1. Introduction
The shoulder is the most mobile joint in the body, allowing rotation across multiple axes, with some capable of full
rotation, as well as enabling arm elevation and overhead reaching. This mobility is facilitated by the rotator cuff, a complex group of muscles and tendons. With repetitive movements, the rotator cuff wears out eventually leading to rotator cuff tears (RCTs). This injury most commonly occurs with aging, but it also affects athletes and individuals in professions that involve frequent shoulder movements, such as manual labor or cleaning, making it one of the most prevalent shoulder injuries. According to [
1], approximately 2 million people in the U.S. consult their physician each year for this condition. RCTs can advance to more serious conditions over time, reinforcing the relevance of early detection. Magnetic resonance imaging (MRI) is the golden standard imaging technique; however, its use is restricted to imaging centers and it does not always provide accurate depiction of the presence and severity of the tears [
2,
3].
With the RTC incidence, synovial fluid (SF) aspirates locally in the injury area [
4,
5]. This accumulation of SF changes the dielectric properties of the shoulder joint [
6]. This change makes microwave imaging a credible alternative to MRI that we need to investigate. In [
7], we introduced an alternative low-cost, portable and non-invasive electromagnetic imaging (EMI) system for the on-site diagnosis of RCTs. At that time, no EMI system for the shoulder existed (to the best of our knowledge, this remains true), so we had to start from scratch. To save time and resources during the initial design phase, we developed a virtual model of the shoulder and the imaging system to study and optimize the EMI system, as described in [
7]. This model is the first step toward Microwave Digital Twin Prototype (MDTP).
The concept of the Digital Twin (DT) was originally proposed by Michael Grieves at the University of Michigan for monitoring Product Lifecycle Management. It involves creating a virtual model of a physical system, which is continuously updated with real-time data from the existing physical system. DTs are not intended for system design. However, in [
8] the same authors introduce the Digital Twin Prototype (DTP), which exists in virtual space and is to be used in what the authors refer to as the creation phase. Since 2002, DTs have been widely used and developed for Industry 4.0 applications [
9,
10,
11]. In the healthcare sector, a comprehensive review of Digital Twin for Health (DT4H) can be found in [
11]. A wide range of applications are already investigated including detecting and monitoring cardiac pathologies, diabetes, breast or oropharyngeal cancers and Alzheimer’s diseases. DT4H often incorporates Machine Learning (ML) in order to enhance the performance of the illness detection as exemplified in [
12] with the COVID-19. Very recently, DTs have been efficiently used for microwave ablation [
13] and imaging purposes [
14]. In this paper, we introduce the concept of Microwave Digital Twin Prototype as a virtual system that mimics the physical one and is capable of predicting the presence of the RCTs. The model not only includes the anthropomorphological model of the shoulder wether it is injured or not, but also the imaging system and the uncertainties due to its use, like noise, positioning errors and errors due to the RCTs itself like the synovial fluid’s variation that depends on the RCT’s severity.
Compared with our previous work [
7], we aim to improve and systematize the detection of RCTs. So far, we have been solving an inverse problem for detecting the presence of RCTs. This process is time-consuming, requires extensive computing resources and is therefore not compatible with a large number of case studies. As an example, the final design consists of 32 ceramic (
) loaded open-ended waveguides. It requires 11 minutes and 27 seconds for image reconstruction of one shoulder model with the use of 480 computing cores. These amount of resources may not always be available and can limit the practical use of the device in real world. In this paper, we are going to address this issue by use of ML algorithms.
The rise of ML has led to the development of valuable tools in various medical applications, such as predicting sports injuries [
15], simplifying medical imaging processes [
16] and advancing stroke medicine [
17]. Further, combining microwave imaging systems with ML algorithms has significantly improved stroke detection, stroke types classifications and localizing affected areas [
18,
19,
20].
The dataset gathering is a crucial component of machine learning algorithms, particularly in medical applications, but it presents numerous challenges and limitations [
21]. For example, insufficient or biased data can result in poor generalization that highly affect the algorithm’s accuracy in making predictions or diagnoses. To enhance generalization, large and diverse training datasets are necessary. Moreover, the effectiveness of ML algorithms heavily relies on the quality and quantity of the data. However, in the real world, obtaining data from patients involves privacy and authorization challenges and is a time-consuming process. Further, the limited available training data significantly impacts the performance of the classifiers. To address this issue, generating synthetic data through numerical simulations or various computer algorithms has emerged as a promising solution in recent years [
22,
23]. In [
24], numerical simulations of a system are performed to investigate how integrating mathematical models with experimental datasets can enhance classification performance. It is important to note that while use of synthetic data can enhance continual and causal learning, it also carries the risk of introducing biases [
25]. It emphasizes the importance of generating a reliable dataset.
In this work, we use numerical modeling to generate a generalizable dataset of scattering parameters. A parametric study is conducted, considering four main categories, outlined in
Table 1. For the classification of injured and healthy models, we utilize a supervised machine learning support vector machine (SVM).
Note that the key indicator in differentiating the healthy and injured shoulder joint is the presence of RCTs because it is the most challenging case. The mean aspirate volume of SF is reported to correlates with the size of the tear. This volume for the small tears is
, for medium tears is
, and for large tears is
[
5]. In this study, we will consider the presence of a small tear in the injured shoulder model. The paper is structured as follows: Section presents the numerical modeling framework, including the numerical modeling of the system, its properties, and our methods for data generation and classification analysis using SVM.
Section 3 discusses the numerical results for various scenarios, and the conclusion is provided in
Section 4.
