3. Results and Discussion
This molecular docking study has investigate by Pyrx program [
11] to thousands compoundsin in the active site of HIV-1 reverse transcriptase. General Speaking Pyrx is a molecular graphics software and a user-friendly interface for the AutoDock suite of molecular docking tools. Virtual screening involves computationally screening a large number of compounds to identify potential drug candidates [
11].
The work has evaluated the binding energetics scores, specifically using the Vina Scores, by Autodock Vina Algorithm [
10], as a parameter to assess the strength of interaction between the compounds and the target protein. The focus was on selecting molecules with high binding energies. Generally, the negative values of binding energy indicate favorable interactions between the compounds and the target protein. Lower (more negative) binding energies generally suggest stronger binding.The study identified eight compounds with notable Vina Scores, suggesting strong binding affinity to HIV-1 reverse transcriptase. Here are the details of the selected compounds along with their corresponding binding energies:
-Tetrophan
Binding Energy: -11.2 kcal/mol
-Imatinib
Binding Energy: -9.8 kcal/mol
-Amentoflavone
Binding Energy: -12.5 kcal/mol
-Alprazolam
Binding Energy: -11.1 kcal/mol
-Bilobetin
Binding Energy: -12.0 kcal/mol
-Bafetinib
Binding Energy: -10.4 kcal/mol
-Nilotinib
Binding Energy: -11.8 kcal/mol
-Ginketin
Binding Energy: -11.8 kcal/mol
The next step involved assessing the toxicity properties of the selected candidates using two prediction servers, namely pKCSM and Pro-Tox-II. The focus was on key toxicity parameters, and pKCSM, a machine learning platform, was utilized firstly for this evaluation.
Three crucial parameters were considered: Max.tolerated dose (human) (log mg/kg/day), Oral Rat Acute Toxicity (LD50) (mol/kg), and Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day).
The results revealed that among the eight candidates, only two demonstrated a lower overall toxicity profile when considering all the predicted toxicity properties analyzed by pKCSM [
12]. These candidates are Amentoflavone, a biflavonoid found in Ginkgo biloba and Hypericum perforatum [
13], and Nilotinib, a tyrosine kinase inhibitor known as AMN107 [
14]. Here are the main toxicity results for these two candidates:
-Amentoflavone:
Max.tolerated dose (human) (log mg/kg/day): 0.438 log mg/kg/day
Oral Rat Acute Toxicity (LD50) (mol/kg): 2.527 mol/kg
Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day): 3.572 log mg/kg_bw/day
-Nilotinib:
Max.tolerated dose (human) (log mg/kg/day): 0.96 log mg/kg/day
Oral Rat Acute Toxicity (LD50) (mol/kg): 2.149 mol/kg
Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day): 2.81 log mg/kg_bw/day
Both Amentoflavone and Nilotinib exhibited high values in the parameters of Max.tolerated dose (human), LD50, and LOAEL, suggesting potential as candidates with lower toxicity concerns. Amentoflavone, being a natural substance, and Nilotinib, a pharmaceutical drug, demonstrated promising toxicity profiles in this analysis.
In the conclusive phase, an in-depth examination of the toxicity properties of Amentoflavone and Nilotinib was conducted using Pro-Tox-II [
15], a virtual lab designed for predicting toxicities of small molecules. Pro-Tox-II is proficient in forecasting various toxicological aspects, including acute toxicity, hepatotoxicity, cytotoxicity, carcinogenicity, mutagenicity, and immunotoxicity. Upon comparison, several noteworthy aspects emerge, which can be succinctly summarized as follows:
-Acute Toxicity (LD50):
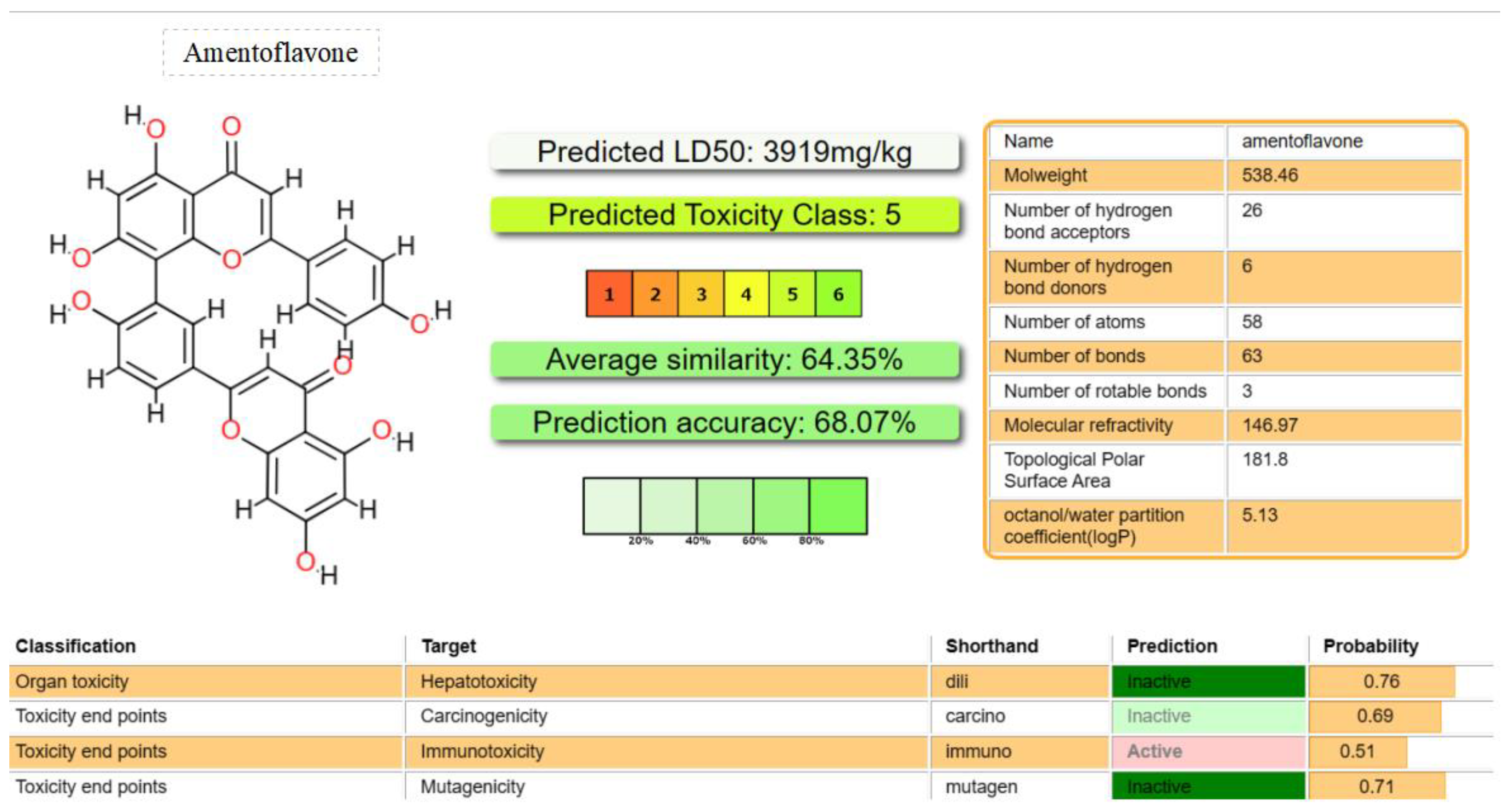
Amentoflavone: Predictive LD50 of 3919 mg/kg.
