1. Introduction
Equine herpesvirus type 1 (EHV-1) (
Varicellovirus equidalpha1) (ICTV,
https://ictv.global/taxonomy) causes rhinopneumonitis, abortion and neurological outbreaks (equine herpesvirus myeloencephalopathy, EHM), which occur independently or synchronously [
38]. Saxegaard (1966) first reported isolation of EHV-1 from a case with neurological signs [
41]. Cases of EHM have been reported with increasing frequency and severity in Europe and the USA [
1,
17,
20,
30]. EHM causes disastrous losses in the equine industry [
20,
25]. The neurological signs ranged from mild ataxia to paraplegia in horses and may be caused by ischemic degeneration resulting from vasculitis, hemorrhage and thrombosis [
22,
30]. A point mutation in the EHV-1 DNA polymerase gene (ORF30) was highly associated with EHM [
36]. The finding that field isolates that are both associated and not associated with EHV-1 neurological disease, and the results of experimental infection and molecular characterization studies support the hypothesis that EHM is strongly correlated with a single nucleotide polymorphism of EHV-1 DNA polymerase gene [
28,
42,
47,
49].
Ab4p is a plaque clone of the neurovirulent strain Ab4 that was isolated from EHM [
16,
43] and has been shown to cause nervous manifestations in experimentally infected horses, hamsters and mice [
3,
15,
16]. We have used hamsters and mice infection models to study the neurovirulence of EHV-1 [
23,
37]. EHV-1 might enter the brain through the nasal mucosa and olfactory bulbs [
18]. An EHV-1 infectious BAC (pAb4pBAC) was established from the Ab4p [
23]. Using this BAC system, UL24 encoded by ORF37 of EHV-1 was reported to be a neurovirulence factor in a mouse infection model [
23]. The Ab4p BAC and Ab4p attB, which is a virus after removal of the BAC sequence from Ab4p BAC, represent important tools for studying EHV-1 virulence and neuropathogenesis.
EHV-1 ORF76 encodes a tegument protein Us9 [
43]. EHV-1 US9 homologues are highly conserved in herpesviruses including herpes simplex virus-1 (HSV-1) [
14,
33,
34], varicella-zoster virus (VZV) [
11], pseudorabies virus (PRV) [
46], simian herpesvirus B [
24], bovine herpesvirus 1 (BHV-1) [
27], feline herpesvirus 1 (FHV-1) [
48], canine herpesvirus [
19], and herpes simplex virus -2 (HSV-2) [
12]. A critical component of the life cycle of alphaherpesviruses is anterograde spread, which is the spread of the virus from the neuron cell body to the axon terminus. US9 has been found to be essential for the anterograde spread of PRV [
31], Bovine herpesvirus 5 (BHV-5) [
10], and Bvoine herpesvirus 1 (BHV-1) [
9], as well as a determinant of neurovirulence and neuroinvasiveness of these viruses. However, it is unclear whether EHV-1 US9 has the same functions.
In the present study, we used Ab4p BAC to construct US9 deletion mutant and revertant viruses to study the functions of EHV1 US9. Our results suggest that US9 is necessary for the anterograde spread of EHV 1 from the olfactory epithelium to the olfactory bulbs.
2. Materials and Methods
2.1. Viruses and Cells
EHV-1 Ab4p BAC and Ab4p attB were used [
23]. Fetal equine kidney (FEK) cells were cultivated with using Dulbecco’s modified Eagle’s medium supplemented 5% fetal bovine serum (FBS). Madin-Darby bovine kidney (MDBK) and Rabbit kidney 13 (RK13) cells were cultivated with using MEM alpha supplemented 5% FBS.
2.2. Construction of Ab4pΔORF76 (US9 Deletion) and Revertant BACs
To construct Ab4pΔORF76 (US9 deletion), we used a RED/ET recombination system (counter-selection BAC modification system, Gene Bridges GmbH, Germany) as described previously [
2,
4,
23]. Primers used were shown in
Table 1. The rpsLneo cassette (rpsLneo gene) was amplified by PCR using primers 1 and 2 and rpsLneo template DNA. The PCR product was used to replace ORF76 in pAb4p BAC, resulting in a recombinant BAC termed pAb4pΔORF76 BAC. For Ab4pΔORF76R construction the rpsLneo gene in the pAb4pΔORF76 BAC was further replaced by ORF76 PCR product amplified using primers 3 and 4 resulting in recombinant BAC termed pAb4pΔORF76R BAC. The constructs were sequenced to confirm the mutation and reversion. Restriction enzyme BamHI digestion patterns of each BAC DNA were examined to confirm absence of extra changes in the viral genomes.
2.3. Recovery of Infectious Ab4p attB, Ab4pΔORF76 and Ab4pΔORF76R Viruses from BAC DNA
BAC DNA was extracted from Eschelichia coli harboring pAb4p BAC, pAb4pΔORF76 BAC and pAb4pΔORF76R BAC one by one using the Nucleo Bond BAC 100 kit (MACHEREY-NAGEL, USA). To recover Ab4p attB, Ab4pΔORF76 and Ab4pΔORF76R viruses without BAC sequence, BAC DNAs were treated with Gateway LR Clonase enzyme mix (Invitrogen, Life Technologies, USA) according to the manufacturer’s instructions. RK13 cells in a 24-well plate of 70-80% confluence were transfected with DNA using lipofectamine 2000 (Invitrogen, Tokyo, Japan) according to the manufacturer’s manual. After 5 days, supernatant was collected to inoculate MDBK cells. The MDBK cells were overlaid by Eagle’s MEM (EMEM) (Nissui, Tokyo, Japan) containing 1.5% of carboxymethylcellulose after 60 min of adsorption, and incubated for 4-5 days at 37 °C. The desired virus plaques were identified and selected under fluorescent microscopy using green fluorescence protein (GFP) fluorescence as a marker. Three rounds of plaque purification purified the recovered viruses.
2.4. Time Course of Viral Growth
The time course of viral growth was determined as described previously (Matsumura et al., 1998) [
32]. MDBK, RK13 and FHK monolayer cells in 24-well plates were inoculated with the indicated viruses at a MOI of 0.1. After 1.5 hr of adsorption, cells were washed three times with EMEM and incubated at 37°C in a 5% CO2 atmosphere in 0.5 mL/well of EMEM with 5% fetal calf serum. At the indicated times, cells were scraped together with culture fluids and centrifuged. The supernatants were used as the extracellular fluid samples. Sedimented cells were washed twice with EMEM, resuspended in 0.5 mL of EMEM, frozen and thawed three times and centrifuged. The obtained supernatants were used as the intracellular fluid samples. Extracellular and intracellular fluid samples were titrated for viral infectivity by plaque assay as previously described [
23].
2.5. Virus Growth Kinetics in Mouse Neurons
Growth kinetics of the viruses were measured in CX (M) mouse neurons (Sumitomo Bakelite, Tokyo, Japan). Neurons were cultured in 24-well plates coated with poly-L-lysine in neuron culture medium (Sumitomo Bakelite, Tokyo, Japan) and infected with the various viruses at 1 MOI. At the indicated times, the supernatant and cells were separately harvested and the titers of examined viruses were calculated using plaque assay as described previously [
49].
2.6. Analysis of Transcription Kinetics by Real-Time RT-PCR
The transcription activity of ORF76 was evaluated by infecting MDBK cells with Ab4p attB, Ab4pΔORF76 and Ab4pΔORF76R at 1 MOI. Infected cells were harvested at 0, 2, 4 and 8 hr post infection. Total RNA was isolated using Nucleospin RNA kit (MACHEREY-NAGEL, USA), 1.5 µg of RNA was heated at 95 °C for 5 min for denaturation, combined with reverse transcriptase master mix (TOYOBO, Osaka, Japan). The mixture was incubated at 30 °C for 10 min, 42 °C for 40 min and then stopped by heating at 99 °C for 5 min. The real-time PCR was carried out in a Thermal Cycler Dice Real Time System (TAKARA) using 12.5 µL of SYBR Premix Ex Taq (TAKARA, Shiga, Japan), 10 µM of ORFs 76, 75 and 67 primers (
Table 1) and 10 ng of cDNA. Relative quantities were measured with the ΔΔCt method [
29].
