1. Introduction
Dengue is a viral disease primarily found in tropical and subtropical areas. Recent estimates suggest that about half of the world’s population resides in areas where infection is possible. Although it has been named for about two centuries, dengue has seen a surge in spread in recent decades, with epidemic outbreaks notably occurring in Central America, South America, and certain areas of Africa and Southeast Asia. Recent outbreaks have also been identified in the United States and Europe [
1,
2]. Dengue is caused by a family of viruses (Den-1, Den-2, Den-3, and Den-4) primarily transmitted through the bite of an infected mosquito (Aedes aegypti or, less commonly, Aedes albopictus). Following the bite, the virus enters the bloodstream, where it can be detected for approximately 2-7 days. If an infected person is bitten by a second mosquito, it can spread the virus to other individuals by subsequently biting them [
3,
4]. To prevent dengue, one should avoid mosquito bites, especially when in areas where the infection is prevalent. Dengue is diagnosed through the observation of symptoms and tests to detect the virus and/or specific antibodies in the blood [
5,
6].
The aim of this short communication is to study this type of infection using in silico technology of Molecular docking using natural molecules against Dengue virus type 1 envelope protein with the objective of finding a molecule theoretically capable of blocking this type of infection [
7,
8]. Nowadays, as is well known, this methodology is very useful and fast, capable of screening many molecules against a specific target of investigation, thus providing a preliminary selection of potential molecules before conducting in vitro and in vivo tests in order to shorten the time and costs in the laboratory [
9,
10].
2. Material and Methods
- Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation was taken from Protein Data Bank (PDB Code:3G77). Docking investigation was performed by Blind Docking methid by Autodock Vina with Pyrx program [
11], using: Grid box Coordinates of binding Center X ( 22.9502), Y(23.9257), Z(28.1043); size_x = 39.2081984878; size_y = 56.9977607989; size_z = 115.869397469.
3. Results and Discussion
Dengue virus is a flavivirus, belonging to the family Flaviviridae, and is primarily transmitted to humans through the bite of infected Aedes mosquitoes, particularly Aedes aegypti and Aedes albopictus. Upon infection, the virus typically manifests in two forms: dengue fever and severe dengue. Dengue fever is characterized by symptoms such as sudden high fever, severe headache, pain behind the eyes, joint and muscle pain, fatigue, nausea, vomiting, and skin rash. While dengue fever is often self-limiting, severe dengue, also known as dengue hemorrhagic fever (DHF) or dengue shock syndrome (DSS), can lead to potentially fatal complications, including severe bleeding, organ damage, and shock [
1,
2,
3,
4,
5].
Dengue is endemic in many tropical and subtropical regions of the world, particularly in urban and semi-urban areas where Aedes mosquitoes thrive. Climate change, urbanization, and globalization have contributed to the spread of the virus to new areas, leading to increased incidence and outbreaks in recent years [
1,
2,
3,
4,
5].
There is currently no specific antiviral treatment for dengue, and management typically involves supportive care to alleviate symptoms and prevent complications [
1,
2,
3,
4,
5].
Efforts to develop a dengue vaccine have been ongoing, with some vaccines showing promising results in clinical trials. However, challenges remain in developing a vaccine that provides effective protection against all four dengue serotypes and is safe for widespread use [
1,
2,
3,
4,
5,
6].
The purpose of this brief communication is to investigate this infection type utilizing in silico technology of Molecular docking with natural molecules targeting Dengue virus type 1 envelope protein, aiming to identify a molecule theoretically capable of inhibiting this infection [
7,
8]. Presently, this methodology is widely recognized for its efficiency and speed, enabling the screening of numerous molecules against a specific investigative target. This process offers a preliminary selection of potential molecules prior to conducting in vitro and in vivo tests, thereby reducing time and costs associated with laboratory research. After conducting several molecular docking experiments, it was observed that Amentoflavone potentially exhibited excellent binding capability with Dengue virus, obtaining a binding energy of -10 kcal/mol.
