C.; Chauvat, F. Responses to oxidative and heavy metal stresses in cyanobacteria: recent advances. Int J Mol Sci 2015, 16, 871–886. [Google Scholar] [CrossRef] [PubMed]
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat Rev Microbiol 2018, 16, 471–483. [Google Scholar] [CrossRef]
- 10- Drury, B.; Scott, J.; Rosi-Marshall, E.J.; Kelly, J.J. Triclosan exposure increases triclosan resistance and influences taxonomic composition of benthic bacterial communities. Environ Sci Technol 2013, 47, 8923–8930. [Google Scholar] [CrossRef]
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Lu, P.L.; Wu, Y.; Ding, H.; Zhan, W. The combined and second exposure effect of copper (II) and chlortetracycline on fresh water algae, Chlorella pyrenoidosa and Microcystis aeruginosa. Environ Toxico Pharmacol 2015, 40, 140–148. [Google Scholar] [CrossRef]
- Metcalf, J.S.; Tischbein, M.; Cox, P.A.; Stommel, E.W. Cyanotoxins and the Nervous System. Toxins (Basel) 2021, 13, 660. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, N.A.; Buniya, H.K. Some Active compounds in Local Isolate of Spirulina laxa G.M. SMITH. M. SMITH. Biochem Cell Arch 2022, 22, 1145–1149. [Google Scholar]
- Hameed, S.G.; Buniya, H.K. Determining the genetic Relationship for some local Isolate of Oscillatoria species present in the local Environment using the Inter Simple Sequence Repeat (ISSR) Technique. Adv Life Sci 2024. (accepted manuscript). [Google Scholar]
- Parvin, M.; Zannat, M.N.; Habib, M.N.B. Two Important Techniques for Isolation of Microalgae. Asian Fish Sci 2007, 20, 117–124. [Google Scholar] [CrossRef]
- Jacobsen, J.H.; Rosgaard, L. One step plasmid construction for generation of knock-out mutants in cyanobacteria: studies of glycogen metabolism in Synechococcus sp. PCC 7002. Photosynth Res 2011, 107, 215–221. [Google Scholar] [CrossRef]
- Bellinger, E.G.; Sigee, D.C. Freshwater Algae: Identification and Use as Bioindicators, 1st ed.John Wiley & Sons Ltd., 2010. [Google Scholar]
- Buniya, H.K.; Ahmaed, M.M.; Mohammed, N.A. Detection of the HEP gene responsible for the production of Hepatotoxin in Blue-green Algae using polymerase chain reaction (PCR) Technique. 2024, Manuscript in press.
- Ahmaed, M.M.; Buniya, H.K. Study of Antibiotic Resistance for some local isolates of Blue-green Algae. Biochem Cell Arch 2022, 22, 711–714. [Google Scholar]
- Bănăduc, D.; Simić, V.; Cianfaglione, K.; Barinova, S.; Afanasyev, S.; Öktener, A.; McCall, G.; Simić, S.; Curtean-Bănăduc, A. ; Öktener, A.; McCall, G.; Simić, S.; Curtean-Bănăduc, A. Freshwater as a Sustainable Resource and Generator of Secondary Resources in the 21st Century: Stressors, Threats, Risks, Management and Protection Strategies, and Conservation Approaches. Int J Environ Res Public Health 2022, 19, 16570. [Google Scholar] [CrossRef]
- Yadav, P.; Singh, R.P.; Rana, S.; Joshi, D.; Kumar, D.; Bhardwaj, N.; Gupta, R.K.; Kumar, A. Mechanisms of Stress Tolerance in Cyanobacteria under Extreme Conditions. Stresses 2022, 2, 531–549. [Google Scholar] [CrossRef]
- Kulik, K.; Lenart-Boroń, A.; Wyrzykowska, K. Impact of Antibiotic Pollution on the Bacterial Population within Surface Water with Special Focus on Mountain Rivers. Water 2023, 15, 975. [Google Scholar] [CrossRef]
- Du, Y.; Wang, J.; Zhu, F.; Mai, D.; Xiang, Z.; Chen, J.; Guo, R. Comprehensive assessment of three typical antibiotics on cyanobacteria (Microcystis aeruginosa): The impact and recovery capability. Ecotoxicol Environ Safe 2018, 160, 84–93. [Google Scholar] [CrossRef]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef]