2.7. Animal Experiments
Briefly, one hundred (four-week-old) specific pathogen free (SPF) male CBA/N1 mice (Japan SLC Corporation, Shizuoka, Japan) were equally divided into four groups; control, Ab4p attB, Ab4pΔORF76 and Ab4p ΔORF76R. Each group were inoculated with 1 × 105 pfu per head of the corresponding virus by the intranasal route. Mice body weight and behavior were observed 3 days before the inoculation to the end of the experiment. For virus isolation and DNA detection, two mice from each group were euthanized every day from 1 to 10 dpi. The brain, olfactory bulbs and lungs were used for virological assay.
2.8. Preparation of Tissues for PCR and Virus Titration
Tissues from euthanized animals were homogenized in EMEM at 10% (w/v), centrifuged at 3,000 rpm for 10 minutes. The supernatant was 10-fold serially diluted in EMEM. A 24-well plate with a confluent MDBK monolayer was inoculated with of 0.1 mL supernatant per well. Plaque assay was used for virus titration. The detection limit in the organ homogenates was 1x102 pfu per gram. DNA was extracted from the organs of infected mice with a Sepagene kit (Sanko Junyaku, Japan). Viral DNA was detected by PCR with the primers for ORF76 and the rpsLneo gene.
2.9. Histopathology and Immunohistochemistry
Tissues were collected, fixed in 10% buffered formalin, dehydrated and embedded in paraffin wax by routine methods, sectioned at 5 μm, stained with hematoxylin and eosin (HE), and examined by light microscopy. Paraffin wax sections were immunolabelled with EHV-1 rabbit antiserum by the avidin-biotin-complex (ABC) immunoperoxidase method with ABC kits (Vector Laboratories, Burlingame, CA, USA) as described previously [
51]. The primary antibody was EHV-1 antiserum (1:1000, Veterinary Microbiology Laboratory at Gifu University) followed by application of a secondary antibody (biotinylated anti-rabbit IgG, DAKO Cytomation, USA). The liquid DAB Substrate Chromogen System (DAKO Cytomation, USA) was used as chromogen and hematoxylin as a counterstain. Tissue sections from an EHV-1 infected mice and sera from a non-immunized rabbit and mice were used as controls.
2.10. Immunofluorescence
Immunofluorescence assay was performed on paraffin wax sections as described previously [
40]. The primary antibody was polyclonal US9 guinea pig serum (1:500, prepared in this study) followed by application of a secondary antibody (anti-guinea pig FITC, Sigma Aldrich, USA). Fluorescent image analysis was examined using Keyence Biozero system (Keyence, Tokyo, Japan).
2.11. Production of Anti-EHV-1 US9-Specific Polyclonal Antibody
ORF76 (US9) was amplified using primers 5 and 6 (
Table 1) which, introduced an EcoRI site directly upstream of the start codon, and a NotI site directly downstream of the US9 stop codon, respectively. The amplified PCR fragment was cloned into the EcoRI and NotI sites of pGEX-6P-1 generating pGST-US9 plasmid. Expression of GST-US9 fusion protein was induced by adding IPTG (isopropyl-β-D-1-thiogalactopyranoside, Takara, Shiga, Japan) to a culture of
E. coli BL21 transformed with the pGST-US9 plasmid. The fusion protein was purified using glutathione-Sepharose 4B beads (GE Healthcare UK Ltd., UK). The purified US9 was made into an emulsion by adding an equal volume of TiterMax
® Gold adjuvant (Funakoshi, Tokyo, Japan) and used to immunize two guinea pigs (Japan SLC Corporation, Shizuoka, Japan). Serum was collected before and after four subcutaneous applications of 100 µg protein at 1-week intervals.
2.12. Western Blotting
For Western blot analyses, RK13 cells were infected with viruses at a multiplicity of 5 pfu/cell and incubated at 37 °C for 1–18 h. Infected cells were harvested and pelleted by centrifugation at 14,000 rpm for 1 min in an Eppendorf centrifuge. Pellets were washed twice with phosphate buffered saline (PBS), resuspended in 100 µL of PBS, mixed with the same volume of sample buffer and heated at 95°C for 5 min. Then the samples were separated by SDS-PAGE and electrotransferred to nitrocellulose membranes (Millipore, MA, USA). The blots were blocked with 5 % low-fat milk in Tris-buffered saline (TBS-T; 10 mM Tris/HCl, pH 8.0, 150 mM NaCl, 0.25 % Tween 20) and incubated for 1 hr with guinea pigs antisera against the US9 gene products at dilutions of 1:1000 in TBS-T. Bound antibody was detected with peroxidase-conjugated anti-guinea pig antibodies (Bethyl, Texas, USA) and visualized by chemiluminescence (Amersham, Japan) and recorded on X-ray films.
2.13. Statistical Analysis
Data was analyzed using One Way ANOVA with Dunn’s multiple comparison test and significance was set at P<0.05 using GraphPad Prism 7 software.
3. Results
3.1. Construction of ORF76 (US9) Deletion Mutant and Its Revertant Virus
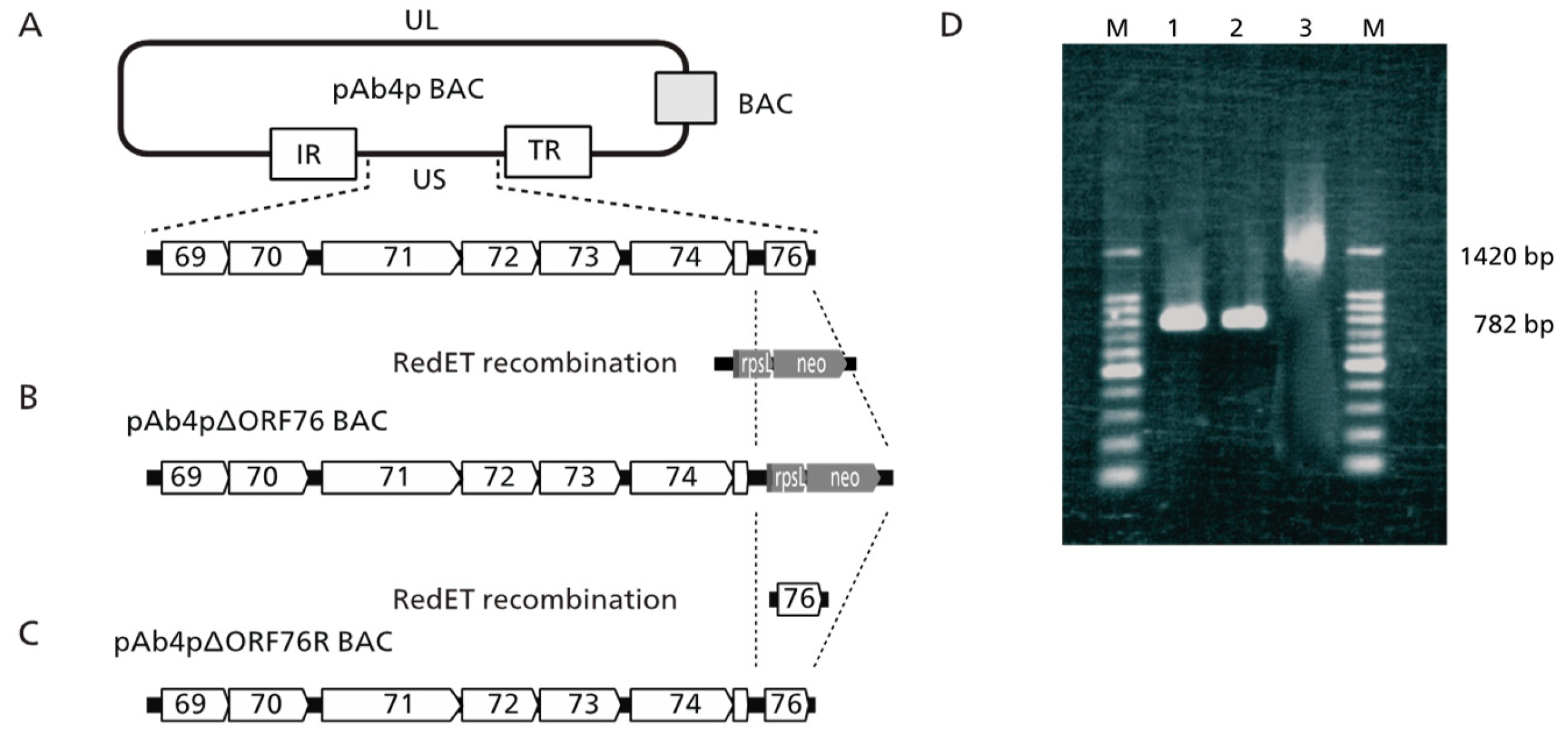
The ORF76 (US9) deletion mutant was constructed using pAb4p BAC with Red mutagenesis molecular recombination. The resulting ORF76 (US9) deletion mutant and revertant BAC plasmids were designated pAb4pΔORF76 (
Figure 1 B) and pAb4p∆ORF76R BAC (
Figure 1C). The correct replacement and genotypes of the generated viruses were confirmed by PCR and sequencing. The 782 bp product, which indicated the presence of ORF76 was detected in cells, infected with Ab4p attB and Ab4pΔORF76R viruses by PCR (
Figure 1D lanes 1 and 2). A PCR product of 1420bp was detected in cells infected with ORF76 deletion mutant Ab4pΔORF76 due to presence of rpsLneo cassette instead of ORF76 (
Figure 1D lane 3). Absence of extra changes in the viral genome were confirmed by BamHI digestion patterns of each BAC DNA (data not shown).