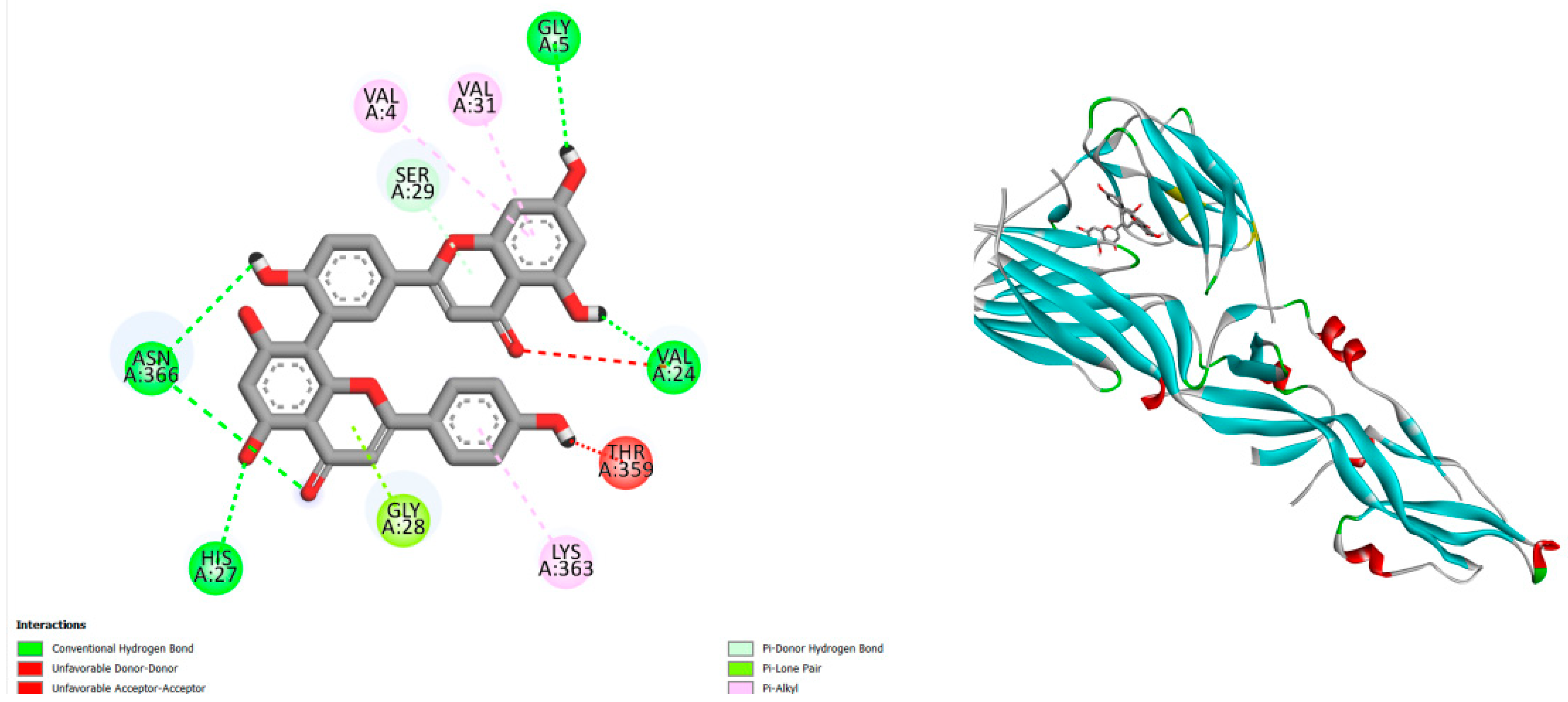
Figure 1.
displays the blind docking outcomes of Structure of Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation in conjunction with docked Amentoflavone -10 kcal mol, within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Amentoflavone. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
Figure 1.
