Submitted:
02 September 2024
Posted:
04 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Reduced-Representation Bisulfite Sequencing
2.2. DNA Methylation Patterns in Muscle
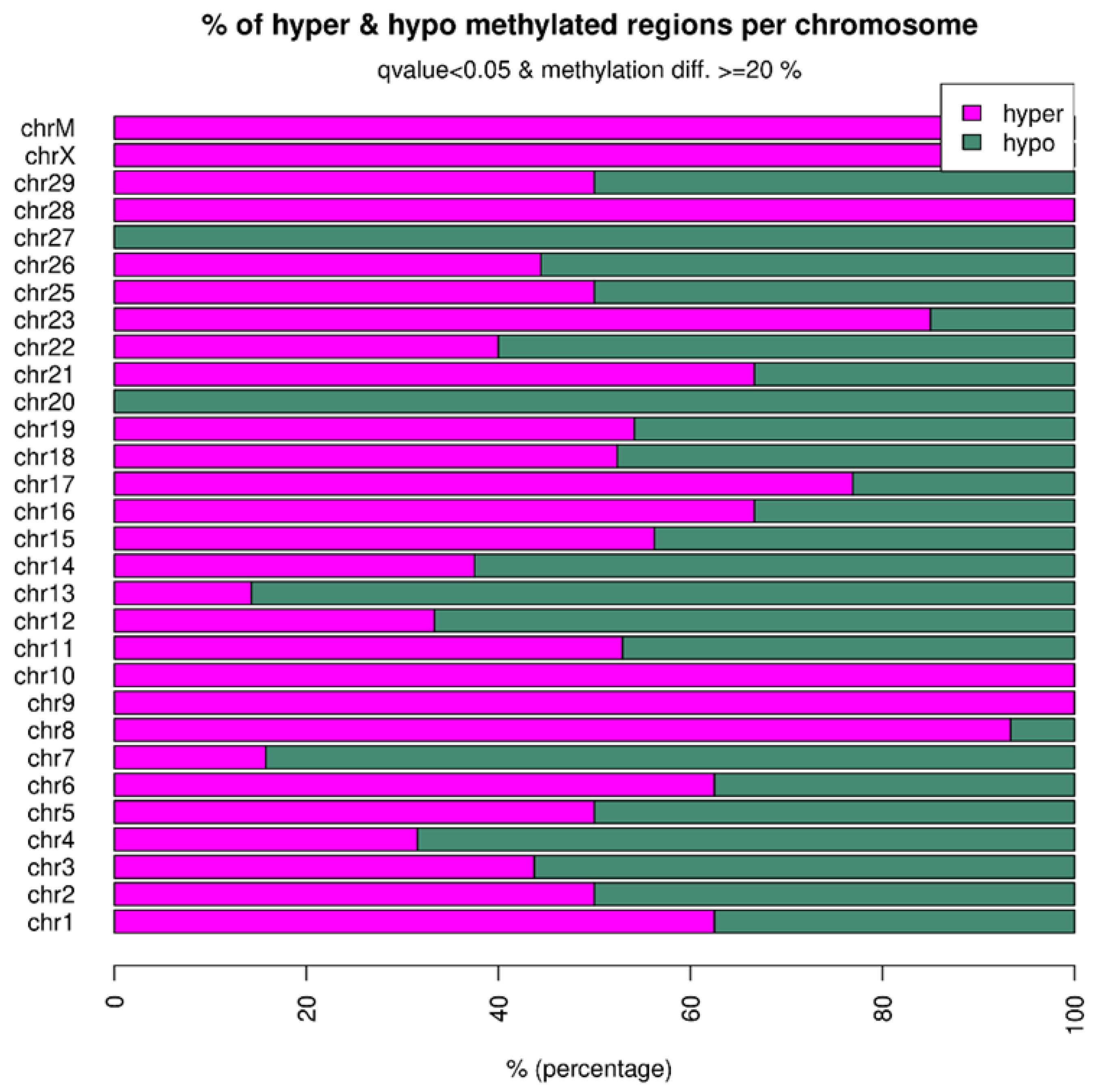
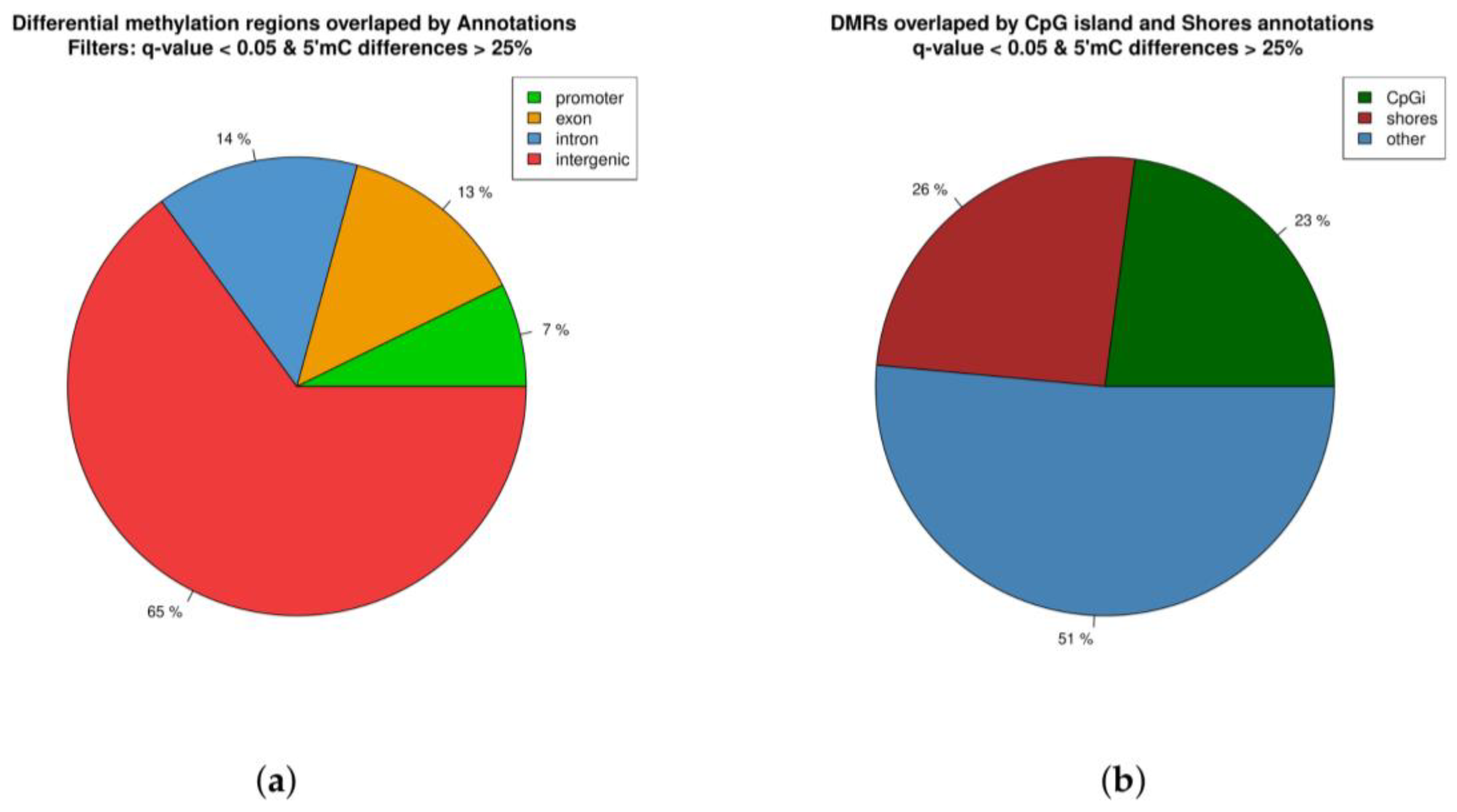
2.3. Distribution of Differentially Methylated Regions across the Genome
2.4. Functional Enrichment Analysis