3.2. Growth of ORF76 (US9) Deletion Mutant in Cultured Cells and Mouse Neurons
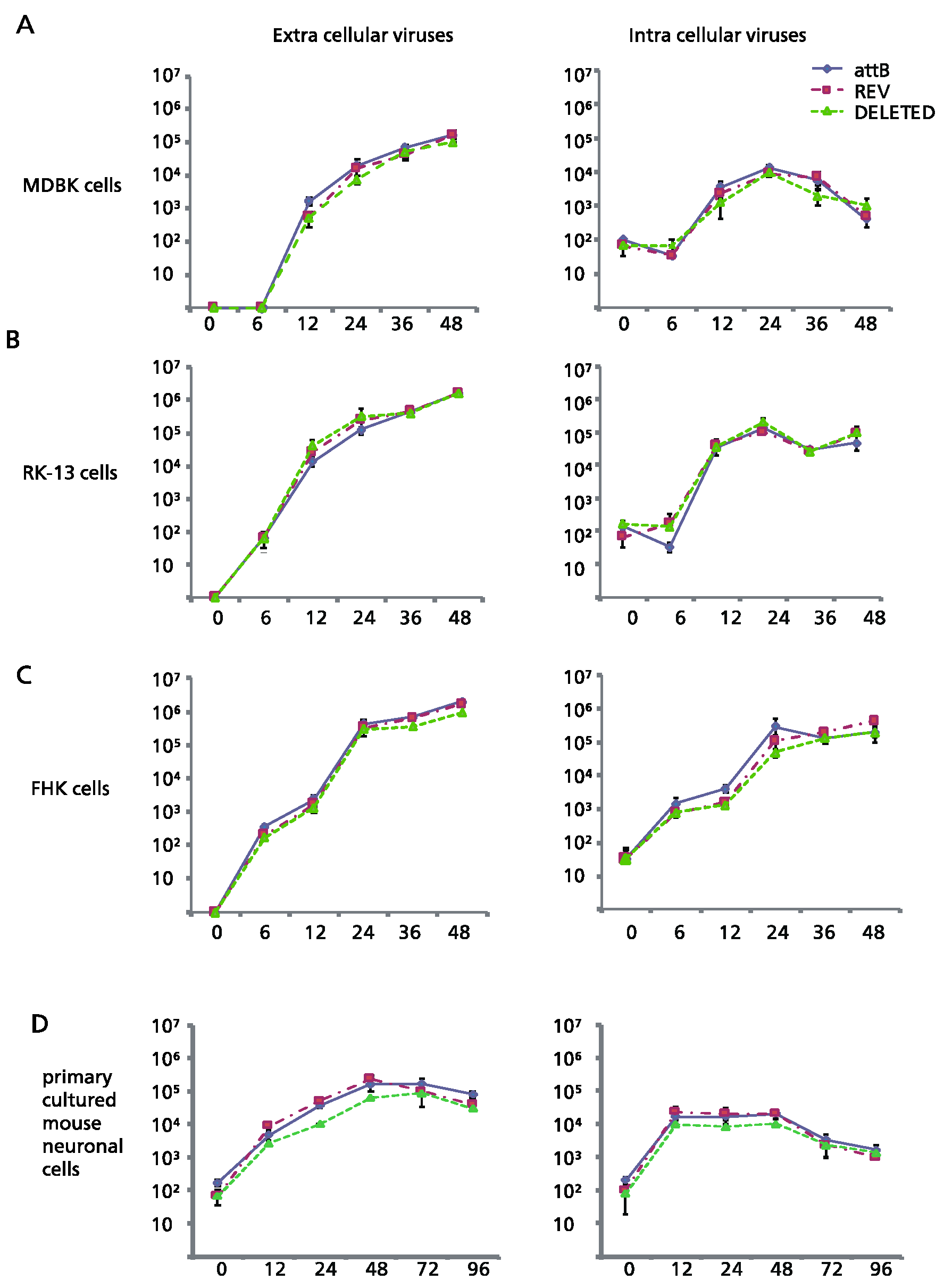
Plaque sizes of the deletion mutant Ab4pΔORF76, the revertant Ab4p∆ORF76R and the neuropathogenic strain Ab4p attB in cultured cells were similar. The infectious progeny yield and time course of Ab4p∆ORF76 were almost the same as those of Ab4p attB and Ab4p∆ORF76R in MDBK cells (
Figure 2A), RK13 cells (
Figure 2B) and FHK cells (
Figure 2C).
In addition, the growth kinetics of Ab4p∆ORF76 in primary cultured mouse neurons were similar to those of Ab4pΔORF76 R and Ab4p attB (
Figure 2D). These results show that ORF76 (US9) is not essential for infectious virus production in cultured cells or mouse neurons.
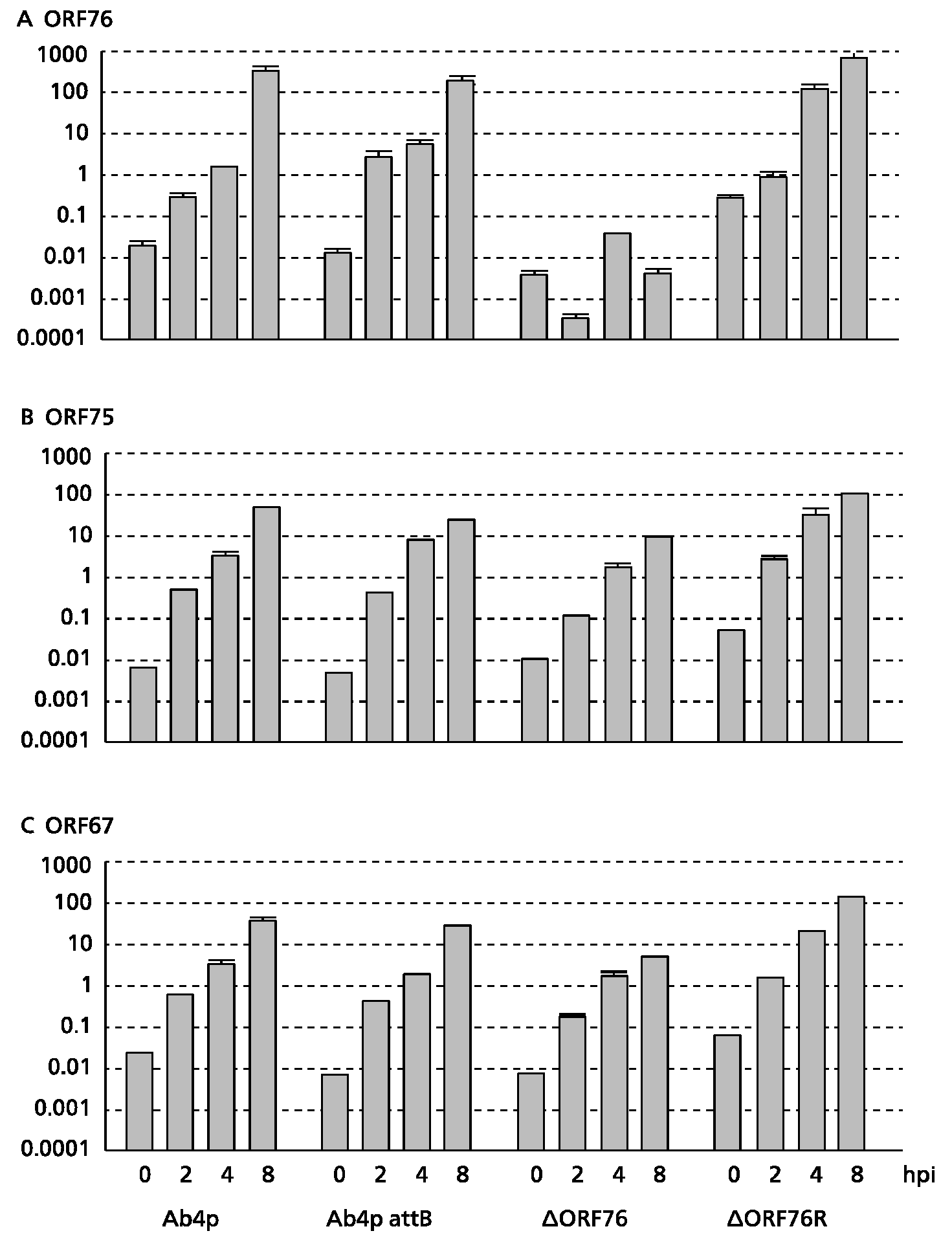
3.3. Effect of ORF76 Deletion on Transcription Activities of Other Genes
The expression levels of the β-actin gene (a control) in MDBK cells were the same for all viruses. No ORF76 transcripts were detected in cells infected with the Ab4p∆ORF76 deletion mutant (
Figure 3C). ORF76 deletion had no effect on the transcription levels of two neighboring ORFs, ORF75 (
Figure 3A) and ORF67 (
Figure 3B) or a distant ORF (ORF30) (data not shown).