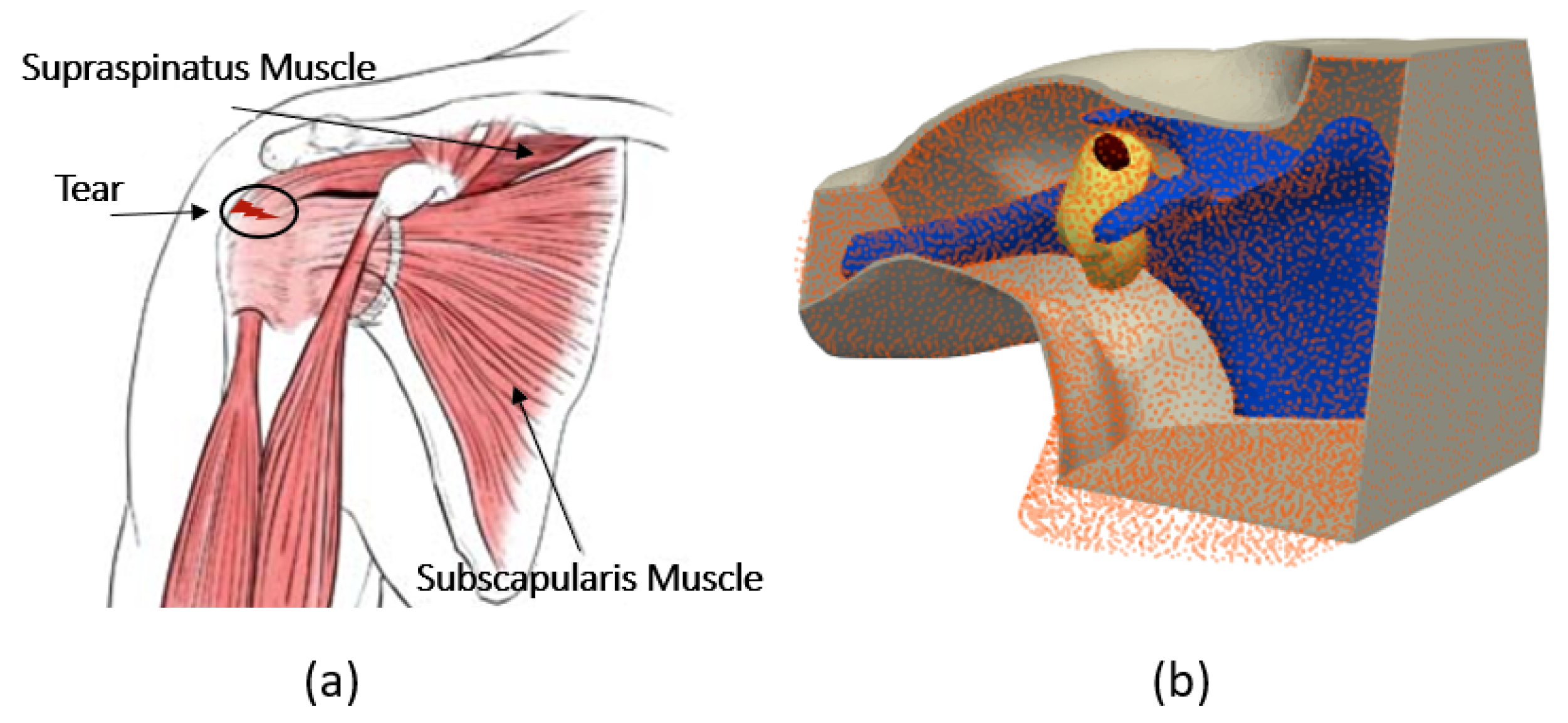
Figure 1.
(a) Anatomy of the shoulder, (b) numerical model of the shoulder.
Figure 1.
(a) Anatomy of the shoulder, (b) numerical model of the shoulder.
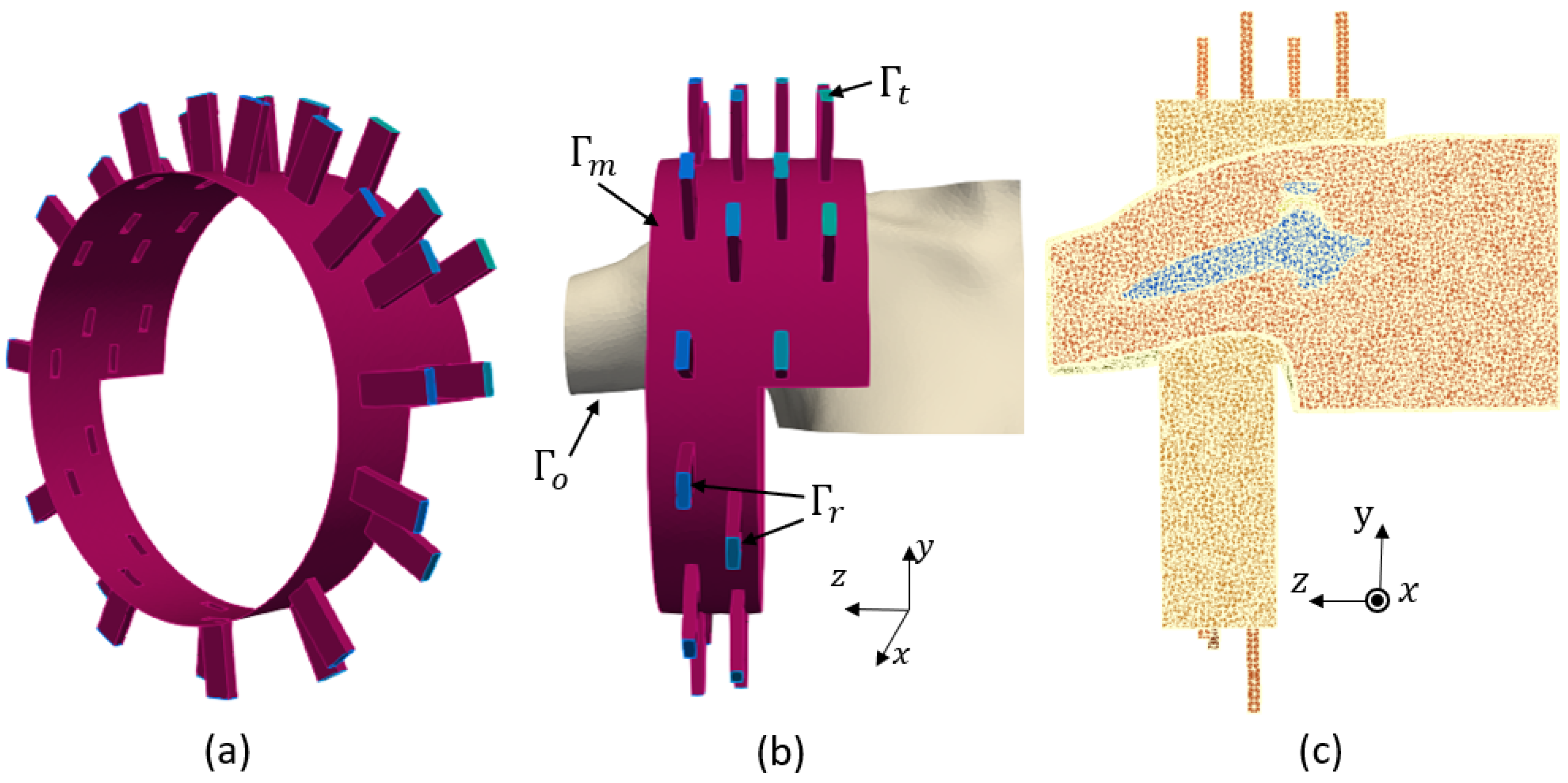
Figure 2.