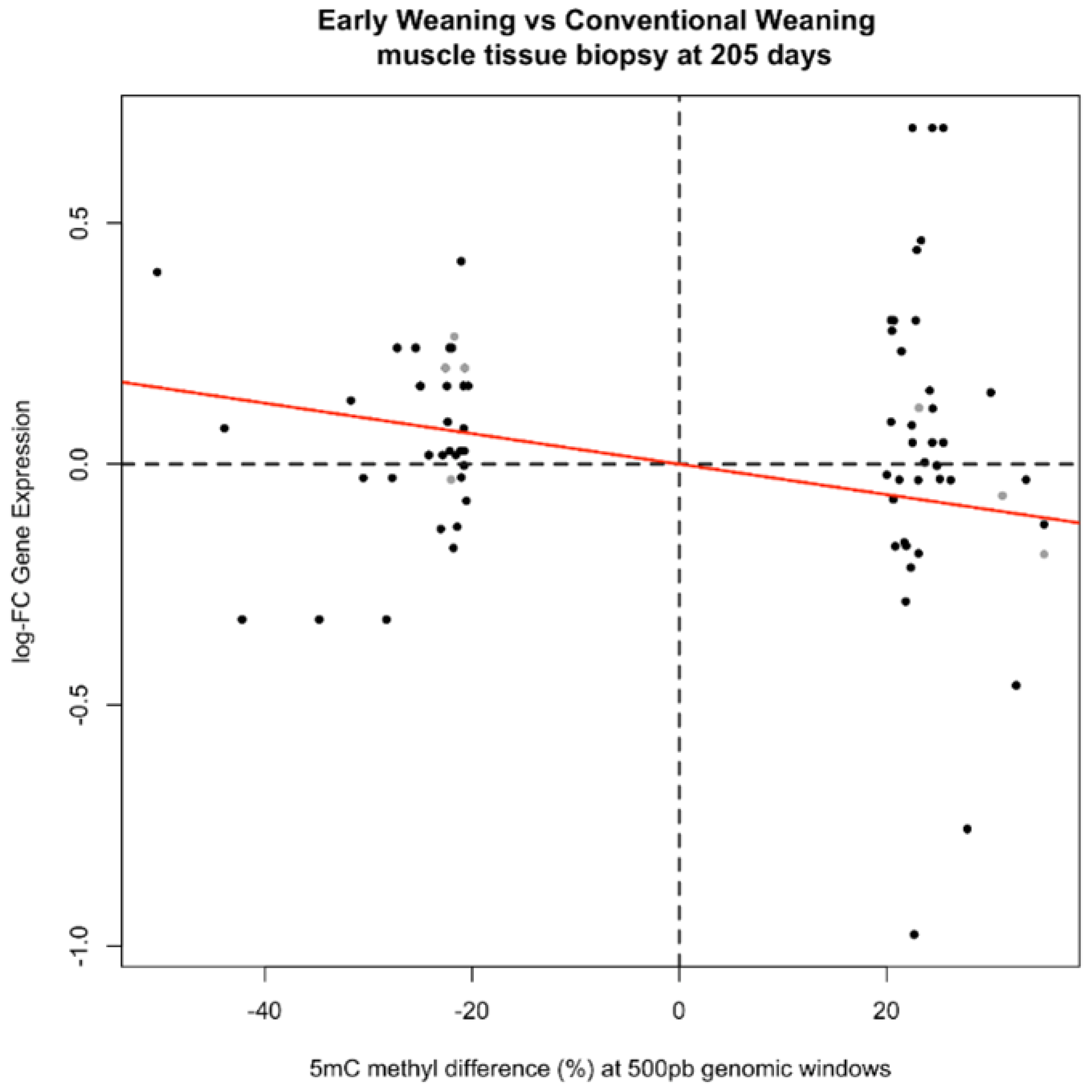
2.5. Relationship between Methylation and Gene Expression
3. Discussion
4. Materials and Methods
4.1. Animals and Collection of Muscle Tissue
4.2. Genomic DNA Extraction, RRBS, Quality Control and Alignment to Reference Genome
4.3. Identification and Characterization of Differentially Methylated Regions and Genes
4.4. Relative Gene Expression
4.5. Relationship between Differentially Methylated Genes and Genes Expression
4.6. Functional Enrichment of Differentially Methylated Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loy, D., Maxwell, D., Rouse, G. Effect of Early Weaning of Beef Calves on Performance and Carcass Quality. 1999 Beef Research Report — Iowa State University, 2000.