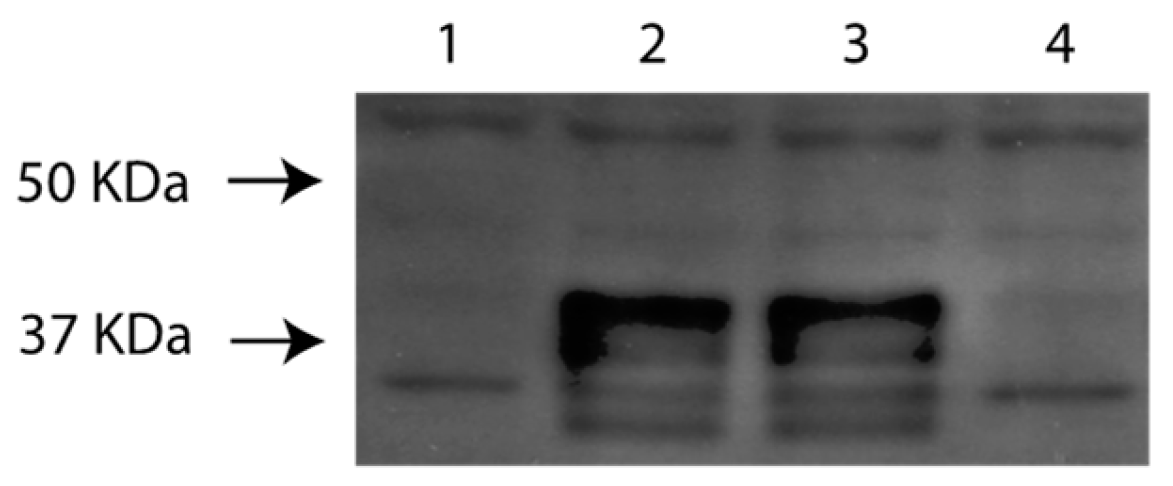
3.4. Identification and Initial Characterization of EHV-1 US9 Protein
The EHV-1 US9-specific antibody reacted with several bands with approximate molecular masses of 35 to 42-kDa in (
Figure 4) on western blots. The EHV-1 Us9-specific bands were absent in mock infected cells and cell infected with Ab4pΔORF76.
3.4. Pathogenicity of ORF76 Deletion Mutant Virus in Mice
Ab4pΔORF76 inoculated mice were apparently healthy, sound and gained body weight with no nervous manifestations throughout the experiment period (
Figure 5). Nervous manifestations including arching back, hyperactivity and paralysis were observed in mice inoculated with Ab4p attB starting from fifth day post infection (dpi) and Ab4pΔORF76R starting from sixth day post infection.
Viruses were recovered from the infected mice lungs, 1 to 5 dpi for Ab4p attB, 1 to 4 dpi for Ab4p∆ORF76R and 1 to 3 dpi for Ab4p∆ORF76. Ab4p attB and Ab4p∆ORF76R viruses could be recovered from olfactory bulbs and brain of infected mice from 3 to 7 dpi. While Ab4p∆ORF76 virus was not recovered from olfactory bulbs and brain at all (
Table 2).
Ab4p attB and Ab4pΔORF76R viruses DNA were detected in olfactory bulbs, brains and lungs of inoculated mice from 1 dpi. While Ab4p∆ORF76 DNA was not detected at all in olfactory bulbs and brains of mice inoculated with Ab4pΔORF76 (
Table 3).
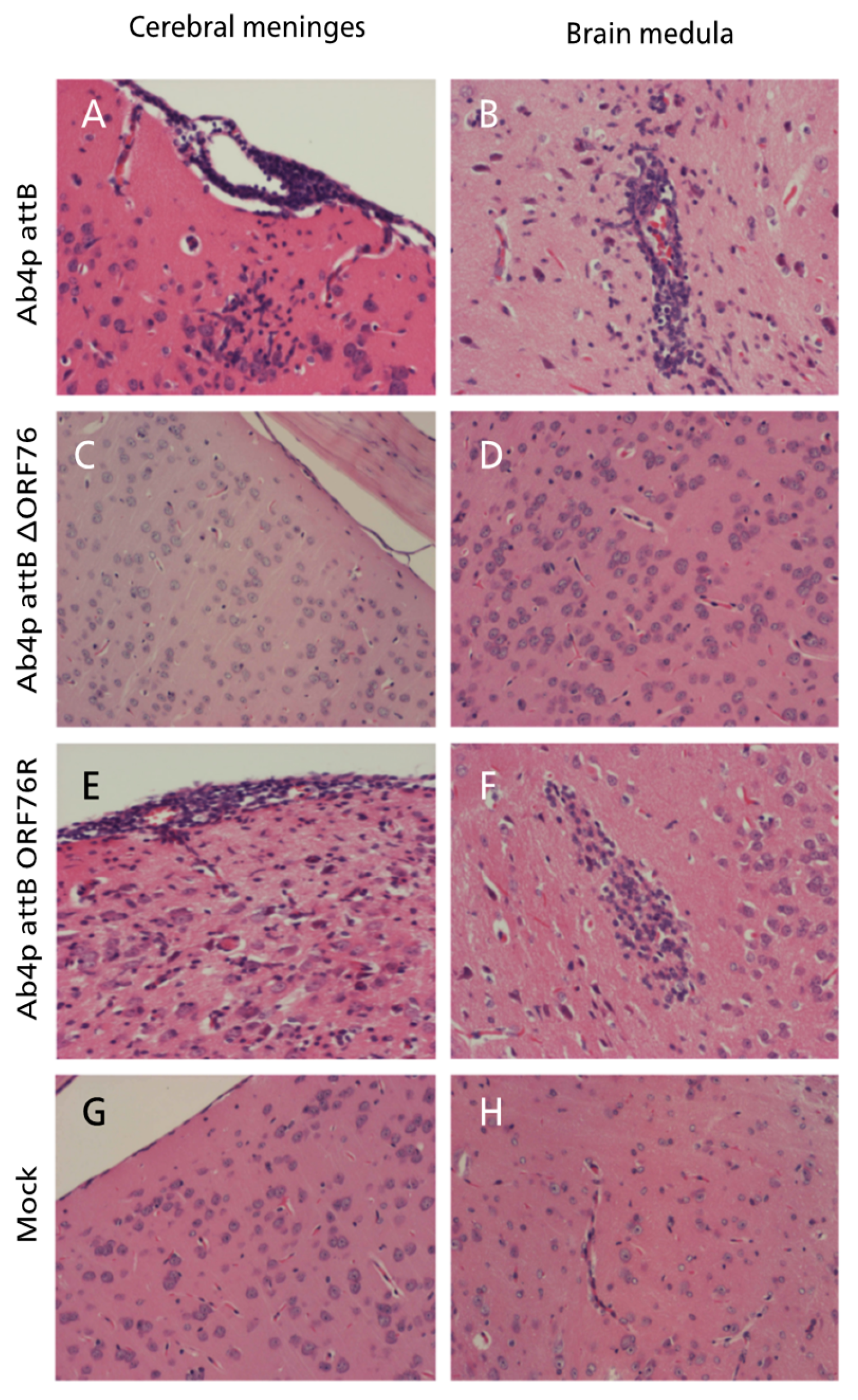
At necropsy, no gross abnormalities were observed in any of the inoculated mice. Histopathological examination of mice infected with each virus revealed mild rhinitis, multiple foci of necrosis of the olfactory epithelial cells along with inflammatory cells infiltrates within the mucosa admixed with the desquamated epithelial cells in the nasal cavity. The olfactory bulbs of mice inoculated with Ab4p attB or Ab4p∆ORF76R showed typical encephalitis at 4 and 6 days post infection respectively, while no pathological changes were recorded in mice inoculated with Ab4pΔORF76. The brains of mice infected with Ab4pΔORF76 showed no significant pathological changes, while the brains of Ab4p attB and Ab4p∆ORF76R infected groups showed lymphocytic meningoencephalitis, consisting of neuronal degeneration and necrosis, perivascular aggregates of mononuclear cells and varying degrees of focal or diffuse gliosis (
Figure 6). No abnormalities were found in other organs except for interstitial pneumonia in all virus-infected groups.