(a): imaging system, (b) boundary conditions, (c) finite element mesh.
Figure 2.
(a): imaging system, (b) boundary conditions, (c) finite element mesh.
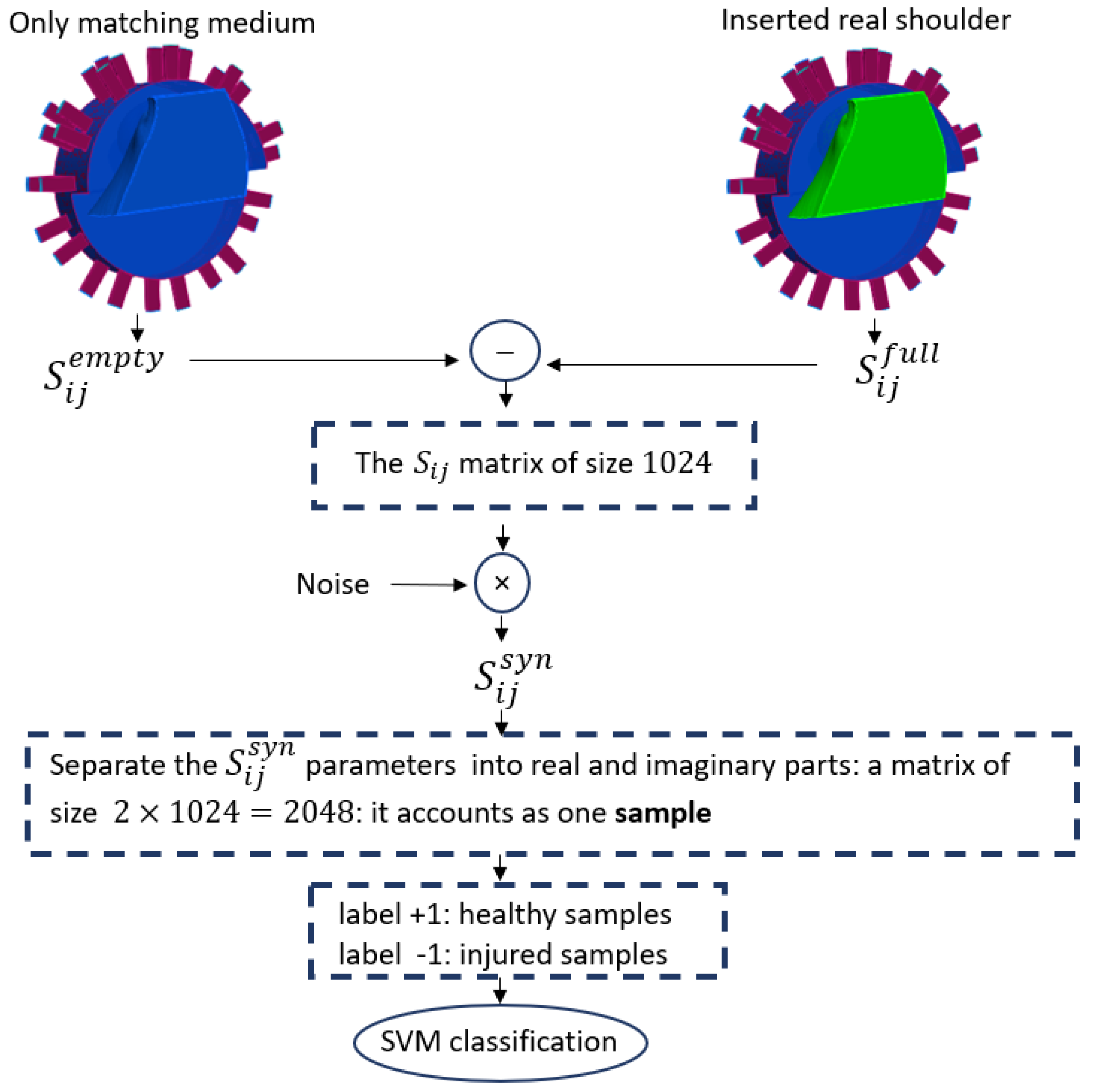
Figure 3.
The workflow of SVM classification.
Figure 3.
The workflow of SVM classification.
