Submitted:
30 August 2024
Posted:
02 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
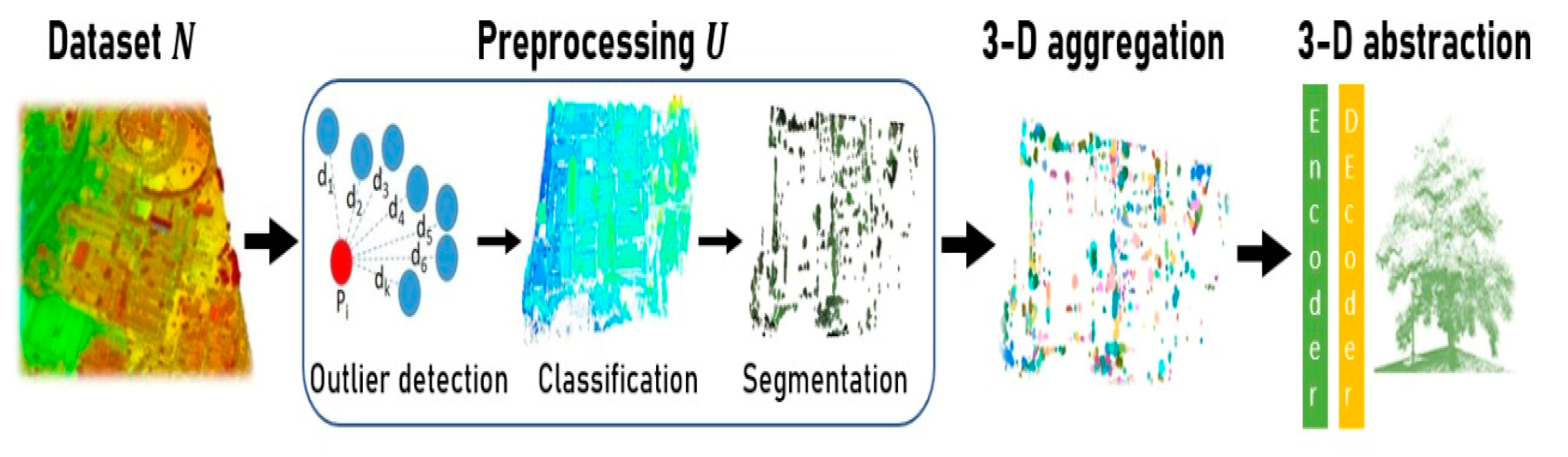
2. Methodology
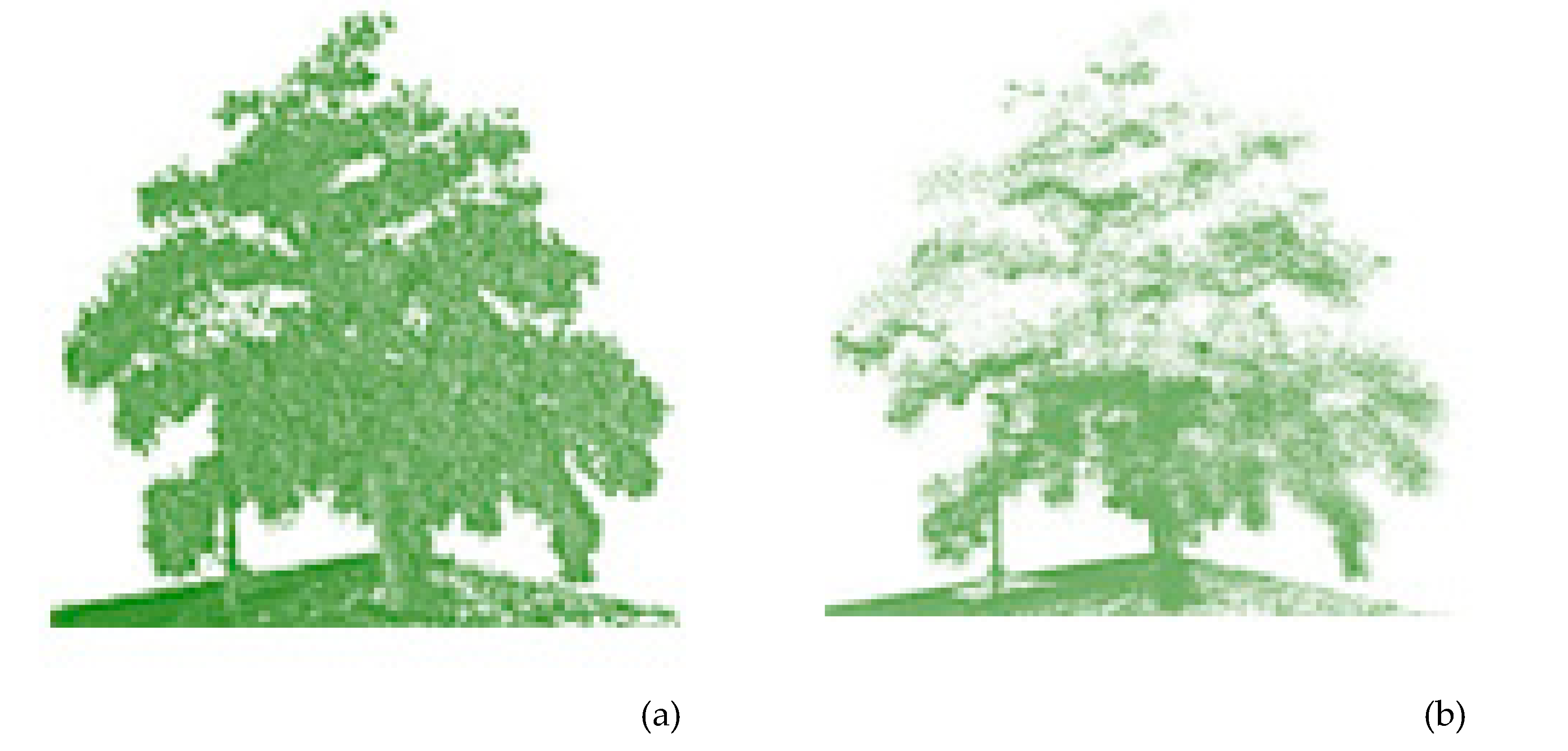
2.1. Classification of the Urban Vegetation
2.2. Segmentation of Individual Trees
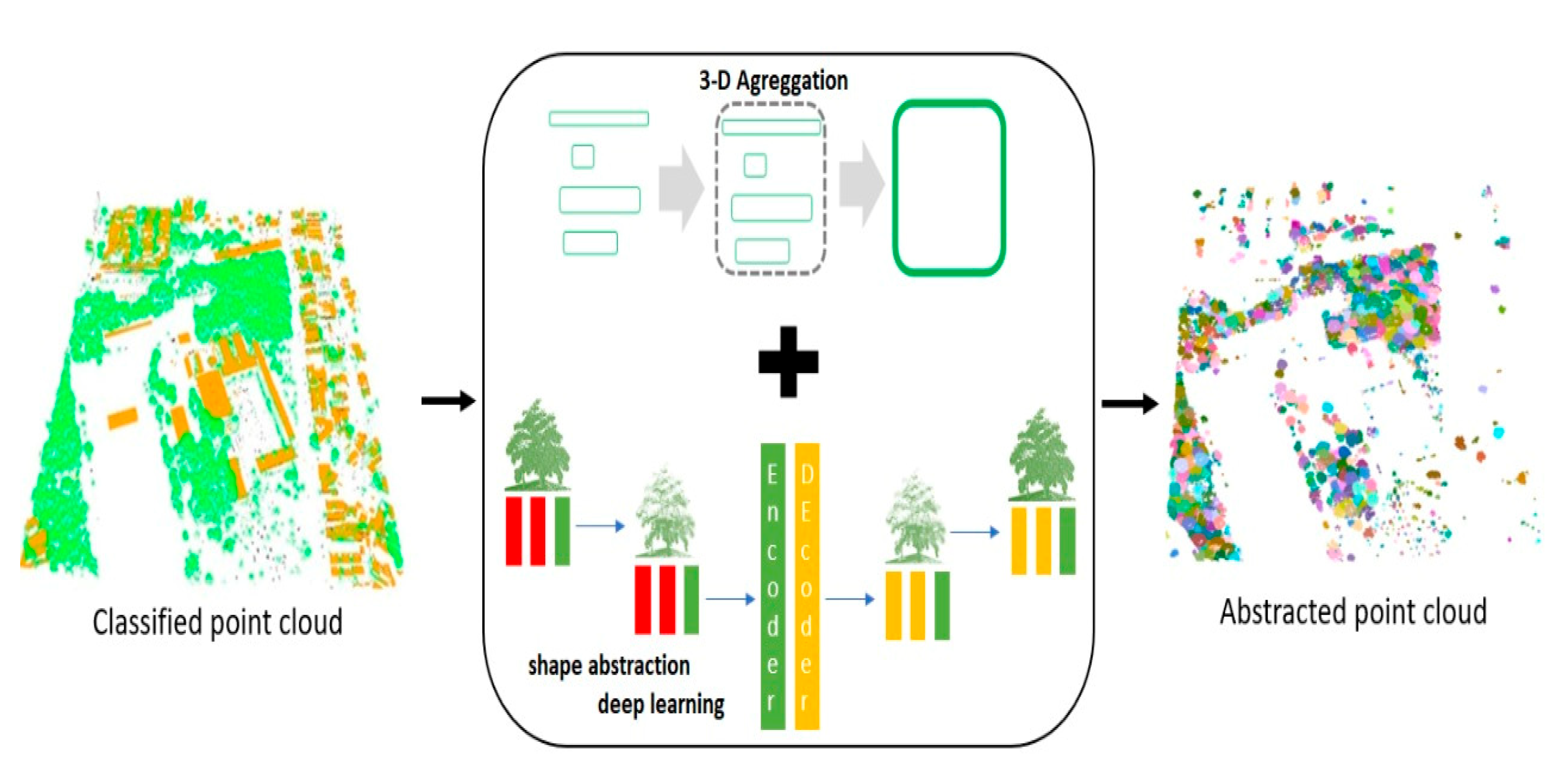
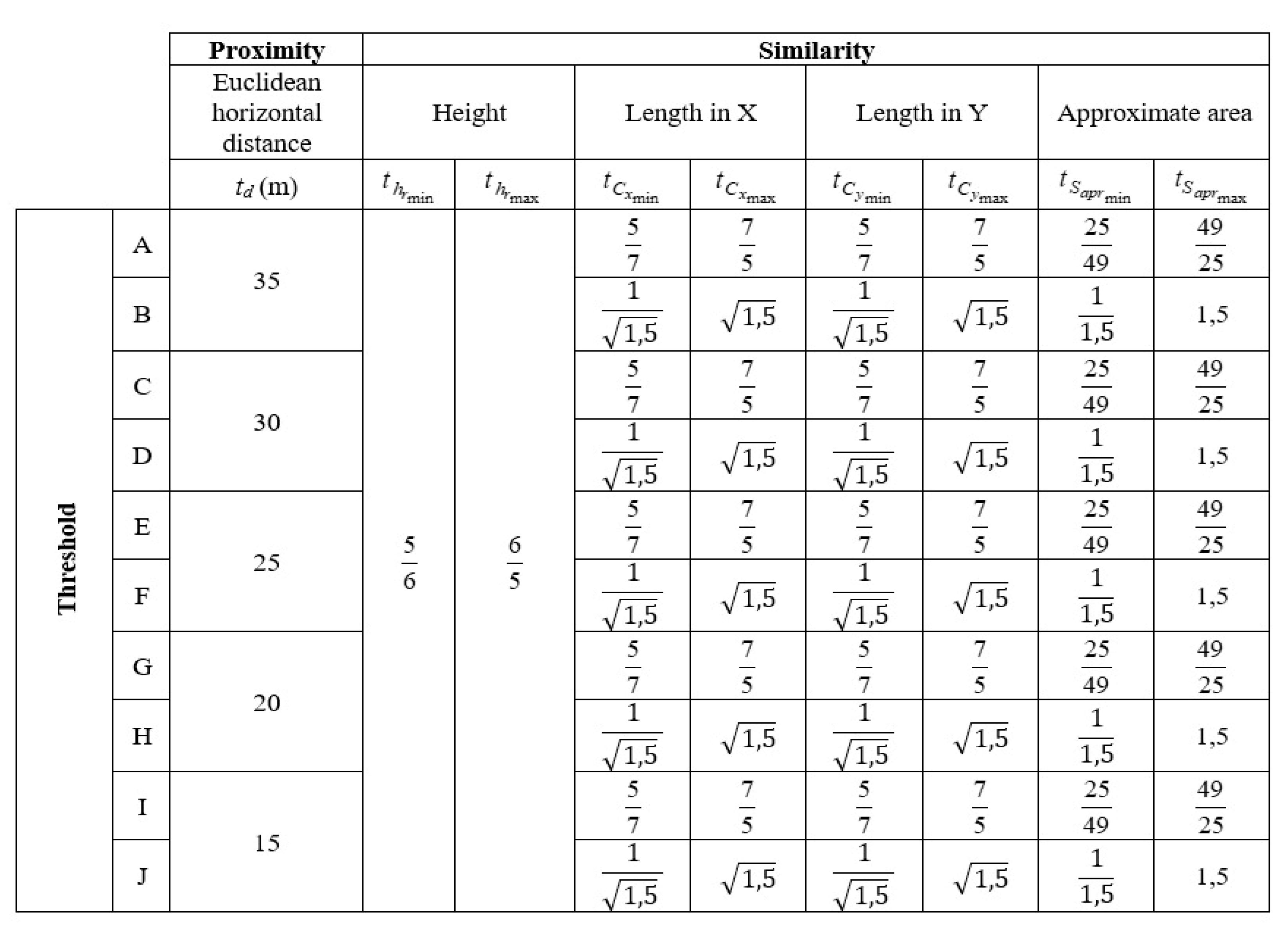
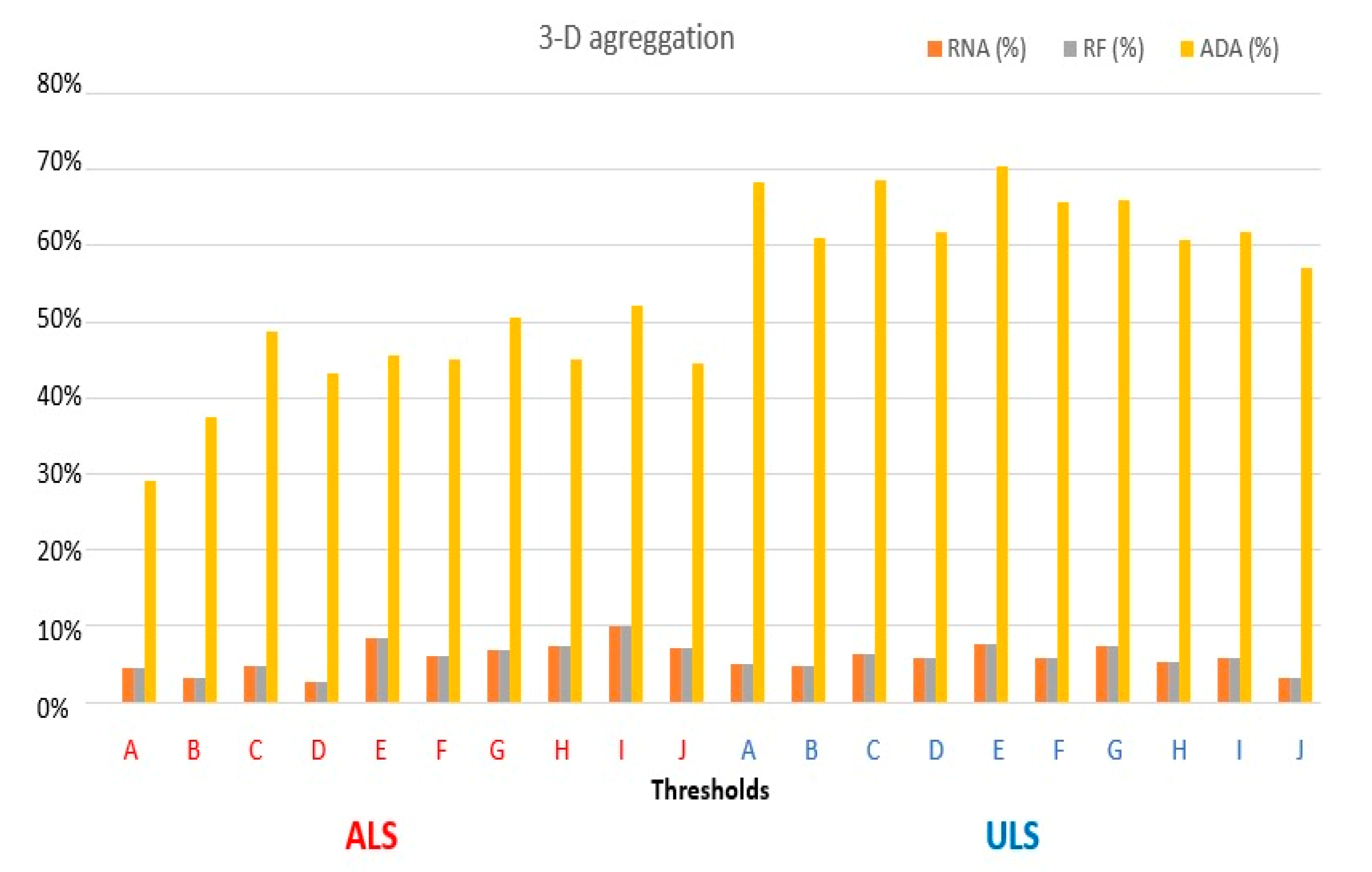
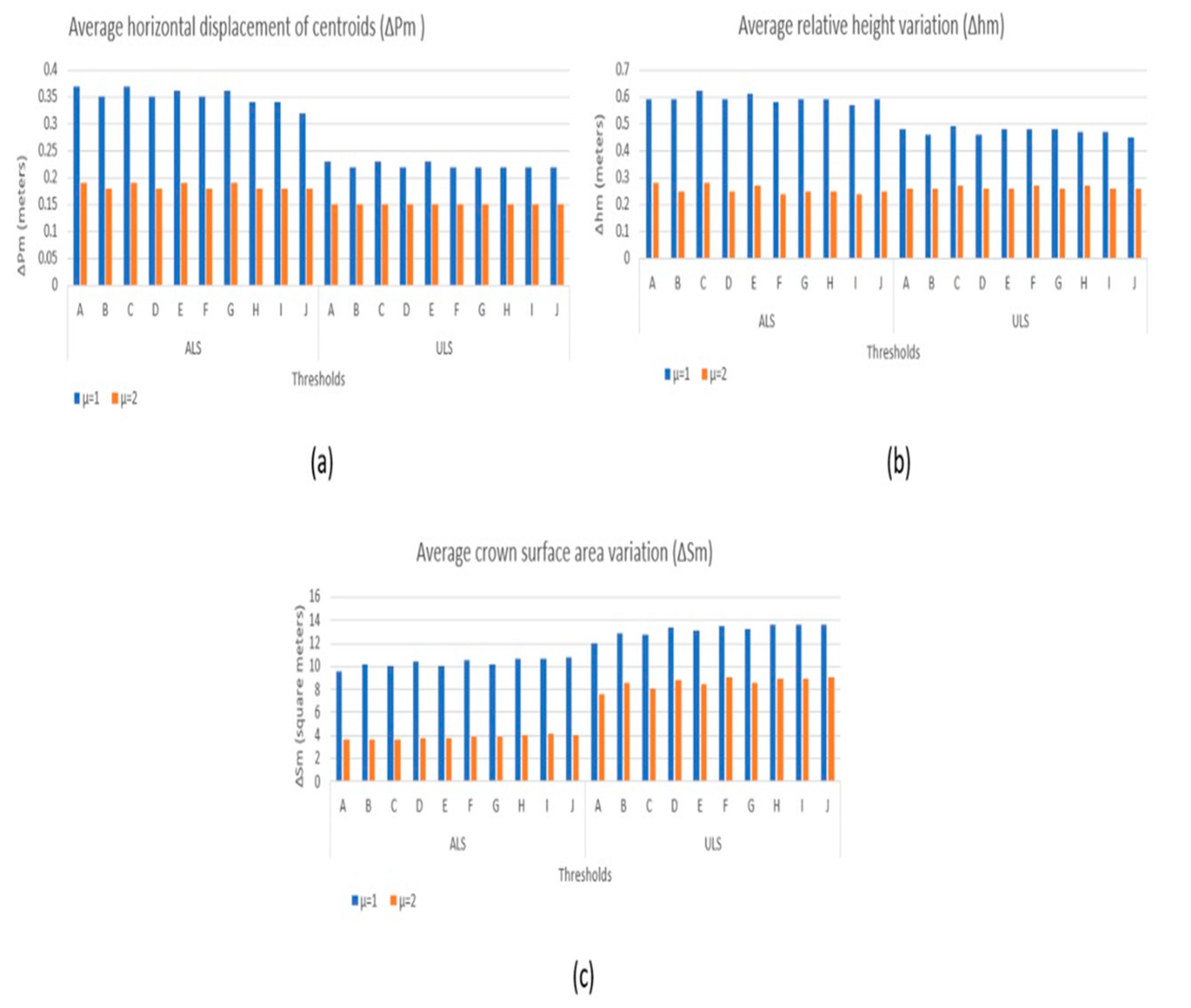