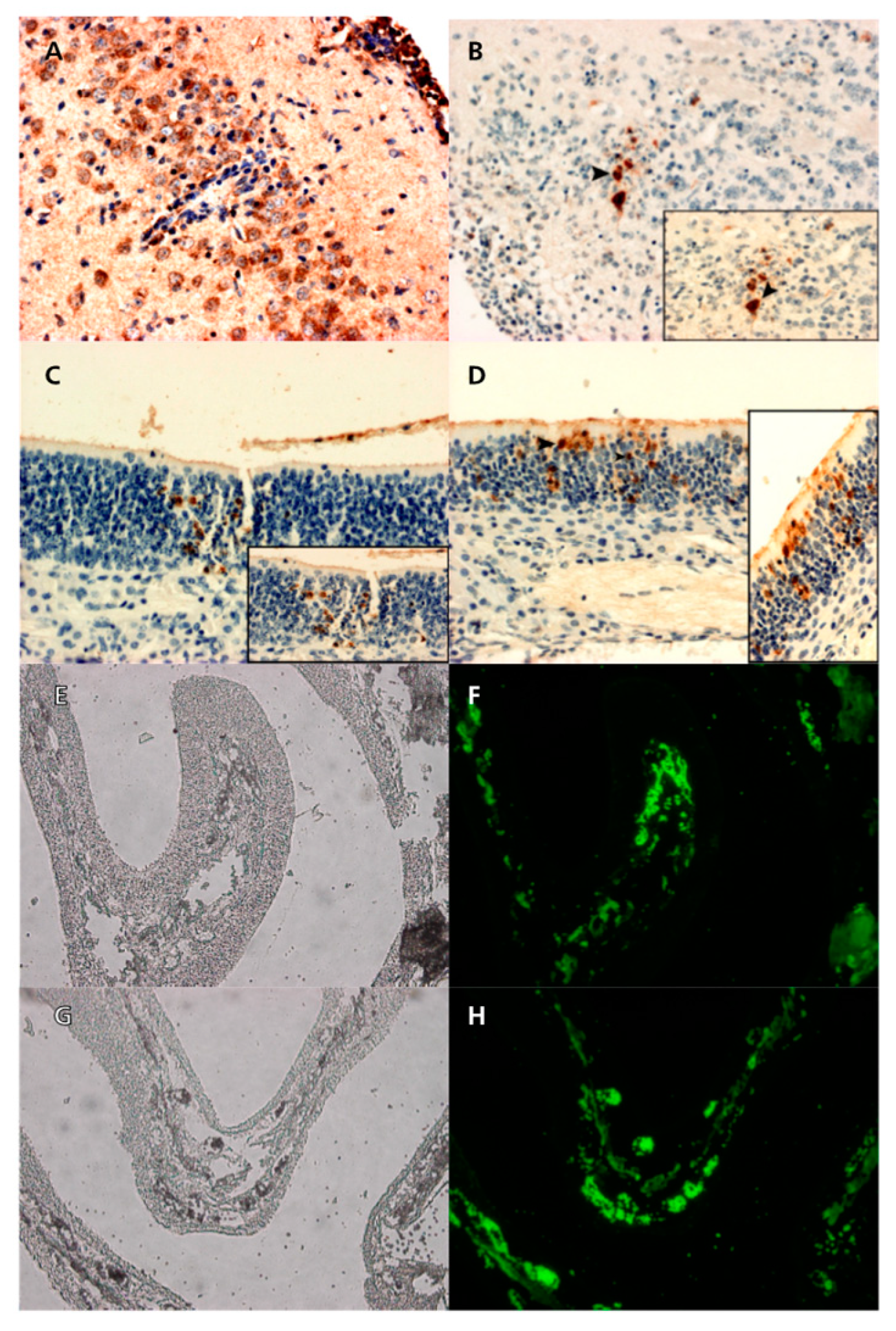
EHV-1 antigens were detected by immunohistochemical and immunofluorescence reactions in the cytoplasm of degenerating olfactory epithelial cells in the nasal cavity of mice infected with each of Ab4p attB, Ab4p∆ORF76R and Ab4pΔORF76. EHV-1 antigen was detected in the brains and olfactory bulbs of mice inoculated with Ab4p attB or Ab4p∆ORF76R (
Figure 7), but not in those of mice infected with Ab4p∆ORF76 (data not shown).
4. Discussion
US9 gene is conserved in most of the alphaherpesviruses. US9 is not essential for replication in cell culture and plays no role in the cell-to-cell spread of HSV-1 [
21,
33], PRV [
7] and BHV-5 [
10]. Our results show that EHV-1 ORF76 (US9) is also not involved in viral replication in cell cultures (RK13, MDBK and FHK) or cultivated primary mouse neuronal cells, and has no role in cell-to-cell spread. The histopathological and immunohistochemical results of this study prove that US9 has an important role in EHV-1 anterograde neuronal transport in the olfactory pathway and so EHV-1 neuropathogenesis.
The EHV-1 US9 gene is 660 nucleotides and encodes a protein of 219 amino acid and 22.287 kDa. Using Western blot assay, US9 polyclonal antibodies reacted with several polypeptides between 35 and 42 kDa. The EHV-1 US9 specific bands were absent in in mock-infected cells and cells infected with the EHV-1 US9 deleted mutant. The molecular masses of the EHV-1 US9-specific bands were higher than expected, which suggests that EHV-1 US9 protein had undergone posttranslational modifications. The US9 ORFs of HSV-1, PRV, BHV-5, and BHV-1 encode proteins with predicted molecular weights of 10.0, 10.8, 13.7, and 14.7 kDa respectively. However, US9-specific polyclonal antibodies precipitated several proteins with molecular weight ranges of 12-25 kDa in HSV-1, 17-20 kDa in PRV, 15-20 kDa in BHV-5 and 28-32 kDa in BHV-1. In each of these viruses, PRV, HSV-1, BHV-5 and BHV-1 US9 is phosphorylated [
5,
6,
9]. It is believed that phosphorylation alters the charge of the SDS coating of the protein, which slows protein migration in the gel, so protein seems of a higher, apparent molecular weight [
39]. Our finding that the EHV-1 US9 has multiple bands of apparent higher molecular mass than expected supports the notion that the alphaherpesvirus Us9 proteins are phosphorylated and suggests that phosphorylation of Us9 has a functional role.
The role of EHV-1 ORF76 (US9) gene in vivo was evaluated in a CBA/N1 mouse infection model. In mice intranasally inoculated with Ab4p attB and Ab4pΔORF76R, the viruses spread in the olfactory pathway and caused histopathological lesions similar to those previously described in wild-type EHV-1 intranasal inoculated mice [
18,
23,
51]. In contrast, in mice intranasally inoculated with the EHV-1 US9 deletion mutant (Ab4pΔORF76), the virus failed to invade the CNS as indicated by several lines of evidence including absence of nervous manifestations, normal body weight gain, no mortalities, normal histopathological finding of the olfactory bulbs and brain, and no virus antigen detection within the CNS by viral isolation, PCR or immunostaining. These results indicate that the ORF76 (US9) plays an essential role in the anterograde spread of EHV-1 virus
In mice intranasally inoculated with BHV-1 and BHV-5 [
8,
26] and EHV-9 [
15,
35], after initial replication in the olfactory mucosa, the viruses are transported to the olfactory bulbs, and then to deeper tissues of the brain through olfactory tract neurons. In the nasal mucosa, the Ab4pΔORF76 replicated efficiently just like Ab4p attB and Ab4pΔORF76R viruses. Immunostaining detected Ab4pΔORF76 antigen in the olfactory epithelium receptor neurons, but not in the bulbs, indicating that Ab4pΔORF76 was not transported to the olfactory bulbs. Our result supports numerous other reports which have shown that Us9 has a role in the anterograde spread of alphaherpesviruses [
7,
10,
13,
44,
45]. Roles of Us9 in vivo might be conserved among alphaherpesviruses.