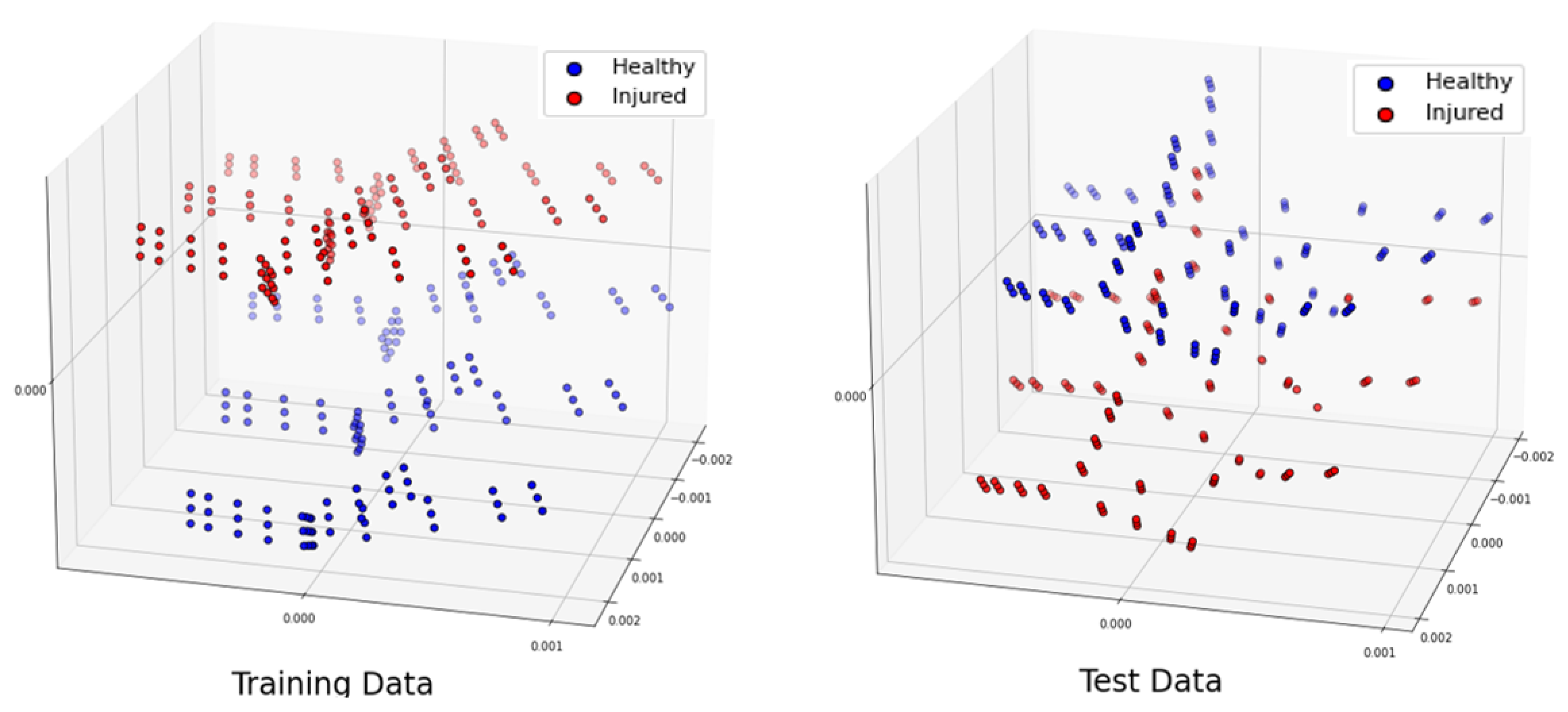
Figure 4.
Projection of the 3 most significant eigenvectors of the training dataset and test dataset when phantoms have a shift of along .
Figure 4.
Projection of the 3 most significant eigenvectors of the training dataset and test dataset when phantoms have a shift of along .
Figure 5.
The translation error of of cm along different axis between training and test dataset phantoms.
Figure 5.
The translation error of of cm along different axis between training and test dataset phantoms.
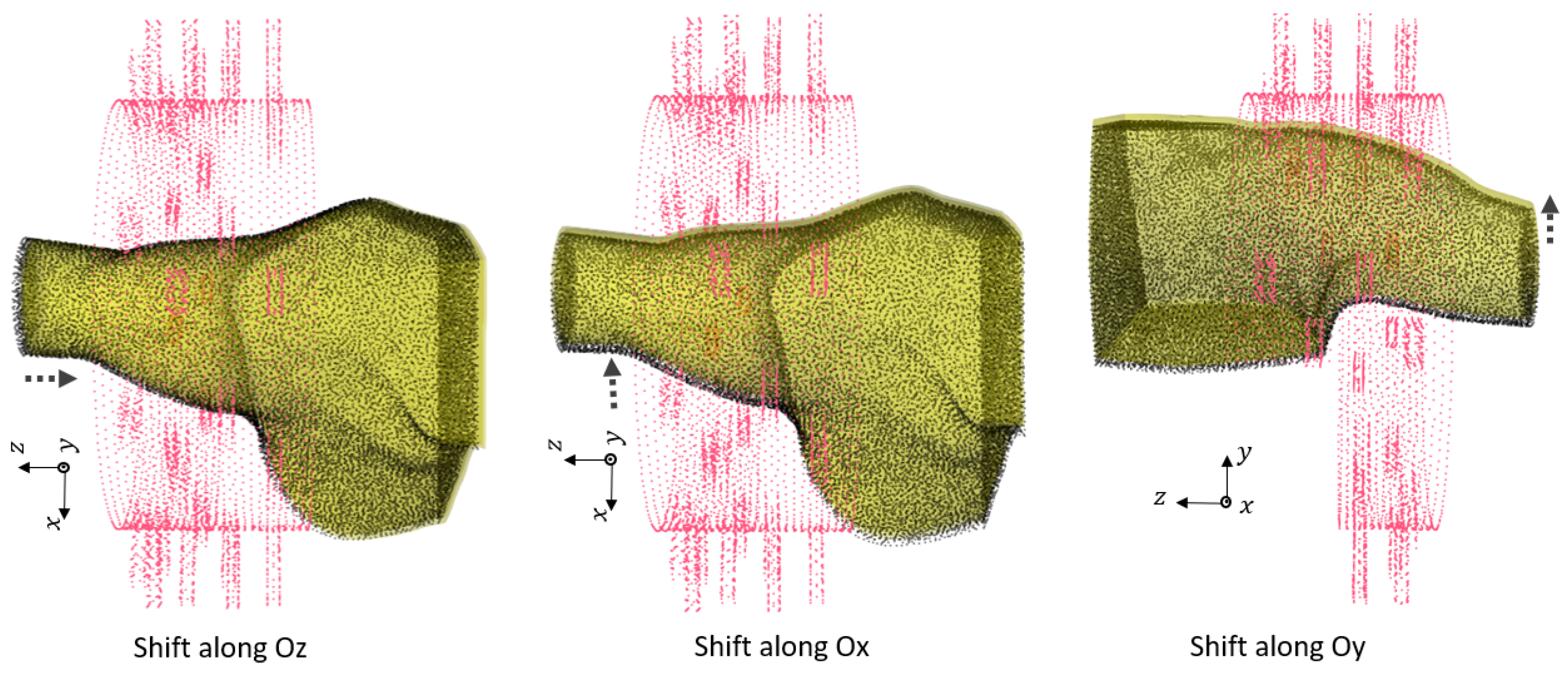
Figure 6.