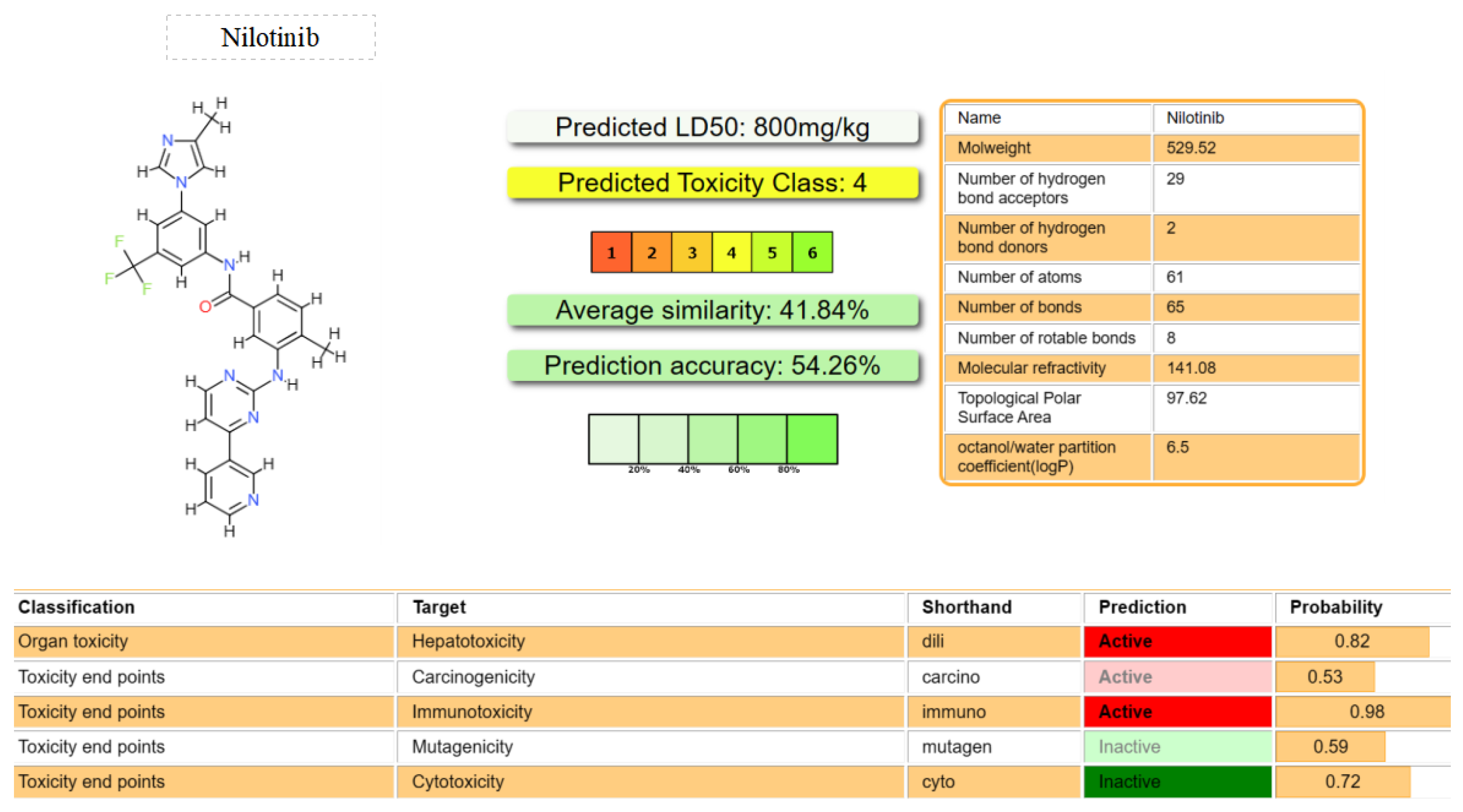
Nilotinib: Predictive LD50 of 800 mg/kg.
-Toxicity Class:
Amentoflavone: Classified as toxicity class 5.
Nilotinib: Classified as toxicity class 4.
Specific Toxicity Predictions for Amentoflavone:
Hepatotoxicity: Probability of approximately 70%, indicating a lower likelihood.
Carcinogenicity: Probability of approximately 70%, suggesting a lower likelihood.
Mutagenicity: Probability of approximately 70%, implying a lower likelihood.
Immunotoxicity: Probability of approximately 50%, indicating a moderate likelihood.
Specific Toxicity Predictions for Nilotinib:
Hepatotoxicity: Probability of over 70%, suggesting a higher likelihood.
Carcinogenicity: Probability of over 70%, indicating a higher likelihood.
Mutagenicity: Probability of less than 70%, suggesting a lower likelihood.
Immunotoxicity: Probability of over 70%, indicating a higher likelihood.
Cytotoxicity: Probability of over 70%, suggesting a higher likelihood.
In summary, Amentoflavone, as a natural substance, exhibits a predictive LD50 that is higher than Nilotinib, and it is classified in a lower toxicity class. Additionally, Amentoflavone shows a lower likelihood of being hepatotoxic, carcinogenic, mutagenic, and moderately immunotoxic compared to Nilotinib, which demonstrates higher probabilities of adverse effects in these categories. These findings provide valuable insights into the comparative toxicity profiles of Amentoflavone and Nilotinib, informing further considerations for their potential use in therapeutic applications.