5. Conclusions
The gene product of EHV-1ORF76 (US9) is not essential for replication of EHV-1 in cell culture or cultivated neurons and is not involved in EHV-1 cell-to-cell spread, but it plays an important role in the anterograde spread of EHV-1.
Author Contributions
Conceptualization, H.F. and H.H.; methodology, M.N., E.N., N.F. and S.K.; software, M.N., E.N., N.F. and S.K.; validation, M.N., E.N., N.F. and S.K.; formal analysis, H.F.; investigation, M.N.; resources, H.F., T.Y., and K.O.; data curation, M.N., N.F., S.K. and H.F; writing—original draft preparation, M.N.; writing—review and editing, A.E., A.S. and H.F.; visualization, M.N., E.N., N.F. and S.K; supervision, H.F., T.Y., K.O. and H.H.; project administration, K.O. and H.F; funding acquisition, H.F All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Japan Society for the Promotion of Science (JSPS), grant numbers 17380181, 21380179 and 18H02343.
Institutional Review Board Statement
The animal study protocol was approved by the Animal Care and Use Committee of GIFU UNIVERSITY (certification number 13045, 15th May, 2014).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
M.N. would like to thank the Egyptian Ministry of Higher Education for supporting him to study abroad.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Allen, G.P. Development of a Real-Time Polymerase Chain Reaction Assay for Rapid Diagnosis of Neuropathogenic Strains of Equine Herpesvirus-1. Journal of Veterinary Diagnostic Investigation 2007, 19, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Ata, E.B.; Zaghawa, A.; Ghazy, A.A.; Elsify, A.; Abdelrahman, K.; Kasem, S.; Nayel, M. Development and characterization of ORF68 negative equine herpes virus type–1, Ab4p strain. Journal of Virological Methods 2018, 261, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Awan, A.R.; Chong, Y.-C.; Field, H.J. The pathogenesis of equine herpesvirus type 1 in the mouse: a new model for studying host responses to the infection. Journal of General Virology 1990, 71, 1131–1140. [Google Scholar] [CrossRef]
- Badr, Y.; Okada, A.; Abo-Sakaya, R.; Beshir, E.; Ohya, K.; Fukushi, H. Equine herpesvirus type 1 ORF51 encoding UL11 as an essential gene for replication in cultured cells. Archives of Virology 2018, 163, 599–607. [Google Scholar] [CrossRef]
- Brandimarti, R.; Roizman, B. Us9, a stable lysine-less herpes simplex virus 1 protein, is ubiquitinated before packaging into virions and associates with proteasomes. Proceedings of the National Academy of Sciences 1997, 94, 13973–13978. [Google Scholar] [CrossRef] [PubMed]
- Brideau, A.; Banfield, B.W.; Enquist, L. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. Journal of Virology 1998, 72, 4560–4570. [Google Scholar] [CrossRef]
- Brideau, A.; Card, J.; Enquist, L. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. Journal of Virology 2000, 74, 834–845. [Google Scholar] [CrossRef]
- Chowdhury, S.; Lee, B.; Mosier, D.; Sur, J.; Osorio, F.; Kennedy, G.; Weiss, M. Neuropathology of bovine herpesvirus type 5 (BHV-5) meningo-encephalitis in a rabbit seizure model. Journal of Comparative Pathology 1997, 117, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Mahmood, S.; Simon, J.; Al-Mubarak, A.; Zhou, Y. The Us9 gene of bovine herpesvirus 1 (BHV-1) effectively complements a Us9-null strain of BHV-5 for anterograde transport, neurovirulence, and neuroinvasiveness in a rabbit model. Journal of Virology 2006, 80, 4396–4405. [Google Scholar] [CrossRef]
- Chowdhury, S.; Onderci, M.; Bhattacharjee, P.; Al-Mubarak, A.; Weiss, M.; Zhou, Y. Bovine herpesvirus 5 (BHV-5) Us9 is essential for BHV-5 neuropathogenesis. Journal of Virology 2002, 76, 3839–3851. [Google Scholar] [CrossRef]
- Davison, A.J.; Scott, J.E. The complete DNA sequence of varicella-zoster virus. Journal of General Virology 1986, 67, 1759–1816. [Google Scholar] [CrossRef] [PubMed]
- Dolan, A.; Jamieson, F.E.; Cunningham, C.; Barnett, B.C.; McGeoch, D.J. The genome sequence of herpes simplex virus type 2. Journal of Virology 1998, 72, 2010–2021. [Google Scholar] [CrossRef]
- Enquist, L.; Tomishima, M.; Gross, S.; Smith, G. Directional spread of an α-herpesvirus in the nervous system. Veterinary Microbiology 2002, 86, 5–16. [Google Scholar] [CrossRef]
- Frame, M.C.; McGeoch, D.J.; Rixon, F.J.; Orr, A.C.; Marsden, H.S. The 10K virion phosphoprotein encoded by gene US9 from herpes simplex virus type 1. Virology 1986, 150, 321–332. [Google Scholar] [CrossRef]
- Fukushi, H.; Taniguchi, A.; Yasuda, K.; Yanai, T.; Masegi, T.; Yamaguchi, T.; Hirai, K. A hamster model of equine herpesvirus 9 induced encephalitis. Journal of Neurovirology 2000, 6, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Gibson, J.; Slater, J.; Field, H. The pathogenicity of Ab4p, the sequenced strain of equine herpesvirus-1, in specific pathogen-free foals. Virology 1992, 189, 317–319. [Google Scholar] [CrossRef] [PubMed]
- Goehring, L.S.; van Maanen, C.; Sloet van Oldruitenborgh-Oosterbaan, M.M. Neurological syndromes among horses in The Netherlands. A 5 year retrospective survey (1999-2004). Veterinary Quarterly 2005, 27, 11–20. [Google Scholar] [CrossRef]
- Gosztonyi, G.; Borchers, K.; Ludwig, H. Pathogenesis of equine herpesvirus-1 infection in the mouse model. APMIS 2009, 117, 10–21. [Google Scholar] [CrossRef]
- Haanes, E.J.; Tomlinson, C.C. Genomic organization of the canine herpesvirus US region. Virus Research 1998, 53, 151–162. [Google Scholar] [CrossRef]
- Henninger, R.W.; Reed, S.M.; Saville, W.J.; Allen, G.P.; Hass, G.F.; Kohn, C.W.; Sofaly, C. Outbreak of neurologic disease caused by equine herpesvirus-1 at a university equestrian center. Journal of Veterinary Internal Medicine 2007, 21, 157–165. [Google Scholar] [PubMed]
- Howard, P.W.; Howard, T.L.; Johnson, D.C. Herpes simplex virus membrane proteins gE/gI and US9 act cooperatively to promote transport of capsids and glycoproteins from neuron cell bodies into proximal axons. Journal of Virology 2013, 87, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.A.; Osburn, B.; Cordy, D.; Kendrick, J. Equine herpesvirus 1 infection of horses: studies on the experimentally induced neurologic disease. American Journal of Veterinary Research 1977, 38, 709–719. [Google Scholar] [PubMed]
- Kasem, S.; Yu, M.H.H.; Yamada, S.; Kodaira, A.; Matsumura, T.; Tsujimura, K.; Madbouly, H.; Yamaguchi, T.; Ohya, K.; Fukushi, H. The ORF37 (UL24) is a neuropathogenicity determinant of equine herpesvirus 1 (EHV-1) in the mouse encephalitis model. Virology 2010, 400, 259–270. [Google Scholar] [CrossRef]
- Killeen, A.M.; Harrington, L.; Wall, L.V.; Kelly, D.C. Nucleotide sequence analysis of a homologue of herpes simplex virus type 1 gene US9 found in the genome of simian herpes B virus. Journal of General Virology 1992, 73, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Kohn, C.W.; Reed, S.M.; Sofaly, C.D.; Henninger, R.W.; Saville, W.J.; Allen, G.P.; Premanadan, C. Transmission of EHV-1 by horses with EHV-1 myeloencephalopathy: Implications for biosecurity and review. Clinical Techniques in Equine Practice 2006, 5, 60–66. [Google Scholar] [CrossRef]