First group of rotations. For : , for : and shift , for : .
Figure 6.
First group of rotations. For : , for : and shift , for : .
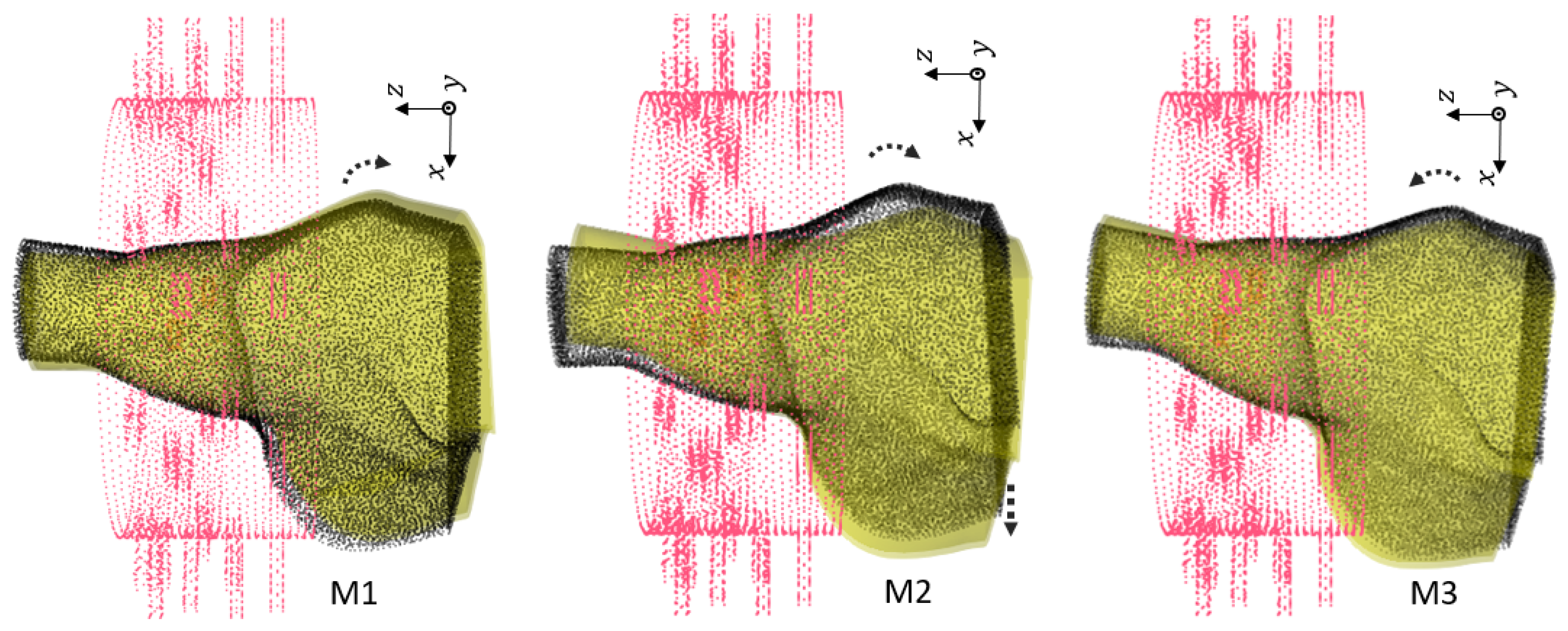
Figure 7.
Second group of rotations. For : . For : and shift . For : and shift .
Figure 7.
Second group of rotations. For : . For : and shift . For : and shift .