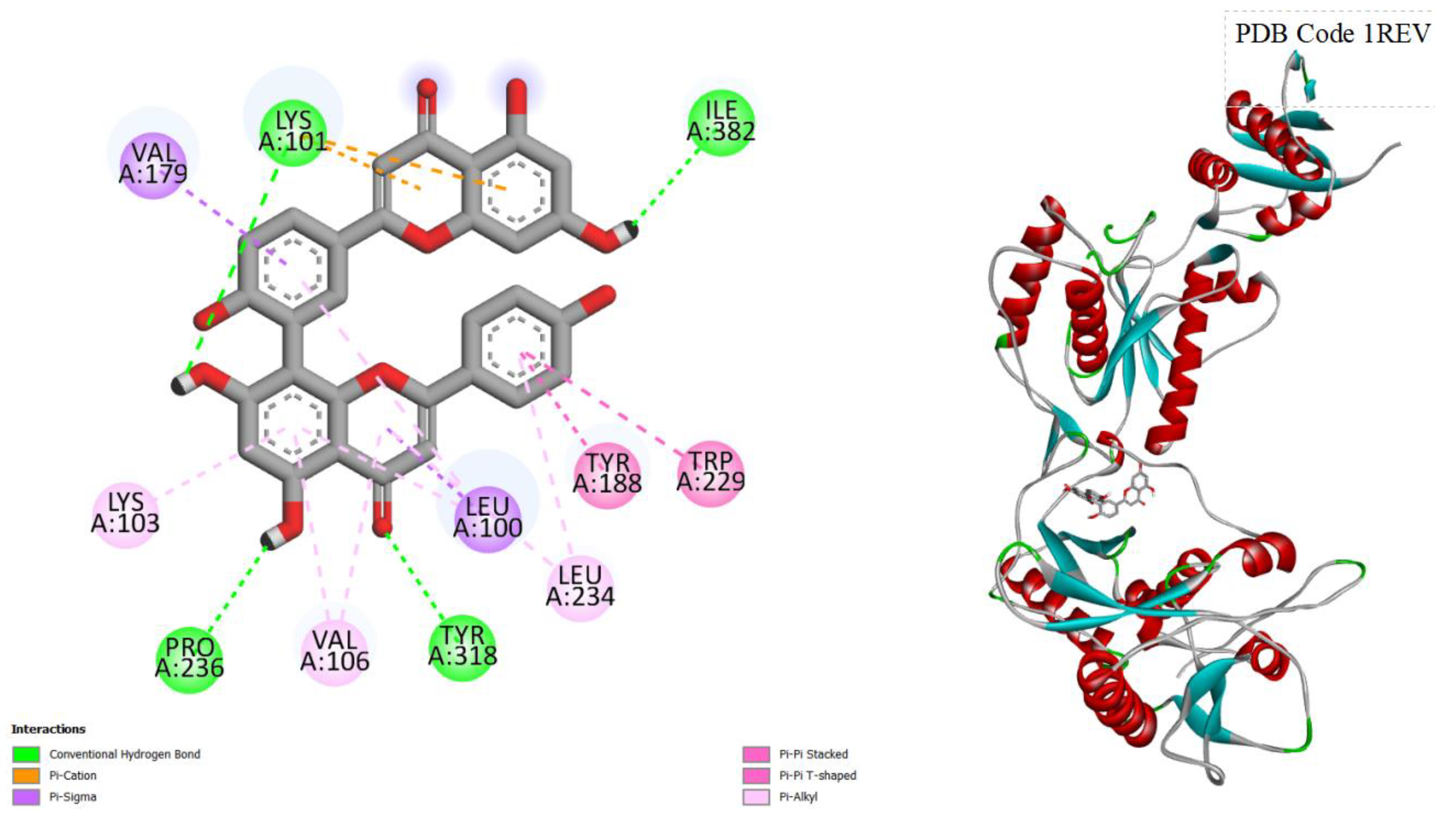
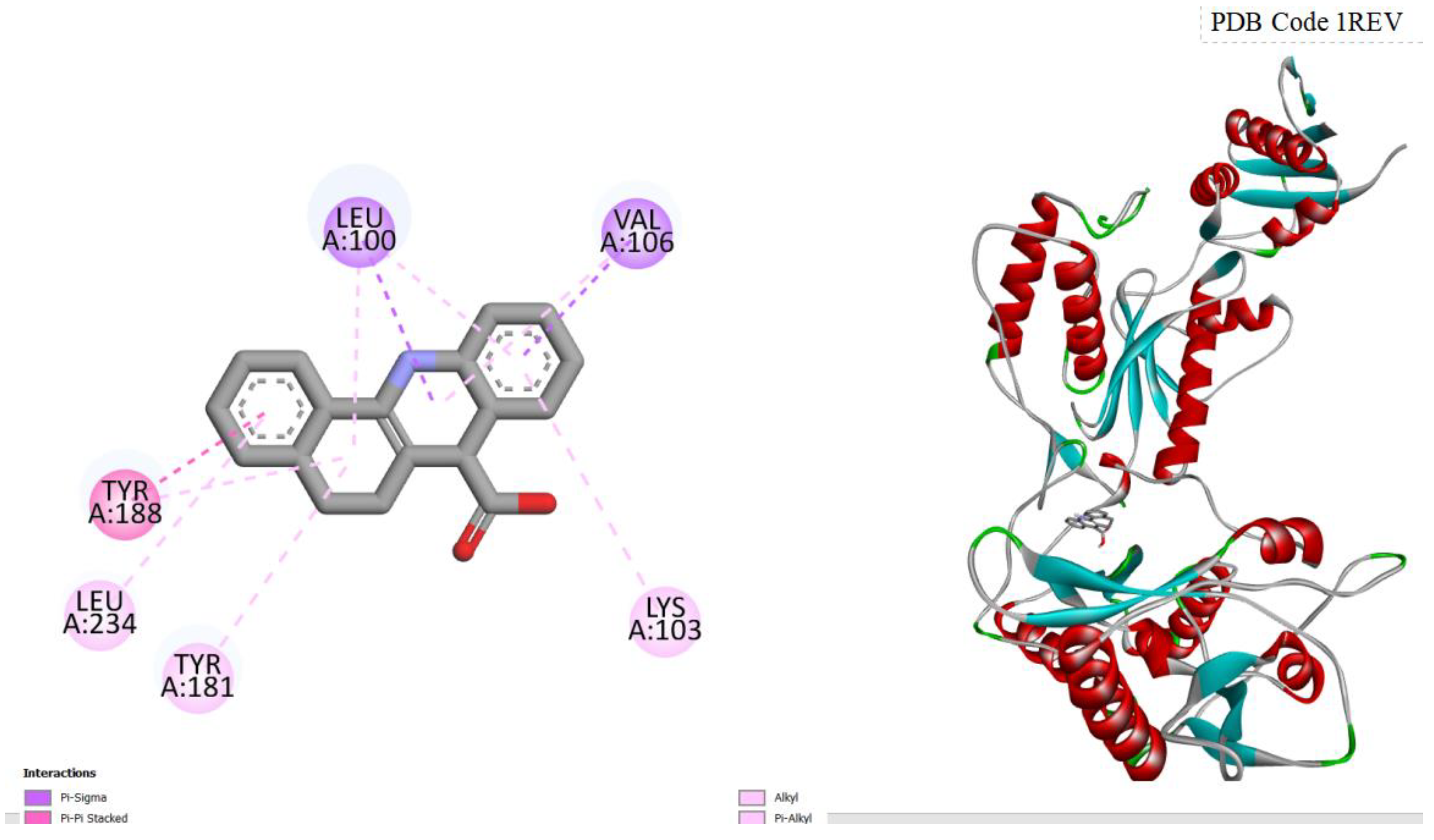
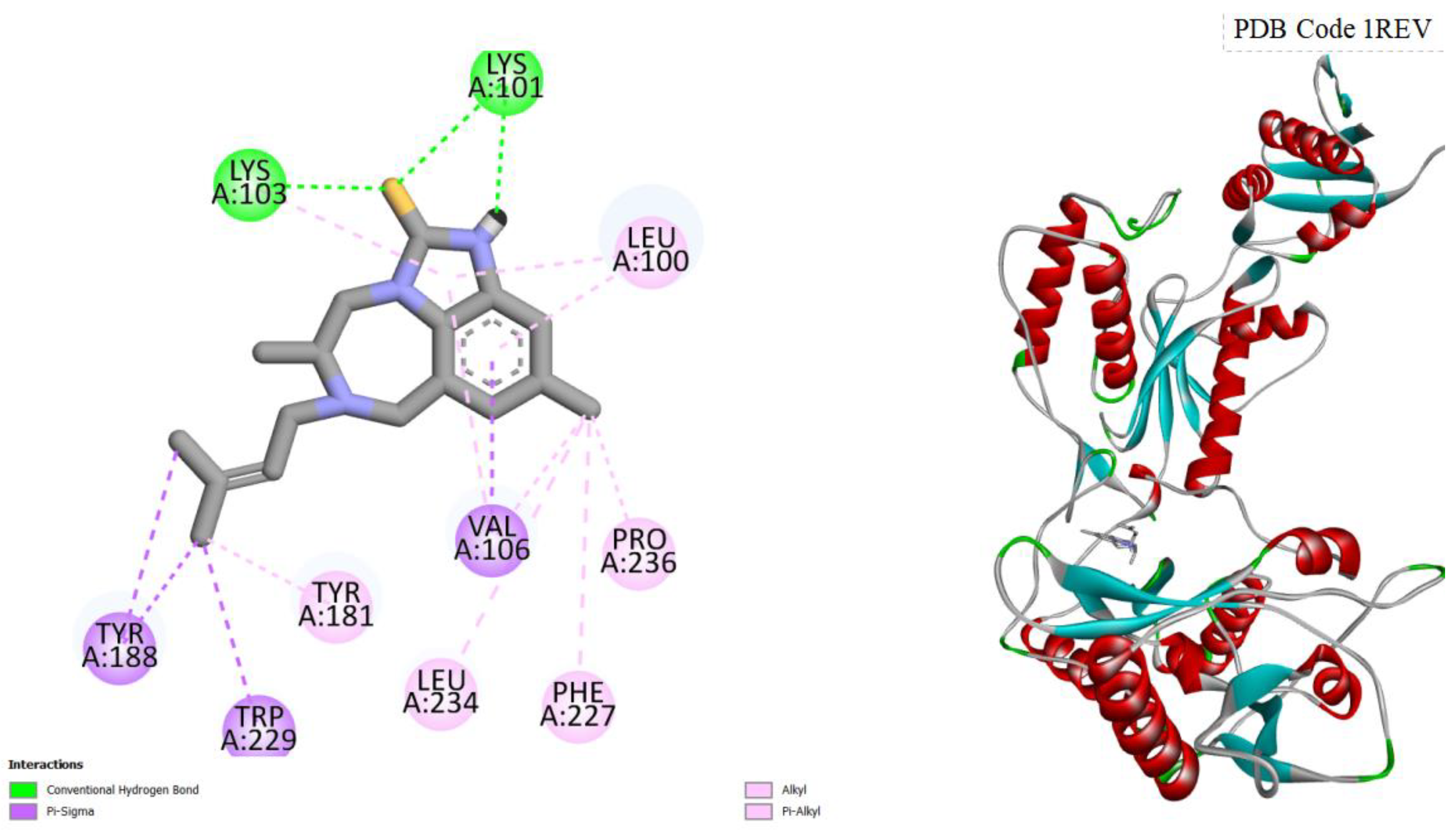
Figure 1.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Amentoflavone within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Amentoflavone. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
Figure 1.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Amentoflavone within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Amentoflavone. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Amentoflavone.