- Lee, B.J.; Weiss, M.L.; Mosier, D.; Chowdhury, S.I. Spread of bovine herpesvirus type 5 (BHV-5) in the rabbit brain after intranasal inoculation. Journal of Neurovirology 1999, 5, 474–484. [Google Scholar] [CrossRef]
- Leung-Tack, P.; Audonnet, J.C.; Riviere, M. The complete DNA sequence and the genetic organization of the short unique region (US) of the bovine herpesvirus type 1 (ST strain). Virology 1994, 199, 409–421. [Google Scholar] [CrossRef]
- Leutenegger, C.; Madigan, J.; Mapes, S.; Thao, M.; Estrada, M.; Pusterla, N. Detection of EHV-1 neuropathogenic strains using real-time PCR in the neural tissue of horses with myeloencephalopathy. Veterinary Record 2008, 162, 688–690. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lunn, D.; Davis-Poynter, N.; Flaminio, M.; Horohov, D.; Osterrieder, K.; Pusterla, N.; Townsend, H. Equine herpesvirus-1 consensus statement. Journal of Veterinary Internal Medicine 2009, 23, 450–461. [Google Scholar] [CrossRef]
- Lyman, M.G.; Kemp, C.D.; Taylor, M.P.; Enquist, L.W. Comparison of the pseudorabies virus Us9 protein with homologs from other veterinary and human alphaherpesviruses. Journal of Virology 2009, 83, 6978–6986. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, T.; Kondo, T.; Sugita, S.; Damiani, A.M.; O’Callaghan, D.J.; Imagawa, H. An equine herpesvirus type 1 recombinant with a deletion in the gE and gI genes is avirulent in young horses. Virology 1998, 242, 68–79. [Google Scholar] [CrossRef] [PubMed]
- McGeoch, D.J.; Dolan, A.; Donald, S.; Rixon, F.J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. Journal of Molecular Biology 1985, 181, 1–13. [Google Scholar] [CrossRef] [PubMed]
- McGraw, H.M.; Awasthi, S.; Wojcechowskyj, J.A.; Friedman, H.M. Anterograde spread of herpes simplex virus type 1 requires glycoprotein E and glycoprotein I but not Us9. Journal of Virology 2009, 83, 8315–8326. [Google Scholar] [CrossRef] [PubMed]
- Narita, M.; Uchimura, A.; Kawanabe, M.; Fukushi, H.; Hirai, K. Invasion and spread of equine herpesvirus 9 in the olfactory pathway of pigs after intranasal inoculation. Journal of Comparative Pathology 2001, 124, 265–272. [Google Scholar] [CrossRef]
- Nugent, J.; Birch-Machin, I.; Smith, K.; Mumford, J.; Swann, Z.; Newton, J.; Bowden, R.; Allen, G.; Davis-Poynter, N. Analysis of equid herpesvirus 1 strain variation reveals a point mutation of the DNA polymerase strongly associated with neuropathogenic versus nonneuropathogenic disease outbreaks. Journal of Virology 2006, 80, 4047–4060. [Google Scholar] [CrossRef]
- Okada, A.; Izume, S.; Ohya, K.; Fukushi, H. Equine herpesvirus type 1 tegument protein VP22 is not essential for pathogenicity in a hamster model, but is required for efficient viral growth in cultured cells. Journal of Veterinary Medical Science 2015, 77, 1293–1297. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Heldens, J.; 2005. Equine herpesviruses 1 (EHV-1) and 4 (EHV-4)--epidemiology, disease and immunoprophylaxis: a brief review. Veterinary Journal 2005, 170, 14–23. [Google Scholar] [CrossRef]
- Peck, S.C. Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant Journal 2006, 45, 512–522. [Google Scholar] [CrossRef]
- Robertson, D.; Savage, K.; Reis-Filho, J.S.; Isacke, C.M. Multiple immunofluorescence labelling of formalin-fixed paraffin-embedded (FFPE) tissue. BMC Cell Biology 2008, 9, 13. [Google Scholar] [CrossRef]
- Saxegaard, F. Isolation and identification of equine rhinopneumonitis virus (equine abortion virus) from cases of abortion and paralysis. Nordic Veterinary Medicine 1966, 18, 504–512. [Google Scholar]
- Smith, K.L.; Allen, G.P.; Branscum, A.J.; Cook, R.F.; Vickers, M.L.; Timoney, P.J.; Balasuriya, U.B. The increased prevalence of neuropathogenic strains of EHV-1 in equine abortions. Veterinary Microbiology 2010, 141, 5–11. [Google Scholar] [CrossRef]
- Telford, E.A.; Watson, M.S.; McBride, K.; Davison, A.J. The DNA sequence of equine herpesvirus-1. Virology 1992, 189, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Tomishima, M.; Enquist, L. A conserved α-herpesvirus protein necessary for axonal localization of viral membrane proteins. Journal of Cell Biology 2001, 154, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Tomishima, M.; Smith, G.; Enquist, L. Sorting and transport of alpha herpesviruses in axons. Traffic 2001, 2, 429–436. [Google Scholar] [CrossRef] [PubMed]
- van Zijl, M.; van der Gulden, H.; de Wind, N.; Gielkens, A.; Berns, A. Identification of two genes in the unique short region of pseudorabies virus; comparison with herpes simplex virus and varicella-zoster virus. Journal of General Virology 2001, 71, 1747–1755. [Google Scholar] [CrossRef]
- Vissani, M.; Becerra, M.; Perglione, C.O.; Tordoya, M.; Miño, S.; Barrandeguy, M. Neuropathogenic and non-neuropathogenic genotypes of Equid Herpesvirus type 1 in Argentina. Veterinary Microbiology 2009, 139, 361–364. [Google Scholar] [CrossRef]
- Willemse, M.J.; Strijdveen, I.G.; van Schooneveld, S.H.; van den Berg, M.C.; Sondermeijer, P.J. Transcriptional analysis of the short segment of the feline herpesvirus type 1 genome and insertional mutagenesis of a unique reading frame. Virology 1995, 208, 704–711. [Google Scholar] [CrossRef]
- Yamada, S.; Matsumura, T.; Tsujimura, K.; Yamaguchi, T.; Ohya, K.; Fukushi, H. Comparison of the growth kinetics of neuropathogenic and nonneuropathogenic equid herpesvirus type 1 (EHV-1) strains in cultured murine neuronal cells and the relevance of the D/N752 coding change in DNA polymerase gene (ORF30). Journal of Veterinary Medical Science 2008, 70, 505–511. [Google Scholar] [CrossRef]
- Yanai, T.; Sakai, T.; Fukushi, H.; Hirai, K.; Narita, M.; Sakai, H.; Masegi, T. Neuropathological study of gazelle herpesvirus 1 (equine herpesvirus 9) infection in Thomson’s gazelles (Gazella thomsoni). Journal of Comparative Pathology 1998, 119, 159–168. [Google Scholar] [CrossRef]
- Yu, M.H.; Kasem, S.G.; Tsujimura, K.; Matsumura, T.; Yanai, T.; Yamaguchi, T.; Ohya, K.; Fukushi, H. Diverse pathogenicity of equine herpesvirus 1 (EHV-1) isolates in CBA mouse model. The Journal of veterinary medical science 2010, 72, 301–306. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
PCR identification of the generated recombinant viruses. Lane 1: Ab4p attB ORF76 (782 bp), lane 2: Ab4pΔORF76R ORF76 (782 bp), lane 3: Ab4pΔORF76 (rpsLneo gene 1420bp), M: Molecular weight marker (100-bp).
Figure 1.
PCR identification of the generated recombinant viruses. Lane 1: Ab4p attB ORF76 (782 bp), lane 2: Ab4pΔORF76R ORF76 (782 bp), lane 3: Ab4pΔORF76 (rpsLneo gene 1420bp), M: Molecular weight marker (100-bp).
Figure 2.
Comparison of the in vitro growth curve of mutant viruses generated by BAC technology. (A) MDBK, (B) RK13, (C) FHK and (D) primary cultured mouse neuronal cells. Cells were infected at an MOI of 0.1 except mouse neuronal cells, which were infected at a MOI of 1. Error bars are standard errors.
Figure 2.
Comparison of the in vitro growth curve of mutant viruses generated by BAC technology. (A) MDBK, (B) RK13, (C) FHK and (D) primary cultured mouse neuronal cells. Cells were infected at an MOI of 0.1 except mouse neuronal cells, which were infected at a MOI of 1. Error bars are standard errors.