2.3. Proposed 3D Aggregation with Abstraction Purposes
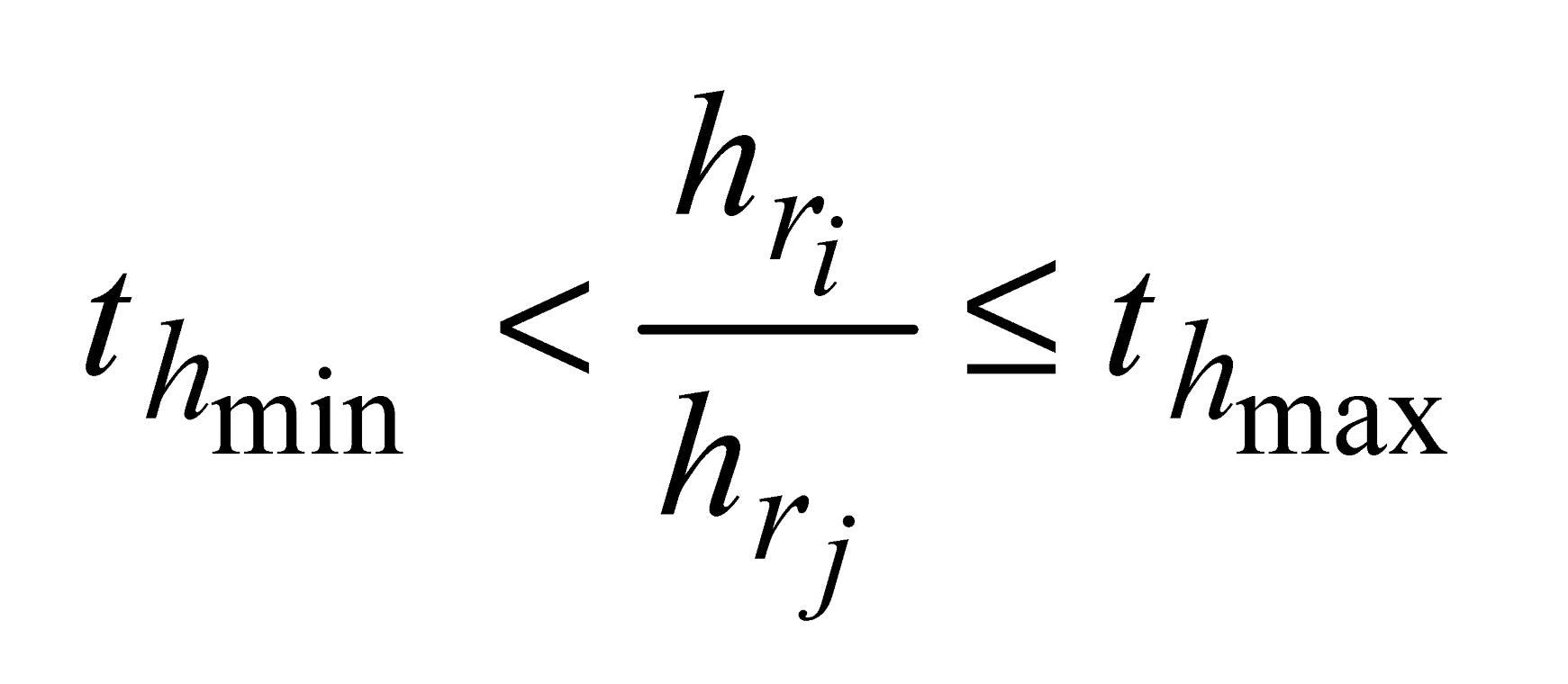
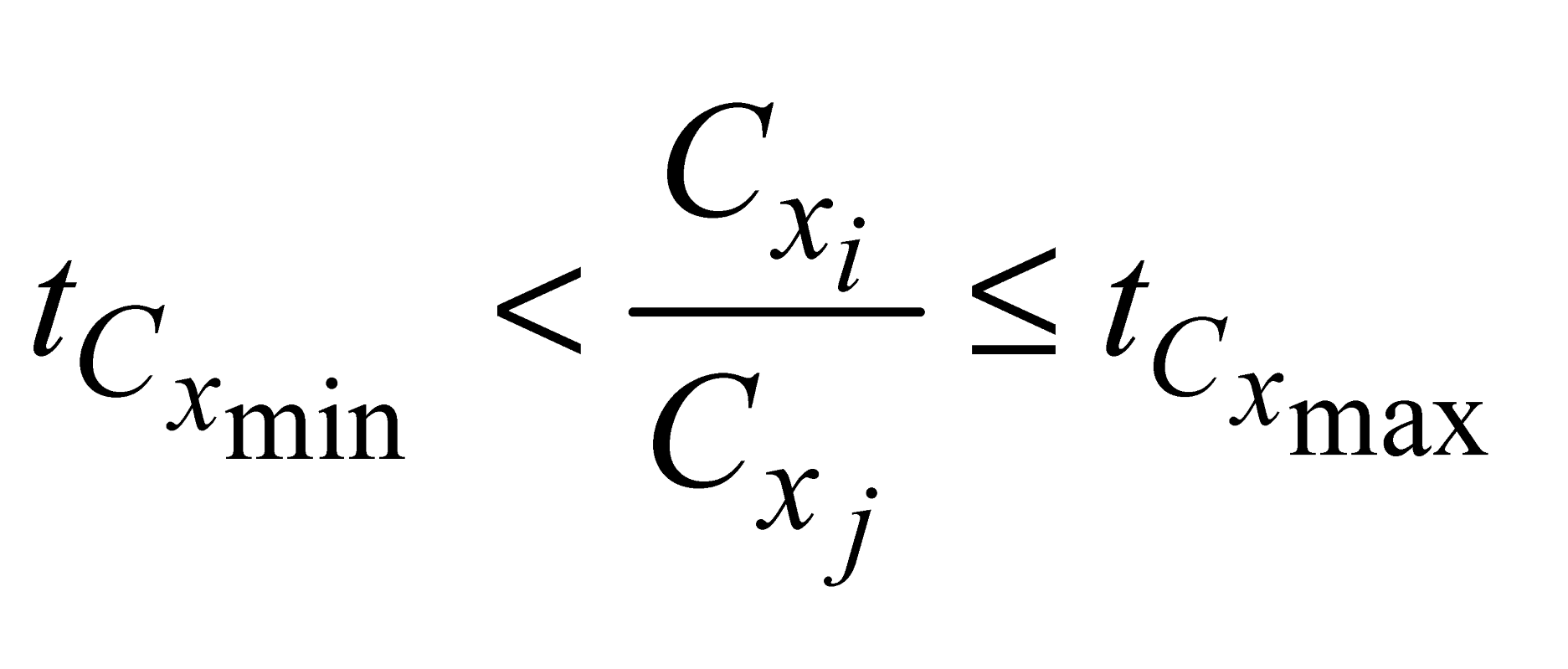
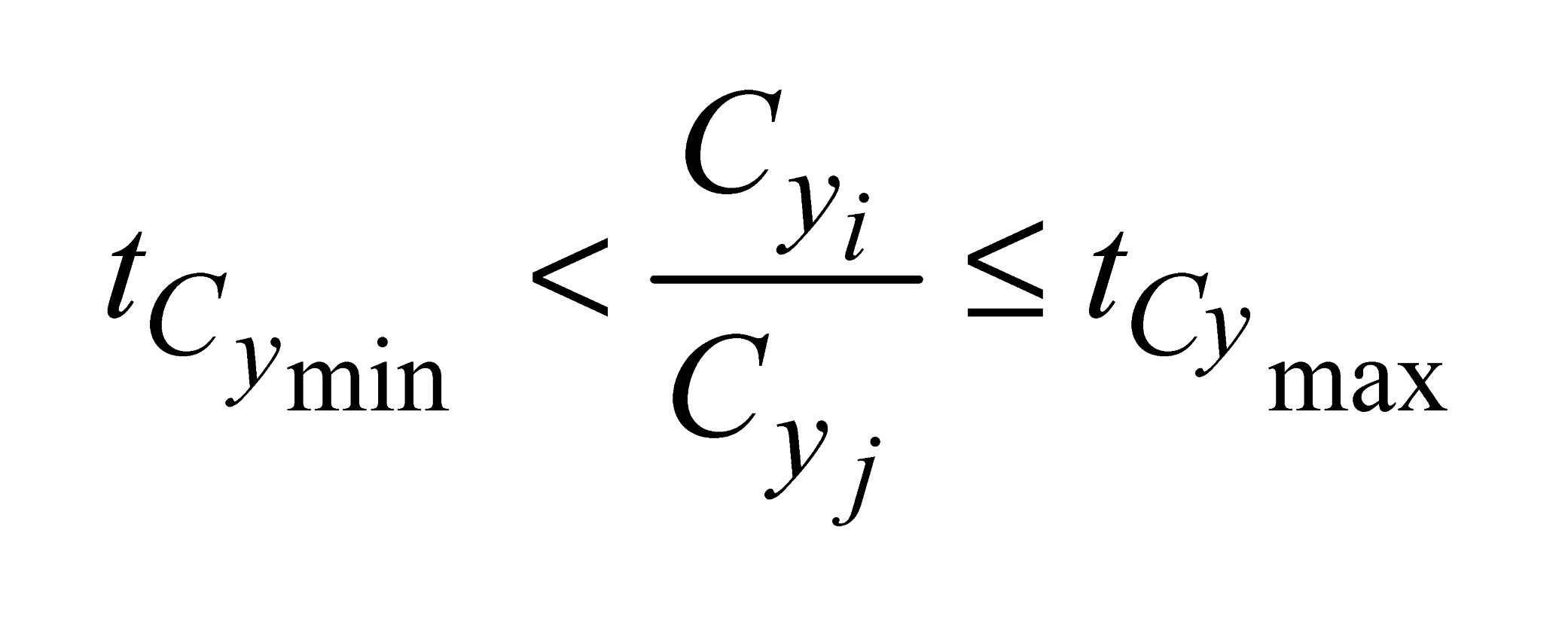
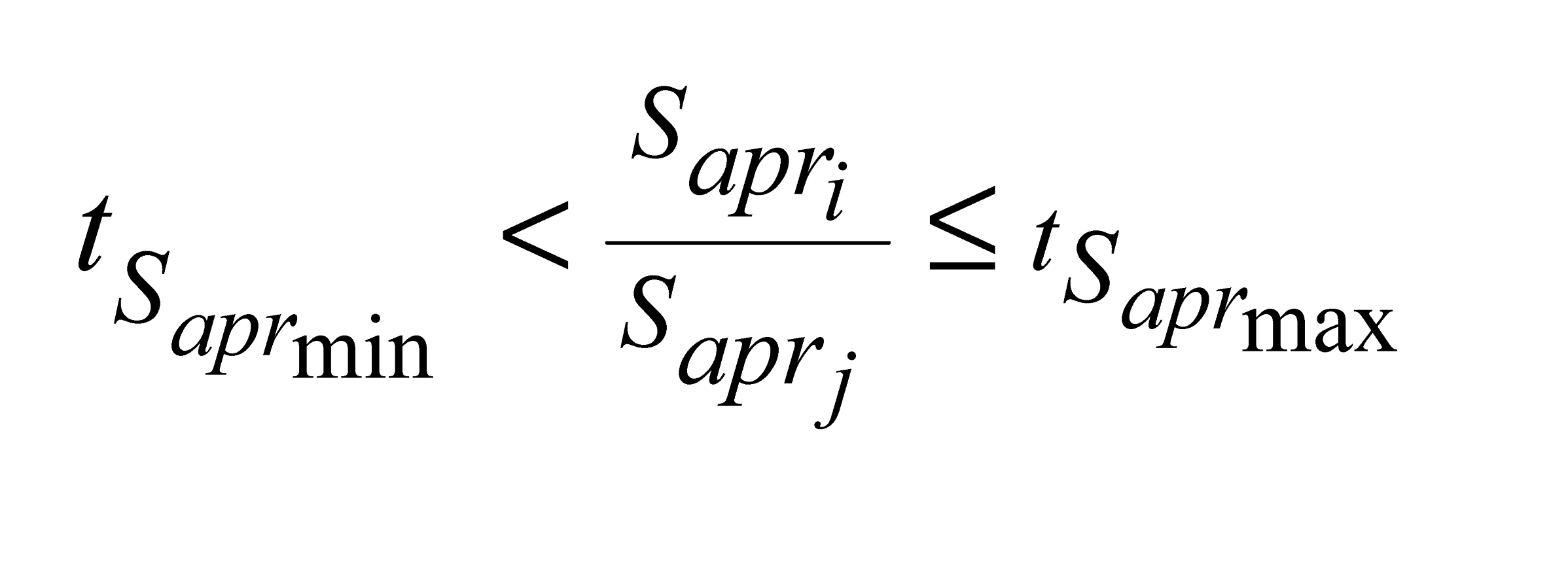
 and
and  are the minimum and maximum threshold values, respectively, for the ratio between the relative tree heights (
are the minimum and maximum threshold values, respectively, for the ratio between the relative tree heights (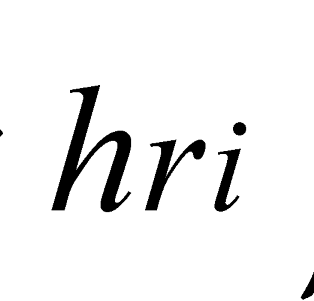 ,