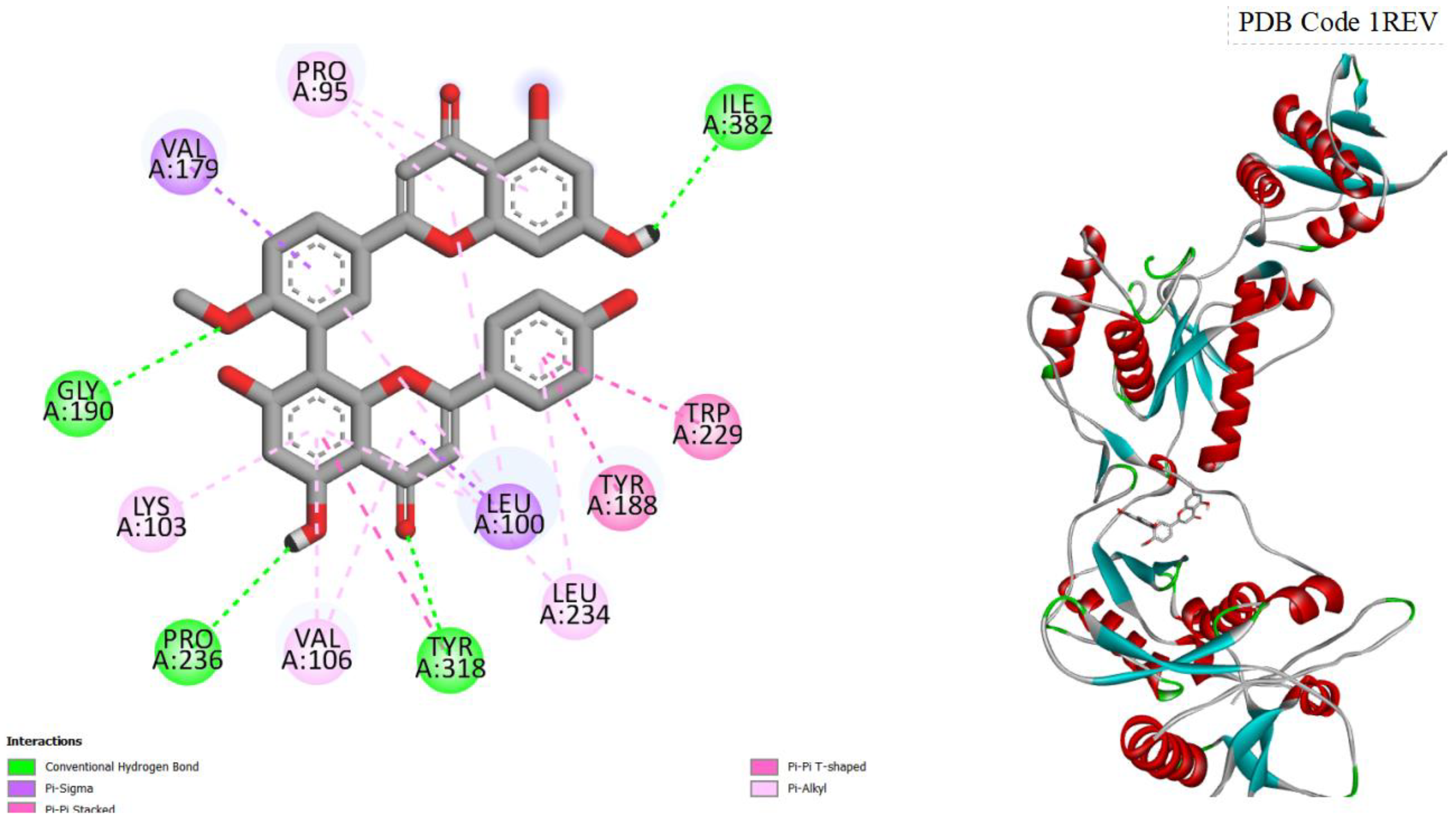
Figure 2.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Bilobetin within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Bilobetin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Bilobetin.
Figure 2.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Bilobetin within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Bilobetin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Bilobetin.