Figure 3.
Transcription activity kinetics of ORF76 (A), ORF75 (B) and ORF67 (C) by real-time RT-PCR. RNA was extracted from infected MDBK cells harvested at 0, 2, 4 and 8 hours post infection (hpi). Relative quantity was evaluated by the crossing point method using the β-actin gene as a control.
Figure 3.
Transcription activity kinetics of ORF76 (A), ORF75 (B) and ORF67 (C) by real-time RT-PCR. RNA was extracted from infected MDBK cells harvested at 0, 2, 4 and 8 hours post infection (hpi). Relative quantity was evaluated by the crossing point method using the β-actin gene as a control.
Figure 4.
Western blotting analysis of the generated recombinant viruses using guinea pig anti Us9 serum detects Us9 polypeptides with relative molecular masses of between 35 and 42 kDa. Markers (lane M) were included to assess the sizes of the Us9 polypeptides. Lane 1: RK 13 Mock, lane 2: Ab4p attB, lane 3: Ab4pΔORF76R, lane 4: Ab4pΔORF76, M: Molecular weight marker.
Figure 4.
Western blotting analysis of the generated recombinant viruses using guinea pig anti Us9 serum detects Us9 polypeptides with relative molecular masses of between 35 and 42 kDa. Markers (lane M) were included to assess the sizes of the Us9 polypeptides. Lane 1: RK 13 Mock, lane 2: Ab4p attB, lane 3: Ab4pΔORF76R, lane 4: Ab4pΔORF76, M: Molecular weight marker.
Figure 5.
Mean body weight curves of control mice and those inoculated with Ab4p attB, Ab4pΔORF76 and Ab4pΔORF76R viruses. Mice were infected intranasally with 1 × 105 PFU of the indicated viruses. Mean body weights were measured from 3 days before inoculation to 11 dpi. Each data point represents the mean relative body weight for the indicated group. Error bars indicate standard errors.
Figure 5.
Mean body weight curves of control mice and those inoculated with Ab4p attB, Ab4pΔORF76 and Ab4pΔORF76R viruses. Mice were infected intranasally with 1 × 105 PFU of the indicated viruses. Mean body weights were measured from 3 days before inoculation to 11 dpi. Each data point represents the mean relative body weight for the indicated group. Error bars indicate standard errors.
Figure 6.
Mouse brain inoculated with Ab4p attB on 5 dpi showing meningitis in the form of mononuclear cells infiltration and congestion together with gliosis and neuronal necrosis at the cerebral cortex (A) and perivascular cuffing with mononuclear inflammatory cells and neuronal necrosis (B). Mouse brain inoculated with Ab4p∆ORF76 on 6 dpi showing normal meninges (C) and cerebral cortex (D). Mouse brain inoculated with Ab4p∆ on 6 dpi showing meningitis in the form of mononuclear cells infiltration and congestion together with neuronal necrosis and congestion at the cerebral cortex(E) and gliosis and neuronal necrosis (F). Mouse brain with mock virus infection on 7 dpi showing normal meninges and brain. Hematoxylin and eosin staining, X200.
Figure 6.
Mouse brain inoculated with Ab4p attB on 5 dpi showing meningitis in the form of mononuclear cells infiltration and congestion together with gliosis and neuronal necrosis at the cerebral cortex (A) and perivascular cuffing with mononuclear inflammatory cells and neuronal necrosis (B). Mouse brain inoculated with Ab4p∆ORF76 on 6 dpi showing normal meninges (C) and cerebral cortex (D). Mouse brain inoculated with Ab4p∆ on 6 dpi showing meningitis in the form of mononuclear cells infiltration and congestion together with neuronal necrosis and congestion at the cerebral cortex(E) and gliosis and neuronal necrosis (F). Mouse brain with mock virus infection on 7 dpi showing normal meninges and brain. Hematoxylin and eosin staining, X200.
Figure 7.
Immunoperoxidase of sections of brains and olfactory epithelium of mice infected with Ab4p attB (A and C), and Ab4p∆ORF76R (B and D). Immunofluorescence of sections of olfactory epithelium of mice infected with Ab4p attB (F), and Ab4p∆ORF76 (H). Visible light images (E, G) and fluorescence images (F, H).
Figure 7.
Immunoperoxidase of sections of brains and olfactory epithelium of mice infected with Ab4p attB (A and C), and Ab4p∆ORF76R (B and D). Immunofluorescence of sections of olfactory epithelium of mice infected with Ab4p attB (F), and Ab4p∆ORF76 (H). Visible light images (E, G) and fluorescence images (F, H).
Table 1.
Sequences of primers used for the construction and analysis of generated mutant viruses.
Table 1.
Sequences of primers used for the construction and analysis of generated mutant viruses.
| Target |
Primer |
Sequence |
rpsL-neo
cassette |
1 |
5’-TTT CCC TCT CAG CGA TCA CTT TTC ACC ACC GAA GAA CAG GCC CTC ATC GGG GCC TGG TGA TGA TGG CGG GAT CG -3’ |
| 2 |
5’-GGG CTG TTG TGG GGT AAA AGG TGG TGT TAC GGA AAC ACG CGT GCC AAG AAT CAG AAG AAC TCG TCA AGA AGG CG-3’ |
rpsL-ORF76 |
3 |
5’-TTT CCC TCT CAG CGA TCA CTT TTC ACC ACC GAA GAA CAG GCC CTC ATC GG -3’ |
| 4 |
5’-GGG CTG TTG TGG GGT AAA AGG TGG TGT TAC GGA AAC ACG CGT GCC AAG AA -3’ |
| ORF76 with EcoRI and NotI |
5 |
5’-ccg gaa ttc ATG GAG AAG GCG GAG GCT GCC GCA-3’ |
| 6 |
5’-aag gaa aaa agc ggc cgc TTA CGG AAA CAC GCG TGC CAA GAA-3’ |
ORF76 |
7 |
5’-CTA CCG TGG AAG CGG TAT GT-3’ |
| 8 |
5’-ATT CTC AGA AGC AGC GGT GT-3’ |
ORF75 |
9 |
5’-CAA CCC TGT CAG AAA CAG CA -3’ |
| 10 |
5’-GGG GGA GGT AGA GTT TCC AG-3’ |
ORF67 |
11 |
5’-TCG GCC CTT ATG TAA TAG CG -3’ |
| 12 |
5’-CTC CTA CTT CAG GCG GTG TC-3’ |
ORF30 |
13 |
5’-gtc agg ccc aca aac ttg at-3’ |
| 14 |
5’-act cgg ttt acg gat tca cg-3’ |
Table 2.
Virus titration and DNA detection in mice organs inoculated with Ab4p attB, Ab4p ΔORF76 and Ab4p ΔORF76R.
Table 2.
Virus titration and DNA detection in mice organs inoculated with Ab4p attB, Ab4p ΔORF76 and Ab4p ΔORF76R.
| Virus |
Organ1
|
Virus titration2 on
Day post inoculation |
| 0 |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
Ab4p
attB |
O |
- |
- |
- |
200 |
2000 |
300 |
100 |
100 |
- |
- |
- |
| B |
- |
- |
- |
100 |
1000 |
100 |
100 |
100 |
- |
- |
- |
| L |
- |
100 |
200 |
100 |
1000 |
100 |
- |
- |
- |
- |
- |
Ab4p
ΔORF76 |
O |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
| B |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
| L |
- |
- |
100 |
100 |
100 |
- |
- |
- |
- |
- |
- |
Ab4p
ΔORF76R |
O |
- |
- |
- |
200 |
300 |
500 |
1000 |
300 |
- |
- |
- |
| B |
- |
- |
- |
100 |
200 |
500 |
2000 |
1000 |
- |
- |
- |
| L |
- |
100 |
300 |
100 |
100 |
- |
- |
- |
- |
- |
- |
Table 3.
Virus DNA detection in mice organs inoculated with Ab4p attB, Ab4p ΔORF76 and Ab4p ΔORF76R.
Table 3.
Virus DNA detection in mice organs inoculated with Ab4p attB, Ab4p ΔORF76 and Ab4p ΔORF76R.
| Virus |
Organ1
|
Virus titration2 on
Day post inoculation |
| 0 |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
Ab4p
attB |
O |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| B |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| L |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Ab4p
ΔORF76 |
O |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
| B |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
| L |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
- |
Ab4p
ΔORF76R |
O |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| B |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| L |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).