, 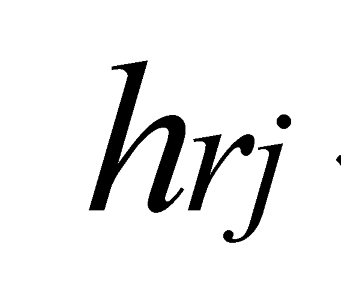 );
); - -
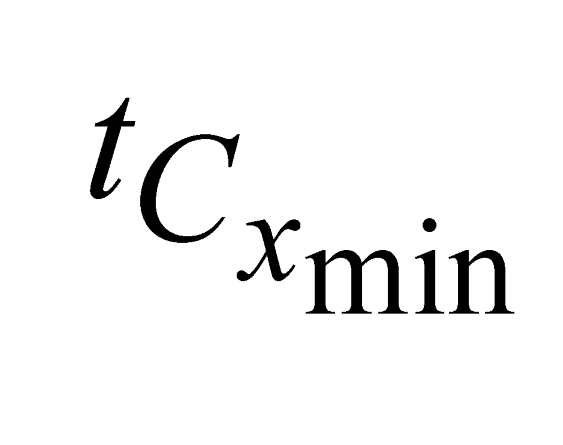 and
and 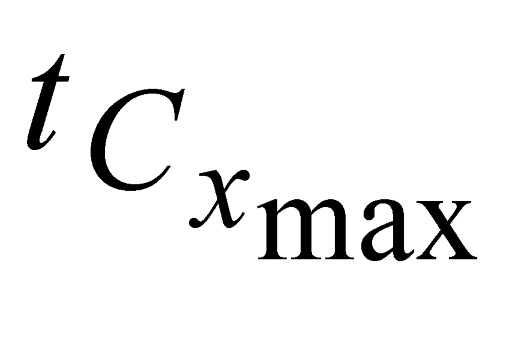 , e are the minimum and maximum threshold values for the ratio between the lengths of the trees in coordinates (
, e are the minimum and maximum threshold values for the ratio between the lengths of the trees in coordinates (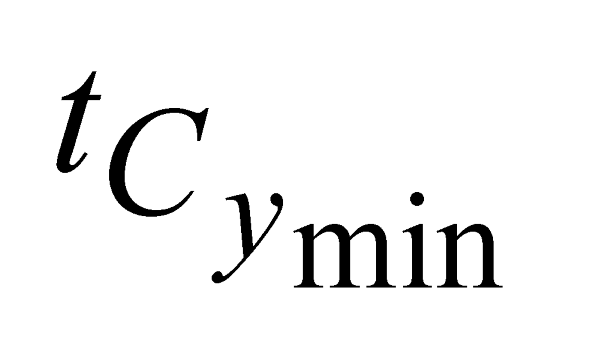 ,