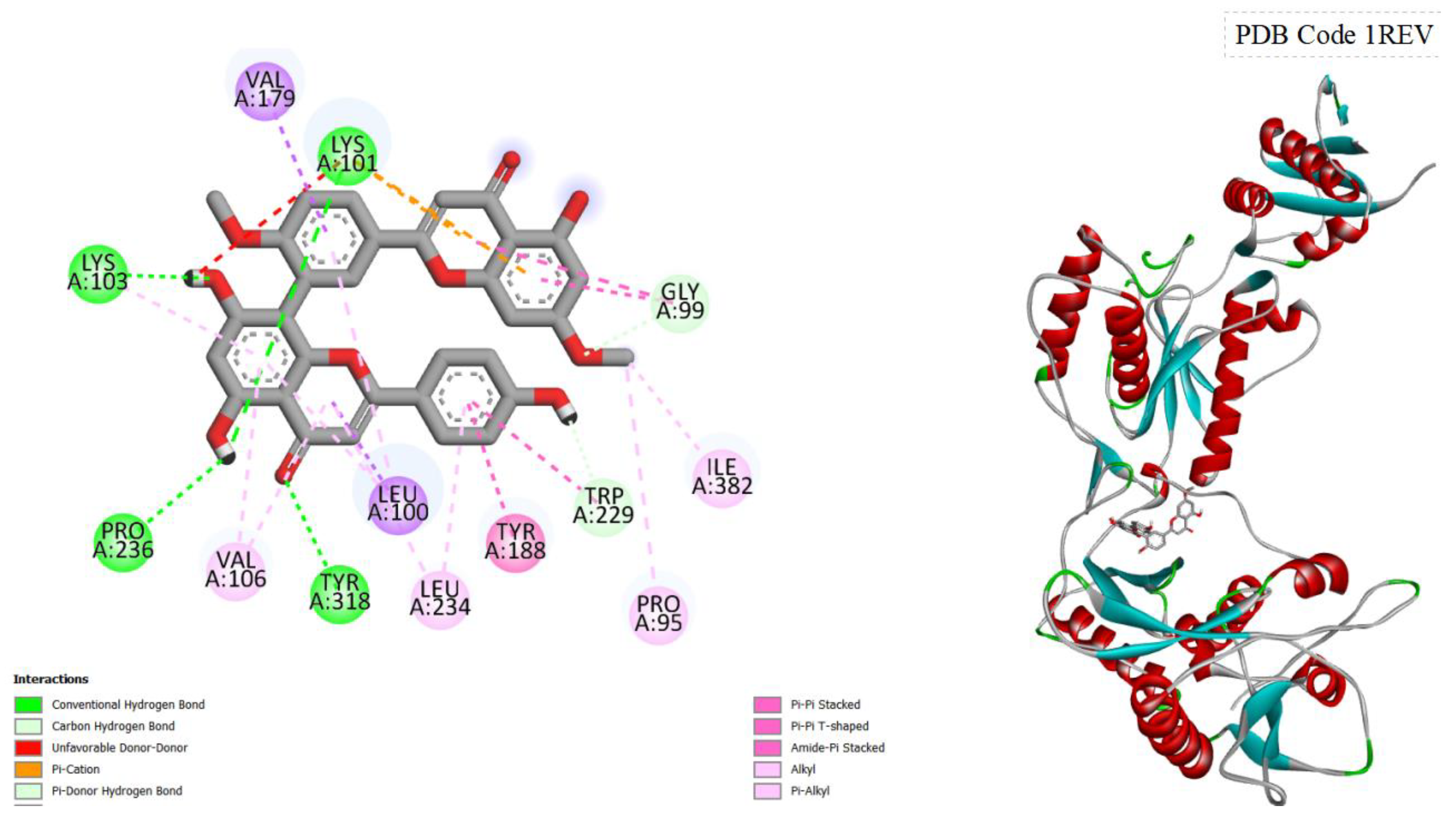
Figure 3.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Ginkgetin within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Ginkgetin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Ginkgetin.
Figure 3.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Ginkgetin within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Ginkgetin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Ginkgetin.
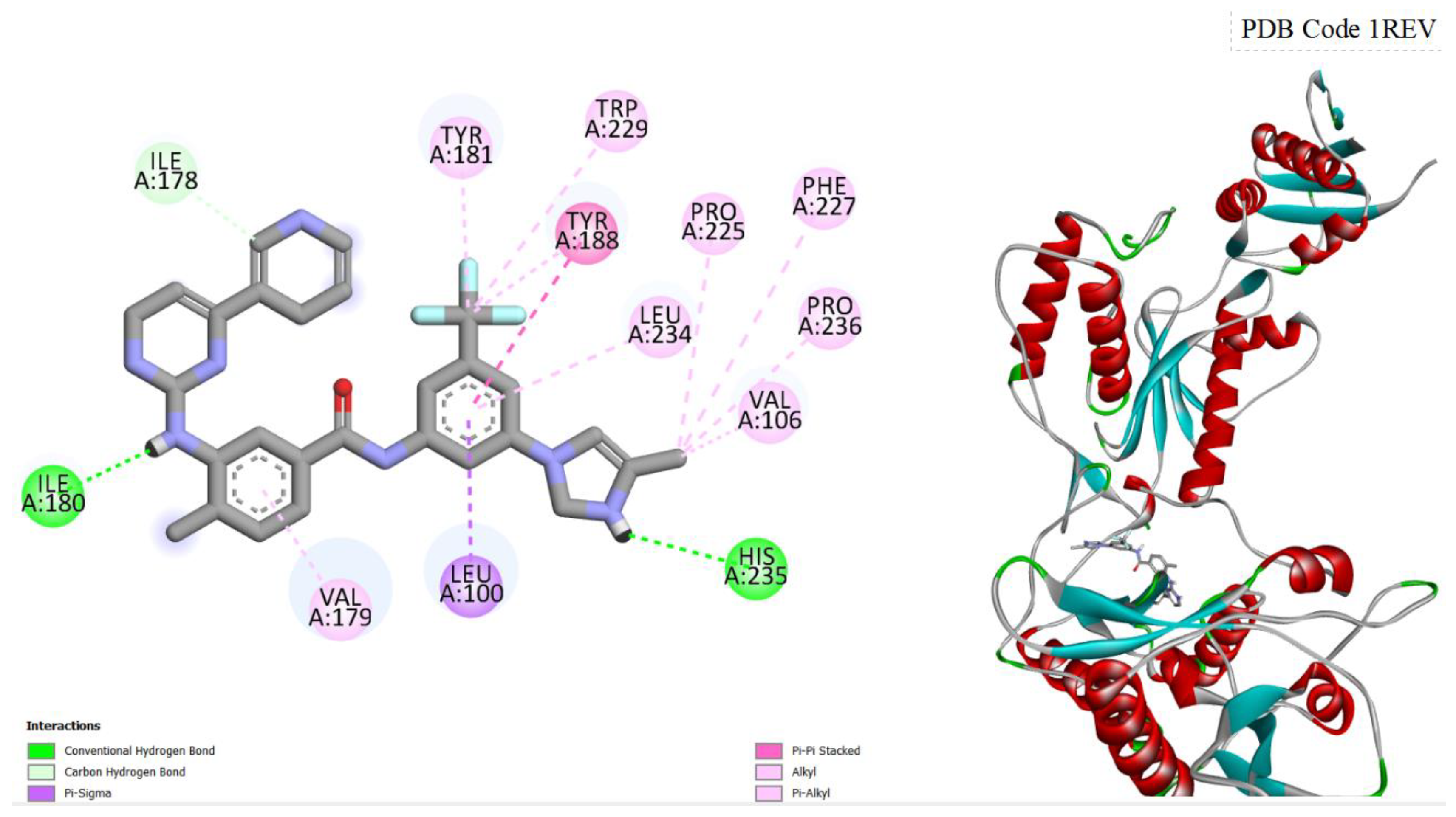
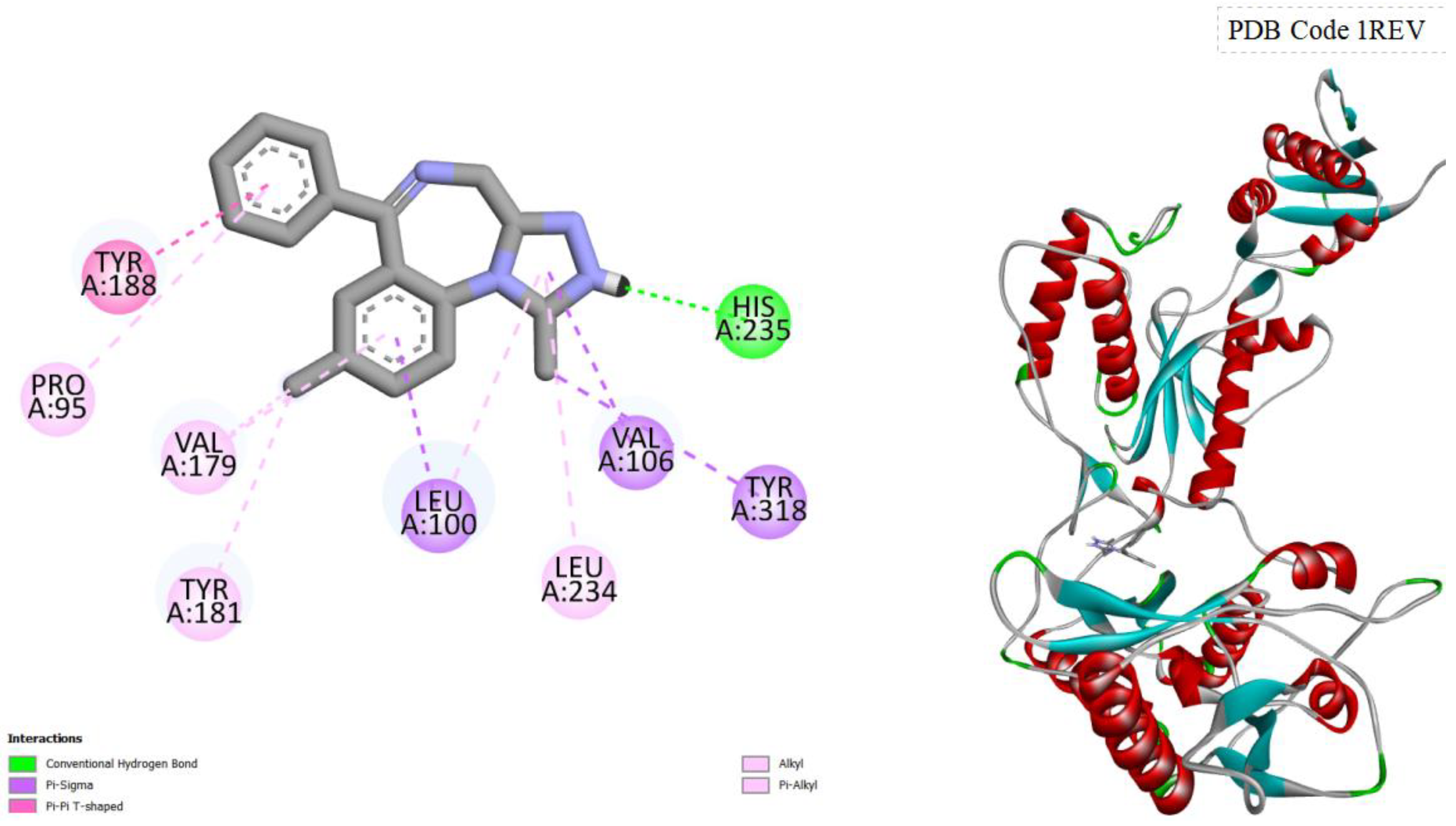
Figure 4.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Nilotinib within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Nilotinib. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Nilotinib.
Figure 4.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Nilotinib within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Nilotinib. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Nilotinib.
Figure 5.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Tetrophan within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Tetrophan . Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Tetrophan.