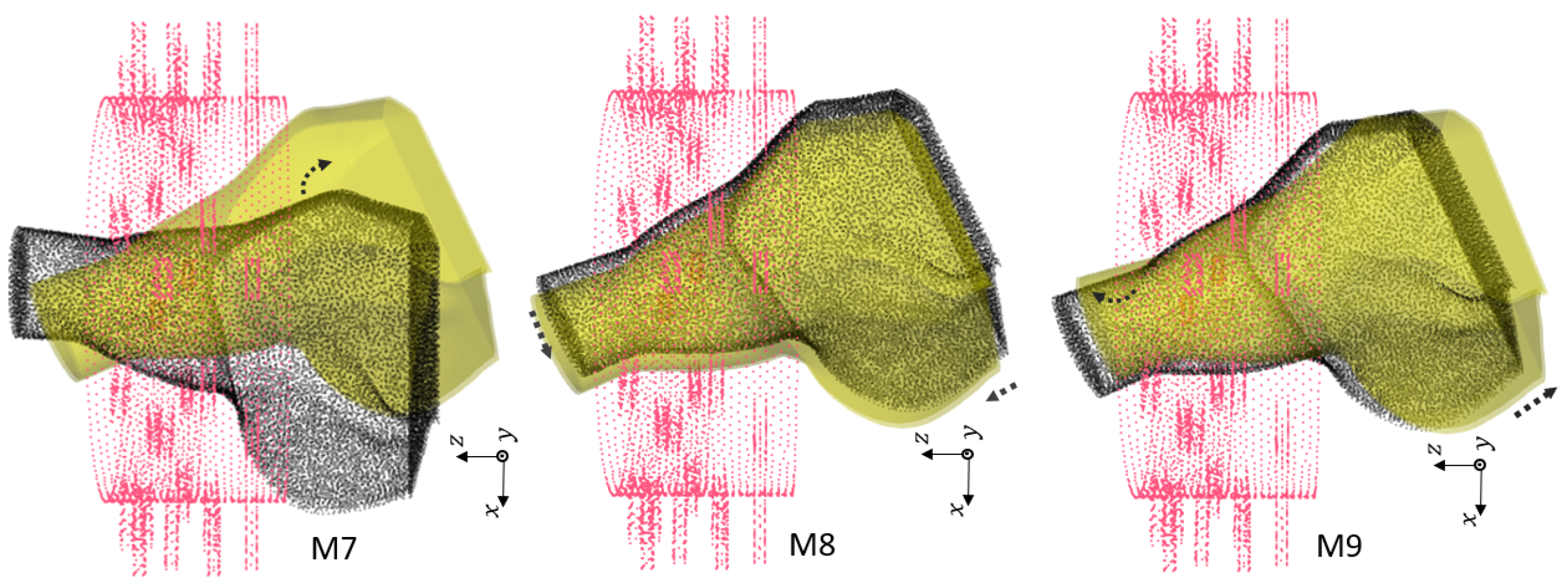
Figure 8.
Third group of rotations. For : . For : , and . For : , and .
Figure 8.
Third group of rotations. For : . For : , and . For : , and .
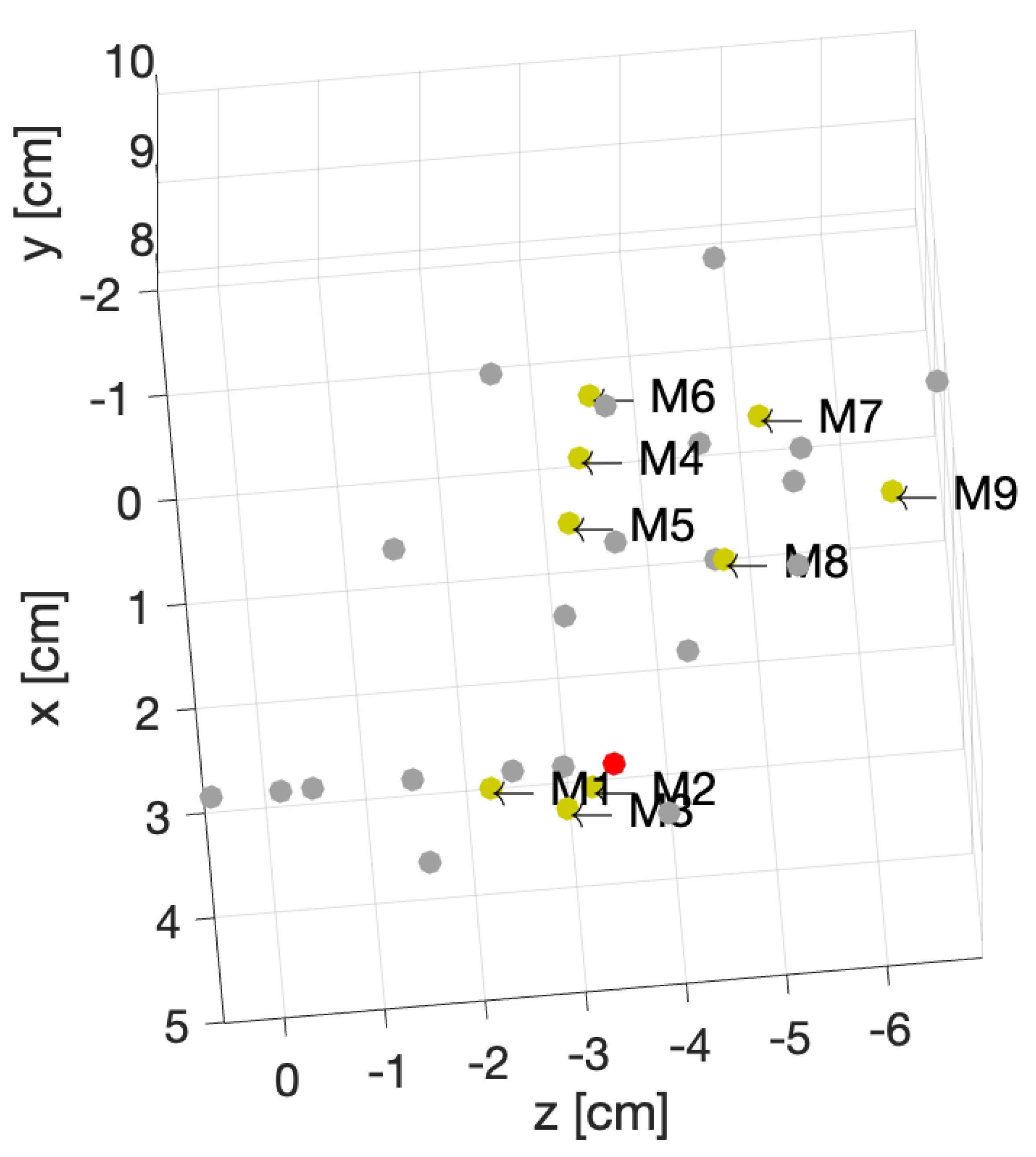
Figure 9.