, 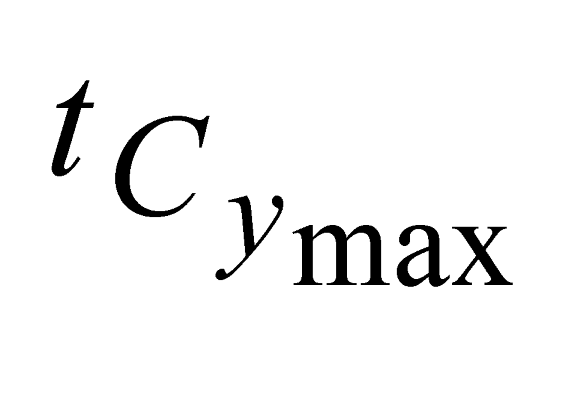 ) and in Y coordinates (
) and in Y coordinates (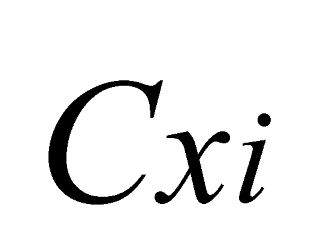 ,
, 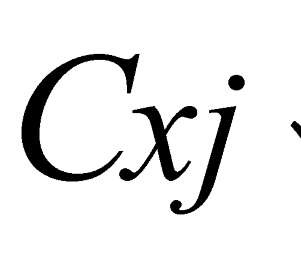 );
);- -
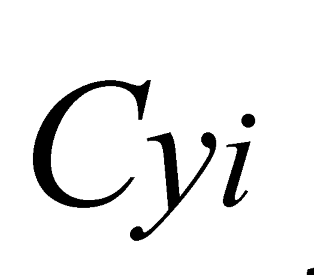 e
e 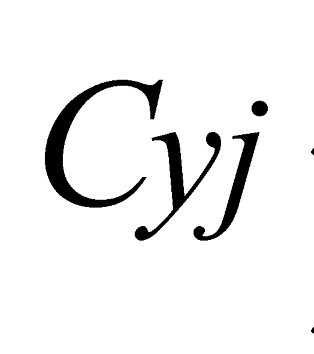 are the minimum and maximum threshold values for the ratio between the approximate areas of the trees (
are the minimum and maximum threshold values for the ratio between the approximate areas of the trees ( ,
,  ).