Figure 5.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Tetrophan within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Tetrophan . Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Tetrophan.
Figure 6.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Alprazolam within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Alprazolam. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Alprazolam.
Figure 6.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Alprazolam within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Alprazolam. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Alprazolam.
Figure 7.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Bafetinib within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Bafetinib. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Bafetinib.
Figure 7.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Bafetinib within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Bafetinib. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Bafetinib.
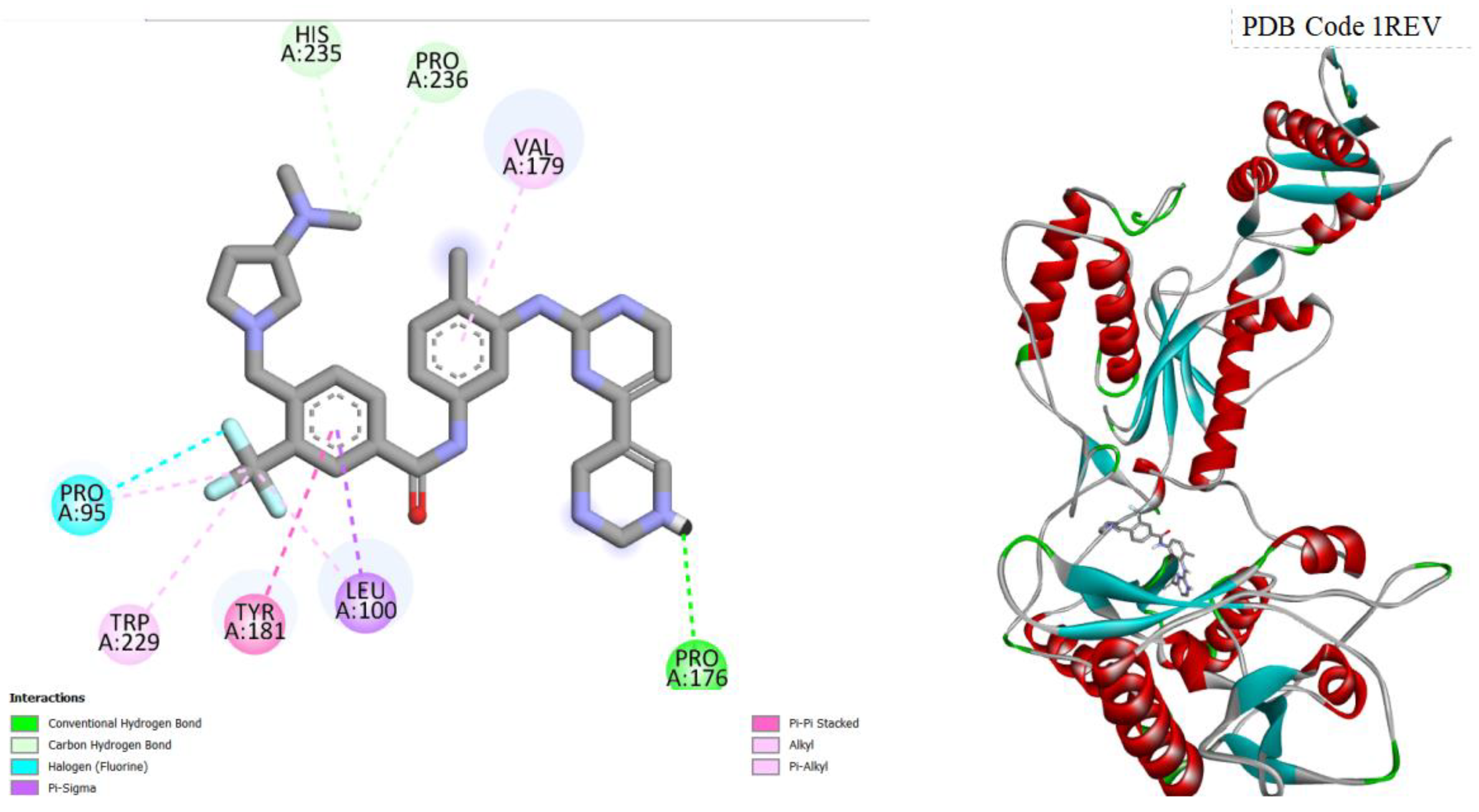
Figure 8.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Imatinib within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Imatinib . Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Imatinib.