Position of the center of the rotations for 31 different phantoms.
Figure 9.
Position of the center of the rotations for 31 different phantoms.
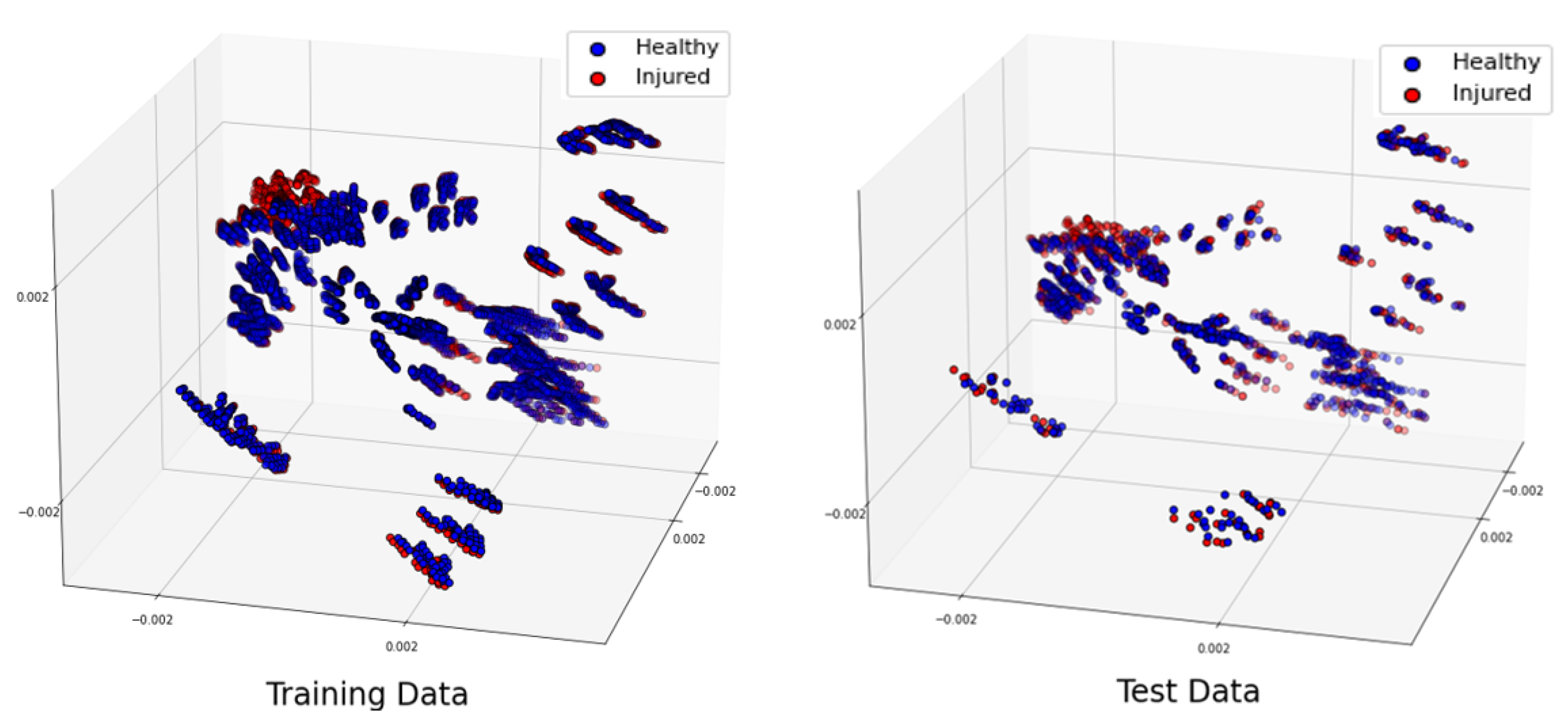
Figure 10.
Projection of the 3 most significant eigenvectors of the large dataset. Training dataset (left), test dataset (right).
Figure 10.
Projection of the 3 most significant eigenvectors of the large dataset. Training dataset (left), test dataset (right).
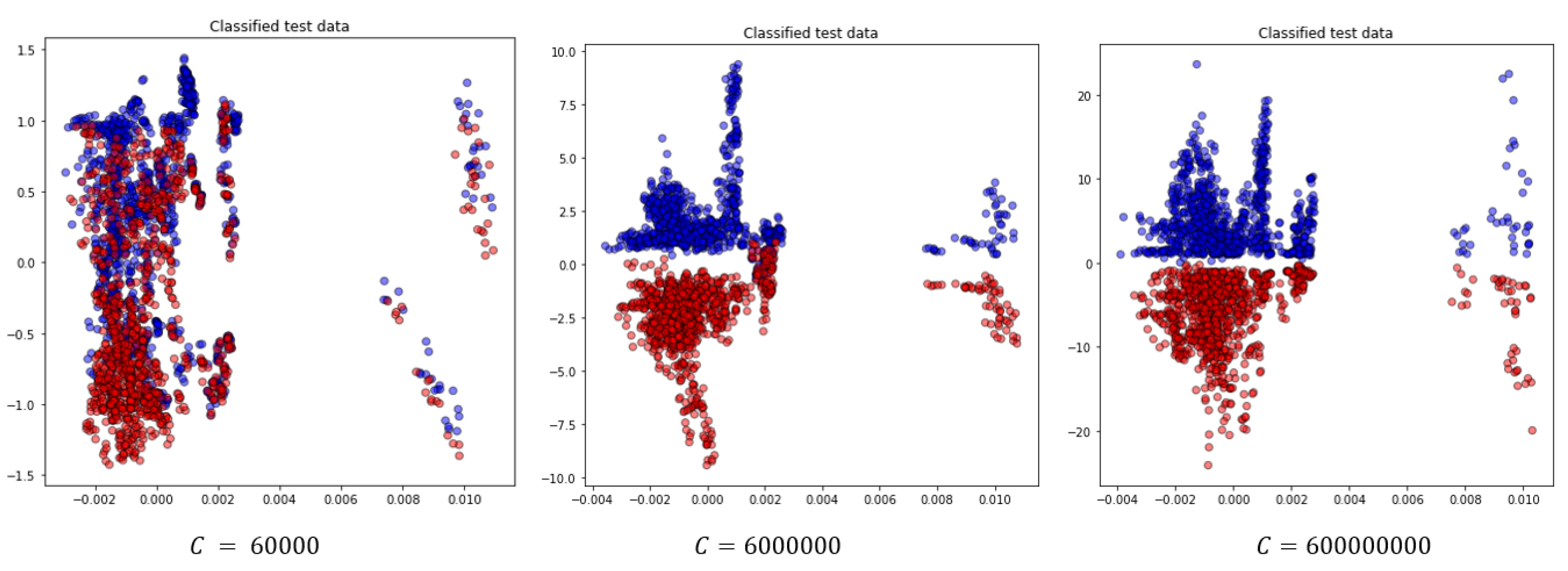
Figure 11.