).
3. Study Area and Materials
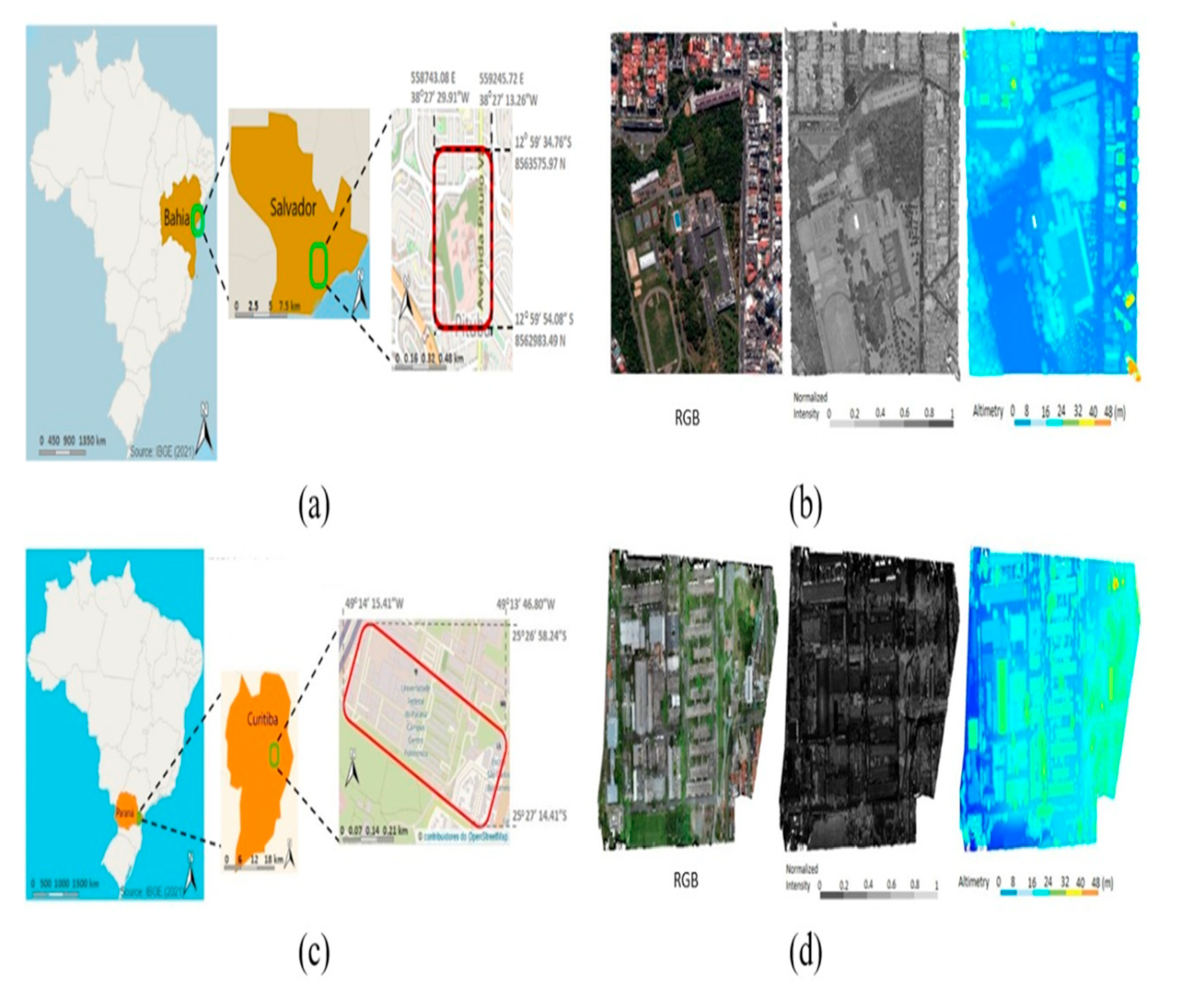
3.1. Study Area
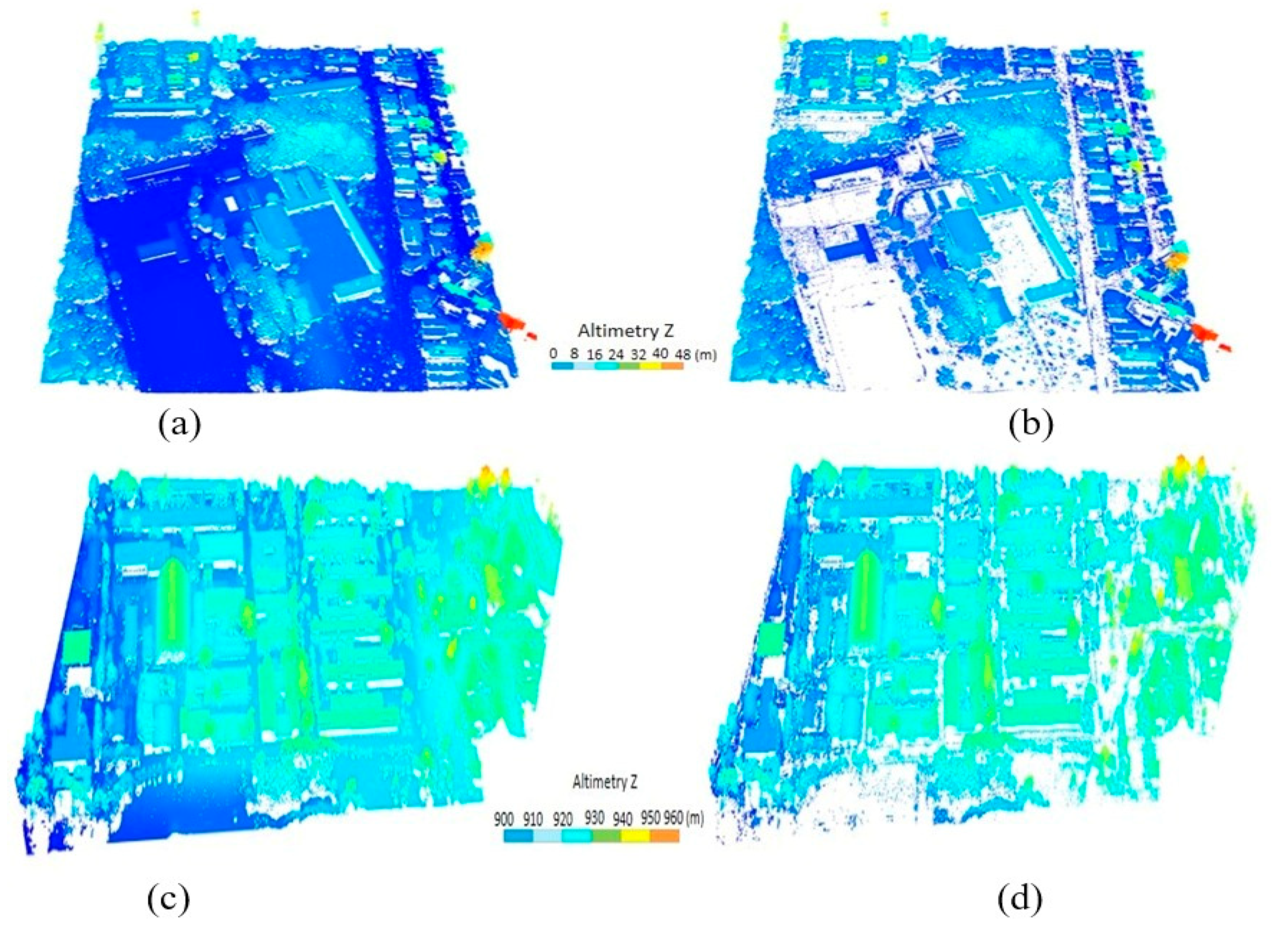
3.2. Data
4. Results
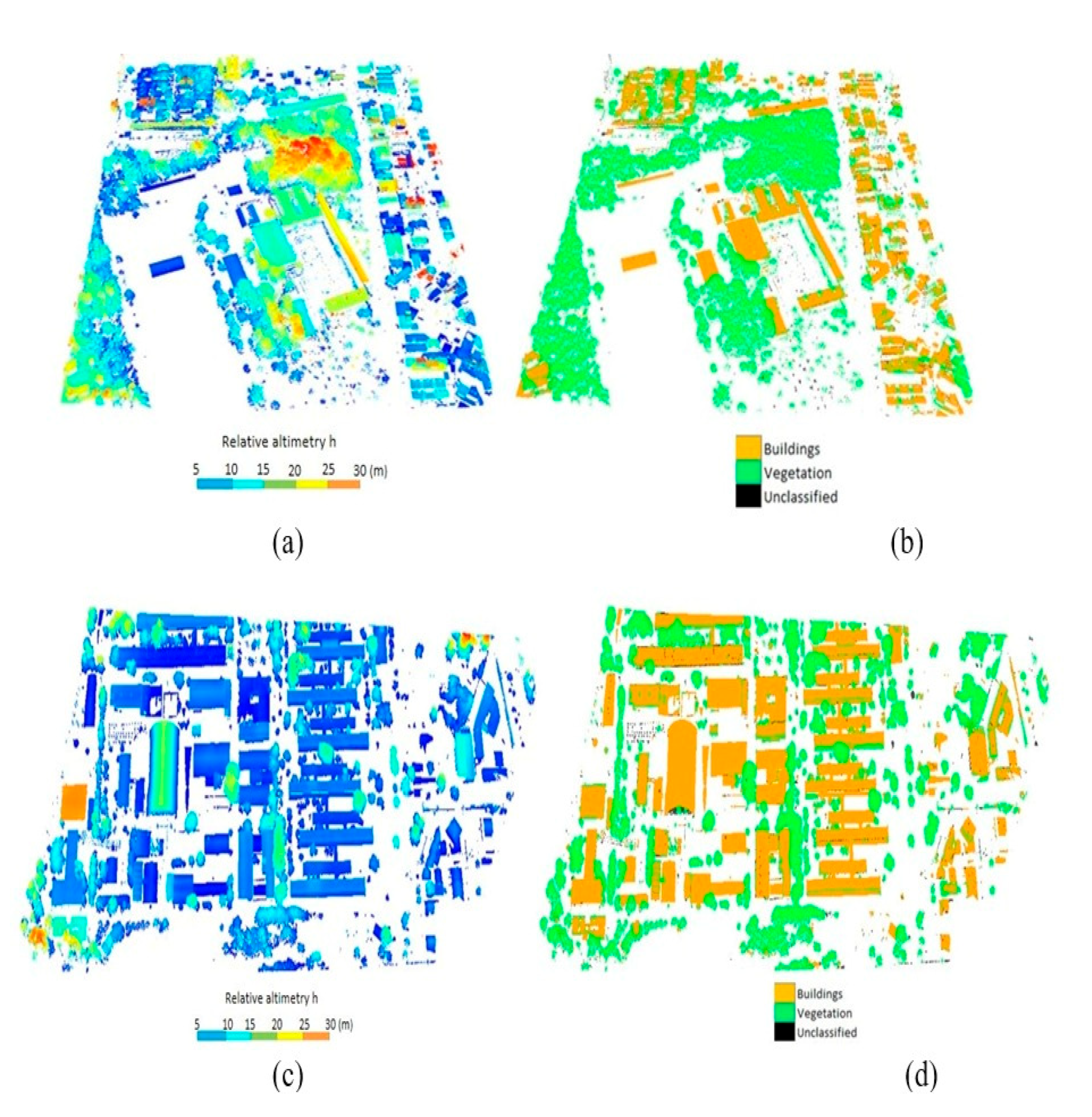
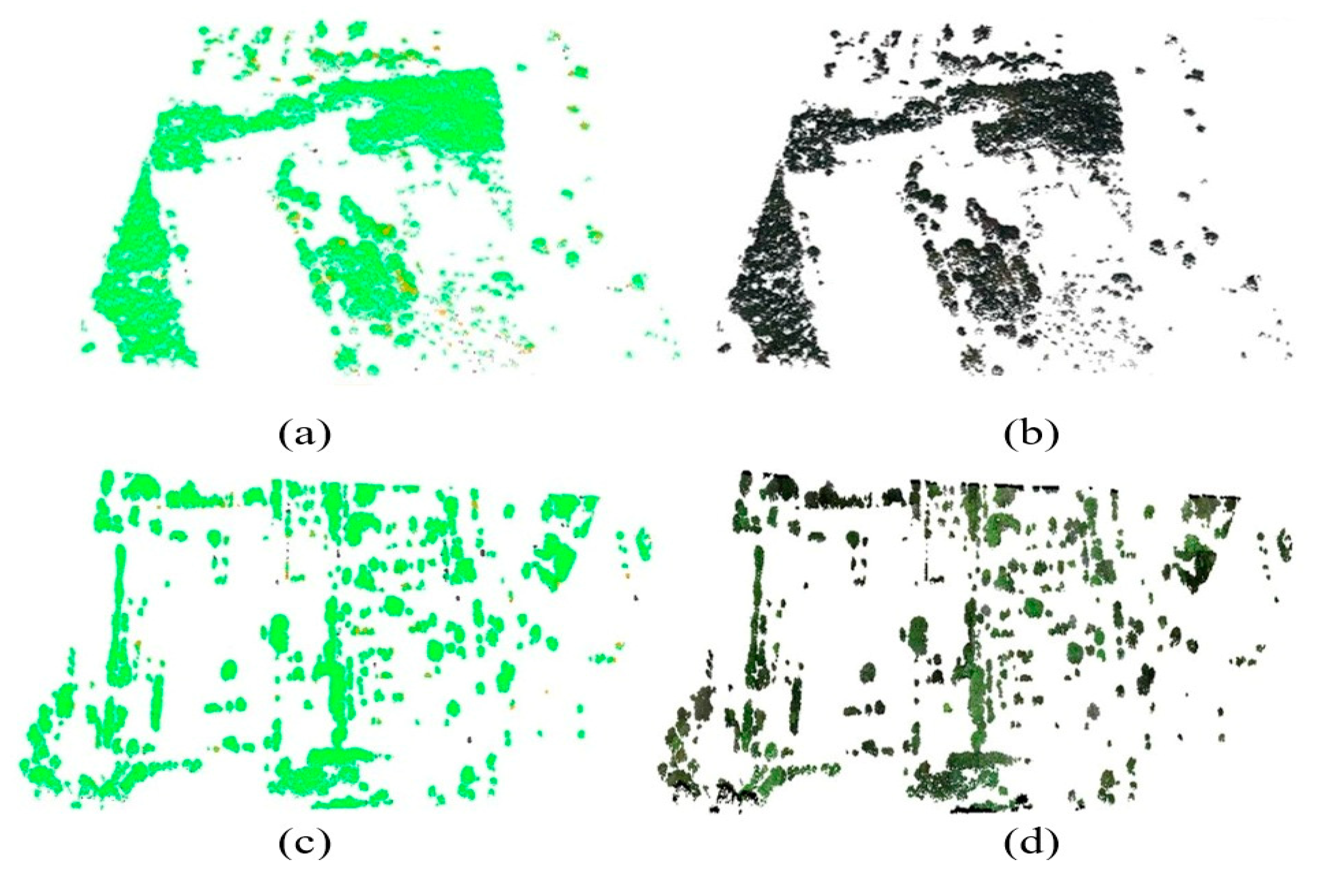
4.1. Classification and Segmentation of the Individual Urban Vegetation
5. Discussion
5.1. Classification and Segmentation of Urban Vegetation
6. Conclusions
Funding
Conflicts of Interest
References
- S. L. Robinson, J. T. Lundholm. Ecosystem services provided by urban spontaneous vegetation, Urban Ecosystems. 15 (3) (2012) 545–557. [CrossRef]
- S. A. Shiflett, L. L. Liang, S. M. Crum, G. L. Feyisa, J. Wang, G. D. Jenerette, Variation in the urban vegetation, surface temperature, air temperature nexus, Science of the Total Environment. 579 (2017) 495– 505. [CrossRef]
- K. Van Ryswyk, N. Prince, M. Ahmed, E. Brisson, J. D. Miller, P. J. Villeneuve. Does urban vegetation reduce temperature and air pollution concentrations? Findings from an environmental monitoring study of the central experimental farm in Ottawa, Canada, Atmospheric Environment. 218 (2019) 116886. [CrossRef]
- R. Simpson. Improved estimates of tree-shade effects on residential energy use, Energy and Buildings. 34 (10) (2002) 1067–1076. [CrossRef]
- S. W. Myint, A. Brazel, G. Okin, A. Buyantuyev. Combined effects of impervious surface and vegetation cover on air temperature variations in a rapidly expanding desert city. GIScience & Remote Sensing. 47 (3) (2010) 301–320.22.
- X. Li, G. Shao, Object-based urban vegetation mapping with highresolution aerial photography as a single data source. International journal of remote sensing 34 (3) (2013) 771–789.