displays the blind docking outcomes of Structure of Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation in conjunction with docked Amentoflavone -10 kcal mol, within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Amentoflavone. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
4. Conclusion
In conclusion, this molecular docking study has identified Amentoflavone as a potential inhibitor of Dengue virus envelope protein, with a notable binding energy of -10 kcal/mol. This finding suggests that Amentoflavone may possess strong affinity and interaction with the target protein, making it a promising candidate for further investigation as a therapeutic agent against Dengue virus infection. The results underscore the importance of computational approaches, such as molecular docking, in drug discovery efforts, particularly for rapidly evolving infectious diseases like Dengue. Future studies should focus on validating the antiviral activity of Amentoflavone through in vitro and in vivo experiments, as well as exploring its safety and pharmacokinetic profile. Overall, Amentoflavone holds potential as a lead compound in the development of novel Dengue virus inhibitors and contributes to the ongoing efforts to combat this global health burden.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Asish, P.R.; Dasgupta, S.; Rachel, G.; Bagepally, B.S.; Kumar, C.P.G. Global prevalence of asymptomatic dengue infections - a systematic review and meta-analysis. Int. J. Infect. Dis. 2023, 134, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Kok, B. H., Lim, H. T., Lim, C. P., Lai, N. S., Leow, C. Y., & Leow, C. H. (2023). Dengue virus infection–a review of pathogenesis, vaccines, diagnosis and therapy. Virus research, 324, 199018.
- Dengue is caused by a family of viruses (Den-1, Den-2, Den-3, and Den-4) primarily transmitted through the bite of an infected mosquito (Aedes aegypti or, less commonly, Aedes albopictus). Following the bite, the virus enters the bloodstream, where it can be detected for approximately 2-7 days. If an infected person is bitten by a second mosquito, it can spread the virus to other individuals by subsequently biting them.
- Adhani, N.R.; Arneliwati, A.; Tampubolon, M.M. The Relationship between Perceptions and Family Behavior in Preventing Dengue Hemorrhagic Fever. Qistina: J. Multidisiplin Indones. 2023, 2, 1398–1406. [Google Scholar] [CrossRef]
- Ho, S.H.; Lim, J.T.; Ong, J.; Hapuarachchi, H.C.; Sim, S.; Ng, L.C. Singapore’s 5 decades of dengue prevention and control—Implications for global dengue control. PLOS Neglected Trop. Dis. 2023, 17, e0011400. [Google Scholar] [CrossRef] [PubMed]
- Waickman, A.T.; Newell, K.; Endy, T.P.; Thomas, S.J. Biologics for dengue prevention: up-to-date. Expert Opin. Biol. Ther. 2022, 23, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Muthukumaran, R.; Sankararamakrishnan, R. Differences in the Membrane-Binding Properties of Flaviviral Nonstructural 1 (NS1) Protein: Comparative Simulations of Zika and Dengue Virus NS1 Proteins in Explicit Bilayers. ACS Bio Med Chem Au 2024, 4, 137–153. [Google Scholar] [CrossRef] [PubMed]
- Guntamadugu, R.; Ramakrishnan, R.; Darala, G.; Kothandan, S. Molecular docking, simulations of animal peptides against the envelope protein of Dengue virus. J. Biomol. Struct. Dyn. 2023, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ling, M.; Lin, Q.; Tang, S.; Wu, J.; Hu, H. Effectiveness Analysis of Multiple Initial States Simulated Annealing Algorithm, a Case Study on the Molecular Docking Tool AutoDock Vina. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 3830–3841. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Tang, S.; Mei, Z.; Wang, L.; Huang, Q.; Hu, H.; Ling, M.; Wu, J. Vina-GPU 2.0: Further Accelerating AutoDock Vina and Its Derivatives with Graphics Processing Units. J. Chem. Inf. Model. 2023, 63, 1982–1998. [Google Scholar] [CrossRef] [PubMed]
- Houshmand, F., & Houshmand, S. (2023). Potentially highly effective drugs for COVID-19: Virtual screening and molecular docking study through PyRx-Vina Approach. Frontiers in Health Informatics, 12, 150.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).