Figure 8.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Imatinib within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Imatinib . Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Imatinib.
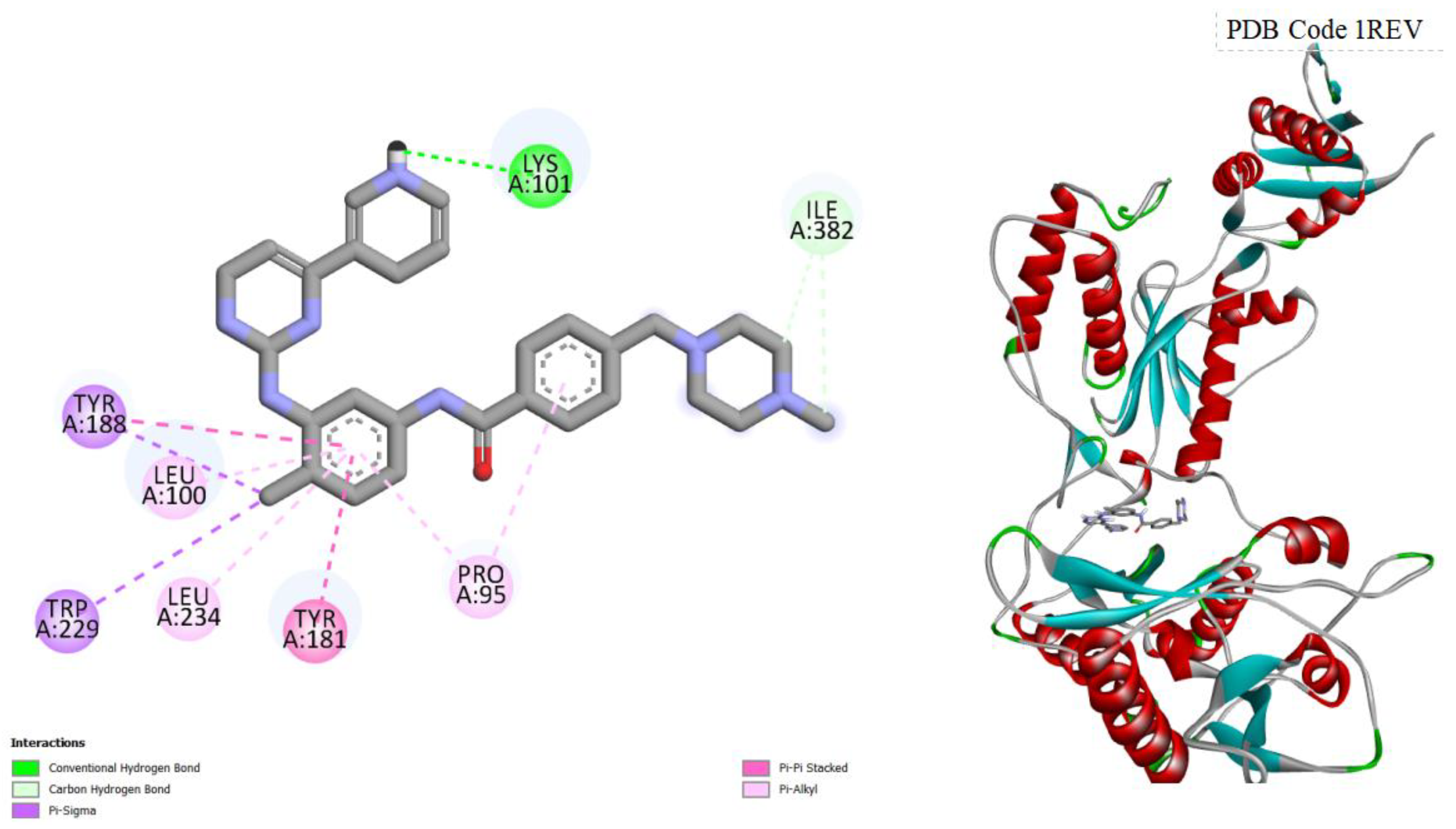
Figure 9.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Crystal Ligand TB9 (4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BE ZO[CD]AZULENE-1-THIONE) within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Crystal Ligand TB9. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Crystal Ligand TB9.
Figure 9.
displays the docking outcomes of HIV-1 Reverse Transcriptase in conjunction with Crystal Ligand TB9 (4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BE ZO[CD]AZULENE-1-THIONE) within the Ligand Binding Site, as analyzed by Autodock Vina through the Pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the target and Crystal Ligand TB9. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Crystal Ligand TB9.