- Urbach, C.; Fastrez, J.; Soumillion, P. A new family of cyanobacterial penicillin-binding proteins, a missing link in the evolution of class A β-lactamases. J Biol Chem 2008, 283, 32516–32526. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, R.; Madhan, K.; Singh, R.N.; Chauhan, A.K.; Nain, L. Developing biochemical and molecular markers for cyanobacterial inoculants. Folia Microbiol 2010, 55, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Xin, R.; Zhang, Y.; Zhang, K.; Yang, Y.; Ma, Y.; Niu, Z. Investigation of the antimicrobial susceptibility patterns of marine cyanobacteria in Bohai Bay: Cyanobacteria may be important hosts of antibiotic resistance genes in marine environment. Sci Total Environ 2024, 909, 168516. [Google Scholar] [CrossRef]
- Jung, A.V.; Le Cann, P.; Roig, B.; Thomas, O.; Baurès, E.; Thomas, M.F. Microbial contamination detection in water resources: interest of current optical methods, trends and needs in the context of climate change. Int J Environ Res Public Health 2014, 11, 4292–4310. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Gillings, M.R. New perspectives on mobile genetic elements: a paradigm shift for managing the antibiotic resistance crisis. Philos Trans R Soc Lond B Biol Sci 2022, 377, 20200462. [Google Scholar] [CrossRef]
- Juwita, S.; Indrawati, A.; Damajanti, R.; Safika Mayasari, N.L.P.I. Multiple antibiotic resistance and virulence factors of Staphylococcus aureus strains isolated from dairy farms in South Sulawesi, Indonesia. Biodiversitas 2022, 23, 1015–1022. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Z.; Lu, T.; et al. Cyanobacterial blooms contribute to the diversity of antibiotic-resistance genes in aquatic ecosystems. Commun Biol 2020, 3, 737. [Google Scholar] [CrossRef]
- Dias, E.; Oliveira, M.; Jones-Dias, D.; Vasconcelos, V.; Ferreira, E.; Manageiro, V.; Caniça, M. Assessing the antibiotic susceptibility of freshwater Cyanobacteria spp. Front Microbiol 2015, 6, 799. [Google Scholar] [CrossRef]
- Hu, L.; Xiao, P.; Jiang, Y.; Dong, M.; Chen, Z.; Li, H.; Hu, Z.; Lei, A.; Wang, J. Transgenerational Epigenetic Inheritance Under Environmental Stress by Genome-Wide DNA Methylation Profiling in Cyanobacterium. Front Microbiol 2018, 9, 1479. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Mao, F.J.; Te, S.H.; He, Y.L.; Gin, K.Y.H. Impacts of Microcystis on the Dissemination of the Antibiotic Resistome in Cyanobacterial Blooms. Acs. Est. Water 2021, 1, 1263–1273. [Google Scholar] [CrossRef]
- Duan, X.; Zhang, C.; Struewing, I.; Li, X.; Allen, J.; Lu, J. Cyanotoxin-encoding genes as powerful predictors of cyanotoxin production during harmful cyanobacterial blooms in an inland freshwater lake: Evaluating a novel early-warning system. Sci Total Environ 2022, 830, 154568. [Google Scholar] [CrossRef] [PubMed]
- Ohdate, K.; Sakata, M.; Maeda, K.; Sakamaki, Y.; Nimura-Matsune, K.; Ohbayashi, R.; Hess, W.R.; Watanabe, S. Discovery of novel replication proteins for large plasmids in cyanobacteria and their potential applications in genetic engineering. Front Microbiol 2024, 15, 1311290. [Google Scholar] [CrossRef]
- Kim, M.J.; Kang, D.; Lee, G.D.; Kim, K.; Kim, J.; Shin, J.H.; Lee, S. Interplays between cyanobacterial blooms and antibiotic resistance genes. Environ Int 2023, 181, 108268. [Google Scholar] [CrossRef] [PubMed]
- Volk, A.; Lee, J. Cyanobacterial blooms: A player in the freshwater environmental resistome with public health relevance? Environ Res 2023, 216, 114612. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Q.; Zhang, J.; Guan, T.; Chen, Y.; Shi, W. Critical roles of cyanobacteria as reservoir and source for antibiotic resistance genes. Environ Int 2020, 144, 106034. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).