- C. Sharma, K. Hara, R. Tateishi. High-resolution vegetation mapping in japan by combining sentinel-2 and landsat 8 based multi-temporal datasets through machine learning and cross-validation approach, Land 6 (3) (2017) 50.
- H. Zhang, A. Eziz, J. Xiao, S. Tao, S. Wang, Z. Tang, J. Zhu, J. Fang. High-resolution vegetation mapping using extreme gradient boosting based on extensive features. Remote Sensing. 11 (12) (2019) 1505.
- H. Guan, Y. Su, T. Hu, J. Chen, Q. Guo, An object-based strategy for improving the accuracy of spatiotemporal satellite imagery fusion for vegetation-mapping applications. Remote Sensing. 11 (24) (2019) 2927.
- T. R. Tooke, N. C. Coops, N. R. Goodwin, J. A. Voogt. Extracting urban vegetation characteristics using spectral mixture analysis and decision tree classifications. Remote Sensing of Environment 113 (2) (2009) 398– 407.
- Q. Feng, J. Liu, J. Gong. UAV remote sensing for urban vegetation mapping using random forest and texture analysis. Remote sensing 7 (1) (2015) 1074–1094.
- G. Matasci, N. C. Coops, D. A. Williams, N. Page. Mapping tree canopies in urban environments using airborne laser scanning (ALS): a Vancouver case study. Forest Ecosystems 5 (1) (2018) 1–9.
- Abdollahi, B. Pradhan. Urban vegetation mapping from aerial imagery using explainable ai (Xai). Sensors 21 (14) (2021) 4738.
- Zhao, H. A. Sander. Assessing the sensitivity of urban ecosystem service maps to input spatial data resolution and method choice. Landscape and urban planning 175 (2018) 11–22.