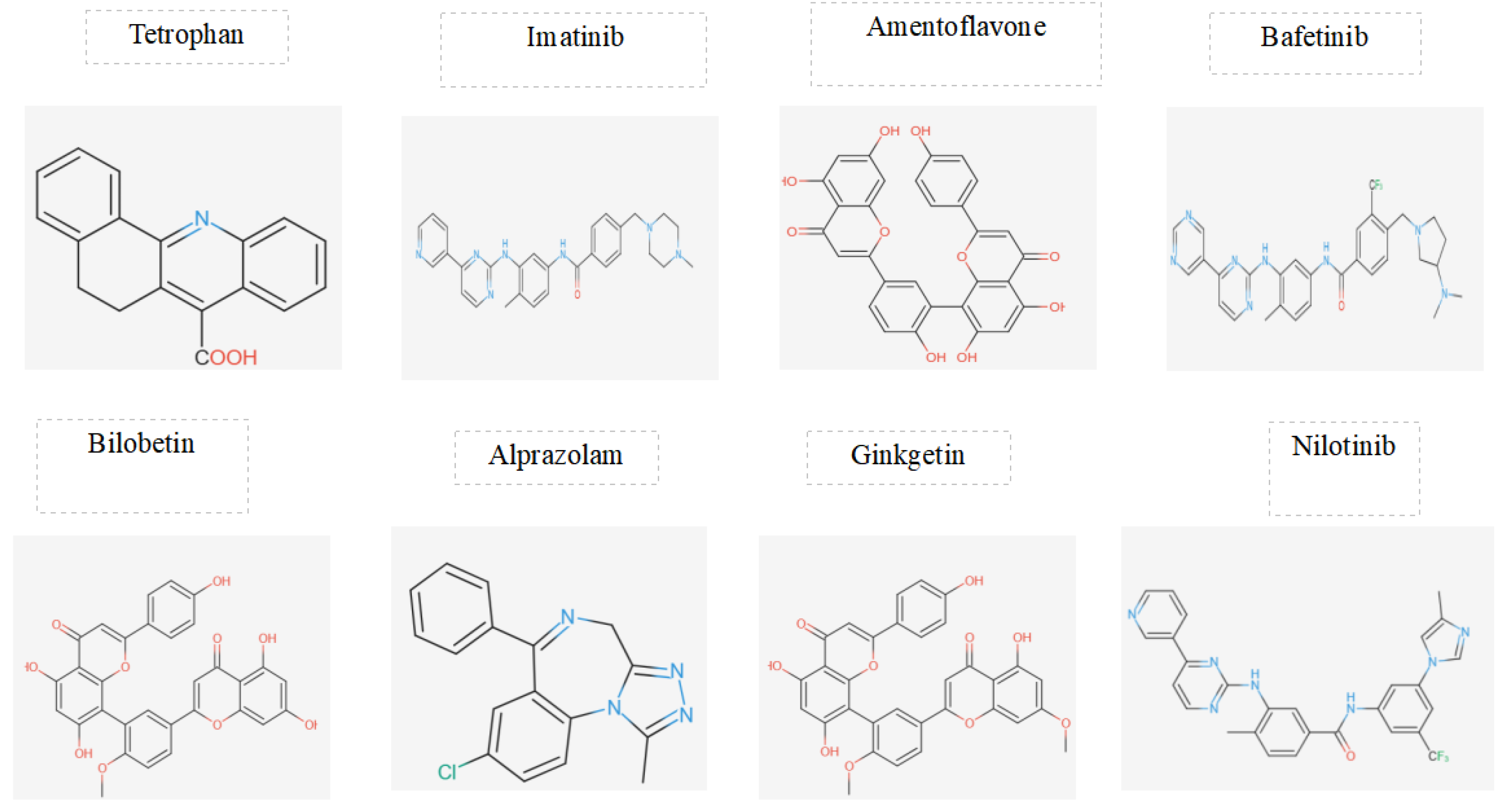
Figure 10.
Comparison 3D Chemical Structures of Best potential candidates investigated for HIV-1 Reverse Transcriptase.
Figure 10.
Comparison 3D Chemical Structures of Best potential candidates investigated for HIV-1 Reverse Transcriptase.
Table 1.
Comparison of Best Binding Energies scores of potential candidates investigated for HIV-1 Reverse Transcriptase, by Autodock Vina through Pyrx program in the Ligand Binding Site pocket.
Table 1.
Comparison of Best Binding Energies scores of potential candidates investigated for HIV-1 Reverse Transcriptase, by Autodock Vina through Pyrx program in the Ligand Binding Site pocket.
| Compounds |
Binding Energies scores (kcal/mol) |
| Crystal Ligand TB9 |
-10.7 |
| Tetrophan |
-11.2 |
| Imatinib |
-9.8 |
| Amentoflavone |
-12.5 |
| Alprazolam |
-11.1 |
| Bilobetin |
-12.0 |
| Bafetinib |
-10.4 |
| Nilotinib |
-11.8 |
| Ginketin |
-11.8 |
Table 2.
Comparison of predicted toxicity parameters of potential candidates investigated for HIV-1 Reverse Transcriptase, by pkCSM Server.
Table 2.
Comparison of predicted toxicity parameters of potential candidates investigated for HIV-1 Reverse Transcriptase, by pkCSM Server.
| Compounds |
AMES toxicity |
Max.
tolerated
dose (human) (log mg/kg/day) |
Oral Rat Acute Toxicity (LD50) (mol/kg) |
Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day) |
Hepatotoxicity |
Skin
Sensitisation |
T.
Pyriformis
toxicity
(log ug/L) |
Minnow toxicity
(log mM) |
| Tetrophan |
No |
0.544 |
2.057 |
1.789 |
yes |
no |
0.285 |
-0.631 |
| Imatinib |
No |
0.317 |
2.9 |
1.409 |
yes |
no |
0.285 |
2.089 |
| Amentoflavone |
No |
0.438 |
2.527 |
3.572 |
yes |
no |
0.285 |
2.685 |
| Bafetinib |
No |
0.383 |
2.85 |
1.321 |
yes |
no |
0.285 |
2.28 |
| Nilotinib |
No |
0.96 |
2.149 |
2.81 |
yes |
no |
0.285 |
5.136 |
| Bilobetin |
No |
0.437 |
2.56 |
2.217 |
yes |
no |
0.273 |
-0.003 |
| Alprazolan |
No |
0.373 |
2.347 |
0.729 |
yes |
no |
0.359 |
-0.448 |
| Ginkgetin |
No |
0.427 |
2.733 |
2.475 |
no |
no |
0.285 |
-2.351 |
Figure 11.
Comparison of predicted toxicity parameters of Amentoflavone investigated for HIV-1 Reverse.
Figure 11.
Comparison of predicted toxicity parameters of Amentoflavone investigated for HIV-1 Reverse.
Transcriptase, using Pro-Tox-II.
Figure 12.
Comparison of predicted toxicity parameters of Nilotinib investigated for HIV-1 Reverse.
Figure 12.
Comparison of predicted toxicity parameters of Nilotinib investigated for HIV-1 Reverse.
Transcriptase, using Pro-Tox-II.
4. Conclusions
In this preliminary study, a succinct molecular docking investigation was conducted using the Pyrx program, screening a multitude of compounds against the active site of HIV-1 reverse transcriptase. Employing the Virtual Screening library, the focus was on evaluating binding energetics scores, with an emphasis on identifying candidates possessing substantial Vina Scores. Eight compounds, namely Tetrophan, Imatinib, Amentoflavone, Alprazolam, Bilobetin, Bafetinib, Nilotinib, and Ginketin, emerged as promising candidates with noteworthy binding energies.
The identification of compounds that exhibit strong binding to HIV-1 reverse transcriptase holds potential for the development of antiretroviral drugs in the battle against HIV infection. Subsequently, the study delved into assessing the toxicity properties of these candidates using pKCSM.
Amentoflavone, a natural biflavonoid found in Ginkgo biloba and Hypericum perforatum, and Nilotinib, a tyrosine kinase inhibitor, displayed lower overall toxicity profiles compared to their counterparts.
Further scrutiny through Pro-Tox-II unveiled Amentoflavone's superior predictive LD50, lower toxicity classification, and diminished probabilities of hepatotoxicity, carcinogenicity, mutagenicity, and moderate immunotoxicity in contrast to Nilotinib. Conversely, Nilotinib, a pharmaceutical drug, exhibited higher probabilities of adverse effects in these categories. To summarize, Amentoflavone and Nilotinib emerge as promising candidates with reduced toxicity concerns.
Amentoflavone, being a natural substance, demonstrates a more favorable toxicity profile than Nilotinib, providing valuable insights for potential therapeutic applications. In vitro and in vivo studies would be necessary to assess their potential as antiretroviral agents.