Projection of the two most significant eigenvectors of classified test data for the random test case for different choice of C parameter.
Figure 11.
Projection of the two most significant eigenvectors of classified test data for the random test case for different choice of C parameter.
Table 1.
Parametric study for detection of RCTs.
Table 1.
Parametric study for detection of RCTs.
| Scenario |
Description |
| Noise level |
Introducing different noise levels in synthetic data |
| Error in value of
|
Dielectric properties variations due to dehydration |
| Localization |
Changes in the location of the shoulder |
| Randomized dataset |
Shuffled training and test dataset |
Table 2.
Complex dielectric properties at 1 GHz.
Table 2.
Complex dielectric properties at 1 GHz.
| Different Tissues |
Value of
|
| Bone cortical |
|
| Tendon |
|
| Muscle |
|
| Skin |
|
| SF |
|
Table 3.
Confusion matrix interpretation
Table 3.
Confusion matrix interpretation
| |
Predicted class |
| |
Healthy |
Injured |
| Actual class |
Healthy |
|
|
| |
Injured |
|
|
Table 4.
The noise levels introduced in each sets of data. The values are in .
Table 4.
The noise levels introduced in each sets of data. The values are in .
| Sample model |
Training dataset |
Test dataset |
| Healthy |
|
|
| Injured |
|
|
Table 5.
Number of generated training and test samples for healthy and injured model. We build the datasets with 36 number of seeds for each noise level.
Table 5.
Number of generated training and test samples for healthy and injured model. We build the datasets with 36 number of seeds for each noise level.
| Sample model |
Training dataset |
Test dataset |
| Healthy |
|
|
| Injured |
|
|
Table 6.
Classification results for different values of C in noise study.
Table 6.
Classification results for different values of C in noise study.
| C |
Acr |
Spec |
Sens |
| 6000 |
|
|
|
| 600000 |
|
|
|
| 6000000 |
|
|
|
Table 7.
The value of at with including the dehydration effect, compared to the original values ( )
Table 7.
The value of at with including the dehydration effect, compared to the original values ( )
| Tissue |
|
Original values based on Table 2
|
|
| Bone cortical |
|
|
|
| Skin |
|
|
|
| Tendon |
|
|
|
| Muscle |
|
|
|
| SF |
|
|
|
Table 8.
Number of generated training and test samples for healthy and injured models. We build the datasets with 36 seeds for each noise level.
Table 8.
Number of generated training and test samples for healthy and injured models. We build the datasets with 36 seeds for each noise level.
| Sample Model |
Total Dataset |
Training Subset |
Test Subset |
| Healthy |
|
900 |
108 |
| Injured, 3 groups of SF value |
|
|
|
Table 9.
Classification accuracy for different values dehydration error, for .
Table 9.
Classification accuracy for different values dehydration error, for .
| Value of SF |
Accuracy |
|
|
|
|
|
|
Table 10.
Number of generated training and test samples for healthy and injured model, for each position of the phantom.
Table 10.
Number of generated training and test samples for healthy and injured model, for each position of the phantom.
| Sample model |
Training dataset |
Test dataset |
| Healthy |
|
|
| Injured |
|
|
Table 11.
Number of generated training and test samples for healthy and injured model, for 31 positioning of the shoulder inside sensing system, due to rotation and shift.
Table 11.
Number of generated training and test samples for healthy and injured model, for 31 positioning of the shoulder inside sensing system, due to rotation and shift.
| Sample model |
Total dataset |
Training subset |
Test subset |
| Healthy |
|
4320 |
144 |
| Injured |
|
4320 |
144 |
Table 12.
Different scenarios for various position of phantom in the imaging system.
Table 12.
Different scenarios for various position of phantom in the imaging system.
| |
Healthy |
Injured |
Accuracy |
Confusion matrix |
| 1 |
M2 |
M2 |
|
|
| 2 |
M2 |
M4 |
|
|
| 4 |
M7 |
M3 |
|
|
| 5 |
M9 |
M6 |
|
|
Table 13.
Confusion matrix for randomized large dataset, with .
Table 13.
Confusion matrix for randomized large dataset, with .
| Models |
Healthy |
Injured |
| Healthy |
1112 |
2 |
| Injured |
0 |
1114 |
Table 14.
The effect of C value in classification for the shuffled large dataset.
Table 14.
The effect of C value in classification for the shuffled large dataset.
| C |
Acr |
Spec |
Sens |
Time (s) |
| 600 |
|
|
|
|
| 6000 |
|
|
|
|
| 60000 |
|
|
|
|
| 600000 |
|
|
|
|
| 6000000 |
|
|
|
|
| 60000000 |
|
|
|
|
| 600000000 |
|
|
|
|