- J. L. Edmondson, I. Stott, Z. G. Davies, K. J. Gaston, J. R. Leake. Soil surface temperatures reveal moderation of the urban heat island effect by trees and shrubs. Scientific Reports 6 (1) (2016) 1–8.23.
- M. A. Rahman, L. M. Stratopoulos, A. Moser-Reischl, T. Z¨olch, K.-H. H¨aberle, T. R¨otzer, H. Pretzsch, S. Pauleit. Traits of trees for cooling urban heat islands: A meta-analysis. Building and Environment 170 (2020) 106606.
- T. Hudak, N. L. Crookston, J. S. Evans, M. J. Falkowski, A. M. Smith, P. E. Gessler, P. Morgan. Regression modeling and mapping of coniferous forest basal area and tree density from discrete-return LiDAR and multispectral satellite data. Canadian Journal of Remote Sensing 32 (2) (2006) 126–138.
- E. Næsset. Airborne laser scanning as a method in operational forest inventory: Status of accuracy assessments accomplished in Scandinavia. Scandinavian Journal of Forest Research 22 (5) (2007) 433–442.
- C. Edson, M. G. Wing, Airborne light detection and ranging (LiDAR) for individual tree stem location, height, and biomass measurements. Remote Sensing 3 (11) (2011) 2494–2528.
- W. Xiao, S. Xu, S. O. Elberink, G. Vosselman. Individual tree crown modeling and change detection from airborne LiDAR data. IEEE Journal of selected topics in applied earth observations and remote sensing 9 (8) (2016) 3467–3477.
- L. Liu, N. C. Coops, N. W. Aven, Y. Pang. Mapping urban tree species using integrated airborne hyperspectral and LiDAR remote sensing data. Remote Sensing of Environment 200 (2017) 170–182.
- Z. Ucar, P. Bettinger, K. Merry, R. Akbulut, J. Siry. Estimation of urban woody vegetation cover using multispectral imagery and LiDAR. Urban Forestry & Urban Greening 29 (2018) 248–260.
- R. Pu, S. Landry. Mapping urban tree species by integrating multiseasonal high resolution pleiades satellite imagery with airborne LiDAR data. Urban Forestry & Urban Greening 53 (2020) 126675.
- M. Munzinger, N. Prechtel, M. Behnisch. Mapping the urban forest in detail: From lidar point clouds to 3D tree models. Urban Forestry & Urban Greening. 74 (2022) 127637.24.
- C. Robinson, U. Demsar, A. B. Moore, A. Buckley, B. Jiang, K. Field, M.-J. Kraak, S. P. Camboim, C. R. Sluter. Geospatial big data and cartography: research challenges and opportunities for making maps that matter. International Journal of Cartography 3 (sup1) (2017) 32– 60.
- F. Poux, The smart point cloud: Structuring 3D intelligent point data (2019).
- M. Kada, 3D building generalization based on half-space modeling. International Archives of Photogrammetry, Remote Sensing and Spatial Information Sciences 36 (2) (2006) 58–64.
- H. Mayer. Scale-spaces for generalization of 3D buildings. International Journal of Geographical Information Science 19 (8-9) (2005) 975–997.
- D. Lee, P. Hardy. Automating generalization–tools and models, in: 22nd ICA Conference Proceedings, La Coruña, Spain, Citeseer, 2005.
- F. Thiemann, M. Sester, 3D-symbolization using adaptive templates. Proceedings of the GICON 27 (2006).
- M. Kada, Scale-dependent simplification of 3D building models based on cell decomposition and primitive instancing, in: International Conference on Spatial Information Theory, Springer, 2007, pp. 222–237.
- Forberg, Generalization of 3D building data based on a scale-space approach, ISPRS Journal of Photogrammetry and Remote Sensing 62 (2) (2007) 104–111.
- R. Guercke, C. Brenner, M. Sester. Generalization of semantically enhanced 3D city models, in: Proceedings of the GeoWeb 2009 Conference, 2009, pp. 28–34.
- W. A. Mackaness, A. Ruas, L. T. Sarjakoski. Generalisation of geographic information: cartographic modelling and applications, Elsevier, 2011.
- H. Fan, L. Meng. A three-step approach of simplifying 3D buildings modeled by CityGML. International Journal of Geographical Information Science 26 (6) (2012) 1091–1107.25.
- S. U. Baig, A. A. Rahman. A three-step strategy for generalization of 3D building models based on CityGML specifications. GeoJournal 78 (6) (2013) 1013–1020.
- C. Niu, J. Li, K. Xu. Im2struct: Recovering 3D shape structure from a single RGB image, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 4521–4529.
- C. Zou, E. Yumer, J. Yang, D. Ceylan, D. Hoiem. 3D-PRNN: Generating shape primitives with recurrent neural networks, in: Proceedings of the IEEE International Conference on Computer Vision, 2017, pp. 900–909.
- S. Tulsiani, H. Su, L. J. Guibas, A. A. Efros, J. Malik. Learning shape abstractions by assembling volumetric primitives, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 2635–2643.
- D. Paschalidou, A. O. Ulusoy, A. Geiger. Superquadrics revisited: Learning 3D shape parsing beyond cuboids, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2019, pp. 10344–10353.
- L. Ortega-C’ordova. Urban vegetation modeling 3D levels of detail (2018).
- P.-S. Wang, Y. Liu, Y.-X. Guo, C.-Y. Sun, X. Tong. O-CNN: Octreebased convolutional neural networks for 3D shape analysis. ACM Transactions On Graphics (TOG) 36 (4) (2017) 1–11.
- H. Thomas, C. R. Qi, J.-E. Deschaud, B. Marcotegui, F. Goulette, L. J. Guibas. Kpconv: Flexible and deformable convolution for point clouds, in: Proceedings of the IEEE/CVF international conference on computer vision, 2019, pp. 6411–6420.
- T. Huang, Y. Liu, 3D point cloud geometry compression on deep learning, in: Proceedings of the 27th ACM international conference on multimedia, 2019, pp. 890–898.
- M. Quach, G. Valenzise, F. Dufaux. Improved deep point cloud geometry compression, in: 2020 IEEE 22nd International Workshop on Multimedia Signal Processing (MMSP), IEEE, 2020, pp. 1–6.26.
- L. Yu, X. Li, C.-W. Fu, D. Cohen-Or, P.-A. Heng. Pu-net: Point cloud upsampling network, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 2790–2799.
- L. Huang, S. Wang, K. Wong, J. Liu, R. Urtasun. Octsqueeze: Octreestructured entropy model for LiDAR compression, in: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2020, pp. 1313–1323.
- L. Wiesmann, Milioto a chen x stachniss c behley j. Deep compression for dense point cloud maps. IEEE Robotics and Automation Letters 6 (2021) 2060–2067.
- W. Li, Q. Guo, M. K. Jakubowski, M. Kelly. A new method for segmenting individual trees from the LiDAR point cloud. Photogrammetric Engineering & Remote Sensing 78 (1) (2012) 75–84.
- R. B. Rusu, Z. C. Marton, N. Blodow, M. Dolha, M. Beetz. Towards 3d point cloud based object maps for household environments. Robotics and Autonomous Systems 56 (11) (2008) 927–941.
- G. Vosselman. Slope based filtering of laser altimetry data. International archives of photogrammetry and remote sensing 33 (B3/2; PART 3) (2000) 935–942.
- J. Shan, S. Aparajithan. Urban DEM generation from raw LiDAR data. Photogrammetric Engineering & Remote Sensing 71 (2) (2005) 217–226.
- K. Zhang, S.-C. Chen, D. Whitman, M.-L. Shyu, J. Yan, C. Zhang. A progressive morphological filter for removing nonground measurements from airborne LiDAR data. IEEE transactions on geoscience and remote sensing 41 (4) (2003) 872–882.
- C.-K. Wang, Y.-H. Tseng. DEM generation from airborne LiDAR data by an adaptive dual-directional slope filter. na, 2010.
- Y. Li, B. Yong, H. Wu, R. An, H. Xu, An improved top-hat filter with sloped brim for extracting ground points from airborne LIDAR point clouds. Remote sensing 6 (12) (2014) 12885–12908.27.
- P. Axelsson, DEM generation from laser scanner data using adaptive TIN models. International archives of photogrammetry and remote sensing 33 (4) (2000) 110–117.
- J. Zhang, X. Lin. Filtering airborne LiDAR data by embedding smoothness-constrained segmentation in progressive tin densification. ISPRS Journal of photogrammetry and remote sensing 81 (2013) 44–59.
- W. Su, Z. Sun, R. Zhong, J. Huang, M. Li, J. Zhu, K. Zhang, H. Wu, D. Zhu. A new hierarchical moving curve-fitting algorithm for filtering LiDAR data for automatic DTM generation. International Journal of Remote Sensing 36 (14) (2015) 3616–3635.
- Z. Hui, Y. Hu, Y. Z. Yevenyo, X. Yu. An improved morphological algorithm for filtering airborne LiDAR point cloud based on multi-level kriging interpolation. Remote Sensing 8 (1) (2016) 35.
- W. Zhang, J. Qi, P. Wan, H. Wang, D. Xie, X. Wang, G. Yan. An easy to-use airborne LiDAR data filtering method based on cloth simulation. Remote sensing 8 (6) (2016) 501.
- M. Isenburg. Laszip: lossless compression of lidar data. Photogrammetric engineering and remote sensing 79 (2) (2013) 209–217.
- J.-R. Roussel, D. Auty, N. C. Coops, P. Tompalski, T. R. Goodbody, A. S. Meador, J.-F. Bourdon, F. De Boissieu, A. Achim. lidr: An r package for analysis of airborne laser scanning (ALS) data. Remote Sensing of Environment 251 (2020) 112061.
- S. C. Popescu, R. H. Wynne. Seeing the trees in the forest. Photogrammetric Engineering & Remote Sensing 70 (5) (2004) 589–604.
- Q. Chen, D. Baldocchi, P. Gong, M. Kelly. Isolating individual trees in a savanna woodland using small footprint LiDAR data. Photogrammetric Engineering & Remote Sensing 72 (8) (2006) 923–932.
- B. Koch, U. Heyder, H. Weinacker. Detection of individual tree crowns in airborne LiDAR data. Photogrammetric Engineering & Remote Sensing 72 (4) (2006) 357–363.28.
- F. Morsdorf, E. Meier, B. Allg¨ower, D. N¨uesch. Clustering in airborne laser scanning raw data for segmentation of single trees. International Archives of the Photogrammetry. Remote Sensing and Spatial Information Sciences 34 (part 3) (2003) W13.
- S.-J. Lee, Y.-C. Chan, D. Komatitsch, B.-S. Huang, J. Tromp. Effects of realistic surface topography on seismic ground motion in the Yangminshan region of Taiwan based upon the spectral-element method and LiDAR DTM, Bulletin of the Seismological Society of America 99 (2A) (2009) 681–693.
- L. S. Delazari, A. L. S. D. Skroch, et al., UFPR Campusmap: a laboratory for a smart city developments. Abstracts of the ICA 1 (2019) 1–2.
- S. Xu, G. Vosselman, S. O. Elberink. Multiple-entity based classification of airborne laser scanning data in urban areas. ISPRS Journal of photogrammetry and remote sensing 88 (2014) 1–15.
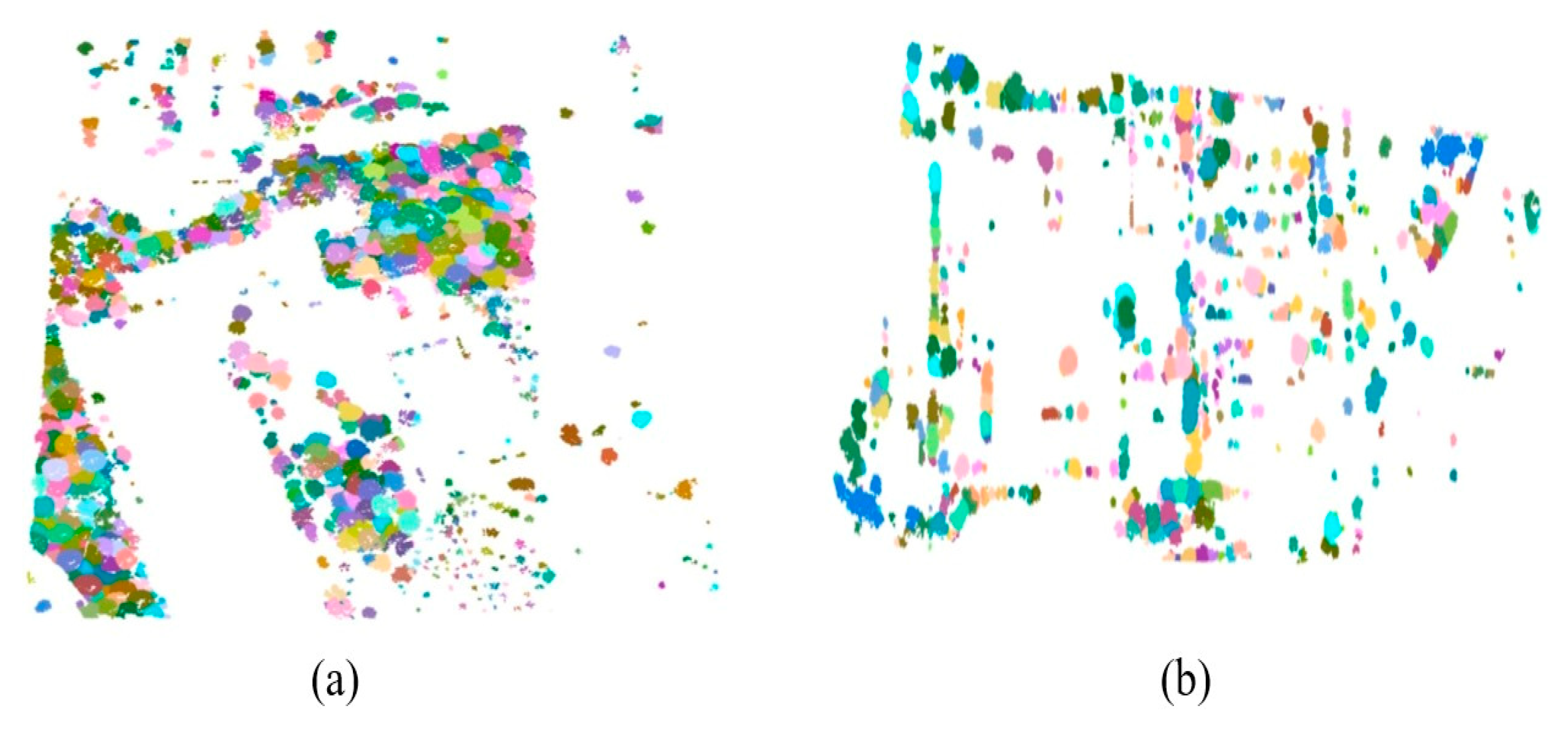
| Label | RGB criteria | Local max. criterion |
| Vegetation | XiG 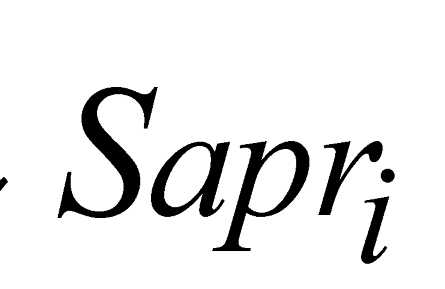 2b-1; XiG > XiR; XiG > XiB 2b-1; XiG > XiR; XiG > XiB
|
Xi is local max. |
| XiG < 2b-1; XiG > XiR; XiG > XiB XiR < 2b-1; XiR < XiG; XiR XiB | ||
XiR 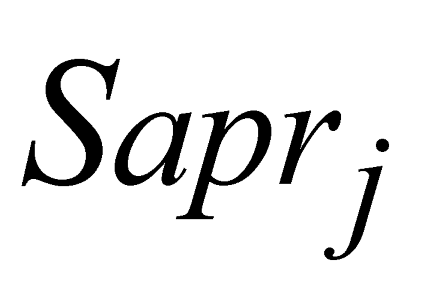 2b-1; XiR < XiG; XiR XiB 2b-1; XiR < XiG; XiR XiB
| ||
| Buildings | XiR 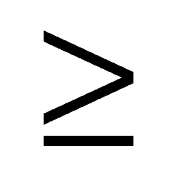 2b-1; XiR > XiG; XiR > XiB 2b-1; XiR > XiG; XiR > XiB
|
Xi is not local max. |
| XiR < 2b-1; XiR > XiG; XiR > XiB XiG 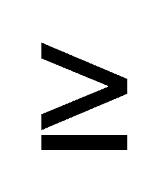 2b-1; XiG < XiR or XiG < XiB 2b-1; XiG < XiR or XiG < XiB
| ||
| XiG < 2b-1; XiG < XiR or XiG < XiB |
| Parameter | Description | Values (m) |
|---|---|---|
| dt1 | bottom limits | 5 |
| dt2 | upper limits | 7 |
| Zu | bottom limits | 15 |
| Speed up | upper limits | 10 |
| hmin | Minimum height for a detected tree | 5 |
| R | Search radius for the local maximum | 5 |
| ALS | Truth reference | |||
|---|---|---|---|---|
| Vegetation | Non-vegetation | Total of points | ||
| Label | Vegetation | 500.164 | 94.225 | 594.389 |
| Non-vegetation | 18.569 | 408.114 | 426.683 | |
| Total | 518.733 | 502.339 | 1.021.072 | |
| ULS | ||||
| Label | Vegetation | 6.455.452 | 1.089.707 | 7.454.159 |
| Non-vegetation | 169.953 | 14.944.094 | 15.114.047 | |
| Total | 6.625.405 | 16.083.801 | 22.659.206 | |
| Data | AD | DA (tree/hectare) | GF (tree/green area) | GD (m) |
|---|---|---|---|---|
| ALS | 851 | 28.65 | 67.33 | 9.73 |
| ULS | 518 | 19.9 | 78.96 | 10.45 |
| Data | Threshold | GF(*) | GD (m) | ANA | AM |
|---|---|---|---|---|---|
| ALS | A | 64.31 | 12.56 | 431 | 382 |
| B | 65.26 | 13.37 | 554 | 271 | |
| C | 64.16 | 14.48 | 457 | 354 | |
| D | 65.58 | 13.94 | 580 | 249 | |
| E | 61.70 | 14.17 | 496 | 284 | |
| F | 63.29 | 14.11 | 625 | 175 | |
| G | 62.65 | 14.64 | 552 | 240 | |
| H | 62.34 | 14.12 | 670 | 118 | |
| I | 60.68 | 14.81 | 625 | 142 | |
| J | 62.57 | 14.07 | 722 | 69 | |
| ULS | A | 75.31 | 17.58 | 311 | 181 |
| B | 75.3 | 16.84 | 397 | 97 | |
| C | 73.93 | 17.63 | 341 | 144 | |
| D | 74.39 | 16 | 409 | 79 | |
| E | 72.68 | 17.81 | 373 | 105 | |
| F | 74.39 | 17.31 | 430 | 58 | |
| G | 73.17 | 17.35 | 405 | 75 | |
| H | 74.84 | 16.79 | 449 | 42 | |
| I | 74.39 | 16.91 | 446 | 42 | |
| J | 76.52 | 16.41 | 482 | 20 |
| Data | Storage space (%) | |
| µ = 1 | µ = 2 | |
| ALS | 72.65 | 45.32 |
| ULS | 98.47 | 96.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