- Restle, J. , Vaz, R. Z., Alves Filho, D. C., Bernardes, R. A. D. L. C., Pascoal, L. L., Senna, D. B. D., & Polli, V. A. Desempenho de vacas Charolês e Nelore desterneiradas aos três ou sete meses. R. Bras. Zootec. 2001, 30(2), 499–507. [CrossRef]
- Beretta, V. , Lobato, J. F. P., & Mielitz Netto, C. G. Produtividade e eficiência biológica de sistemas de produção de gado de corte de ciclo completo no Rio Grande de Sul. R. Bras. Zootec. 2002, 31(2 suppl), 991–1001. [CrossRef]
- Almeida, L. S. P. D. , & Lobato, J. F. P. Efeito da idade de desmame e suplementação no desenvolvimento de novilhas de corte. R. Bras. Zootec. 2004, 33(6 suppl 2), 2086–2094. [CrossRef]
- De Menezes, L.F.G. , Restle J., Pascoal L.L., Brondani I.L., Rosa J.R.P., Pizzuti L.A., Dalla Chieza E. Fontes energéticas para suplementação de bezerros desmamados precocemente, mantidos em pastagem de capim-elefante (Pennisetum purpureum, SCHUM.). C. Anim. Bras. 2008, 9(1), 30-42.
- Scheffler J.M., McCann M.A., Greiner S.P., Jiang H., Hanigan M.D., Bridges G.A., ... & Gerrard D.E. Early metabolic imprinting events increase marbling scores in fed cattle. J. Anim. Sci. 2014. 92(1), 320-324. [CrossRef]
- Gasser, C.L. , Behlke E.J., Grum D.E., Day M.L. Effect of timing of feeding a high concentrate diet on growth and attainment of puberty in early-weaned heifers. J. Anim. Sci. 2006, 84(11), 3118-3122. [CrossRef]
- Reddy, K.E. , Jeong J., Baek Y.C., Oh Y.K., Kim M., So K.M., Kim M.J., Kim D.W., Park S.K., Lee H.J. Early weaning of calves after different dietary regimens affects later rumen development, growth, and carcass traits in Hanwoo cattle. Asian-Australas J. Anim. Sci. 2017, 30(10):1425-1434. [CrossRef]
- Veland, N. , Chen T. Mechanisms of DNA Methylation and Demethylation During Mammalian Development. In Handbook of Epigenetics. 2nd ed. [s.l.]. 2017, Elsevier. p. 11–24. [CrossRef]
- Curradi, M. , Izzo A., Badaracco G., Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Molecular and cellular biology. 2002, 22(9), 3157 3173. [CrossRef]
- Niciura S.C.M., Saraiva N.Z. Epigenética: bases moleculares, efeitos na fisiologia e na patologia, e implicações para a produção animal e a vegetal. 2nd ed. São Carlos, SP: Embrapa Pecuária Sudeste; Brasília, DF: Embrapa Informação Tecnológica. 2015. http://www.alice.cnptia.embrapa.br/handle/doc/1077512.
- Zhang, N. Epigenetic modulation of DNA methylation by nutrition and its mechanisms in animals. Animal Nutrition. 2015, 1(3), 144–151. [Google Scholar] [CrossRef] [PubMed]
- McKay, J. A. , & Mathers, J. C. Diet induced epigenetic changes and their implications for health: Nutrition, epigenetics and health. Acta Physiologica. 2011, 202(2), 103–118. [CrossRef]
- Feil, R. , & Fraga, M. F. Epigenetics and the environment: emerging patterns and implications. Nature Reviews Genetics. 2012, 13(2), 97–109. [CrossRef]
- Mentch, S. J. , & Locasale, J. W. One-carbon metabolism and epigenetics: understanding the specificity. Annals of the New York Academy of Sciences. 2016, 1363(1), 91–98. [CrossRef]
- Silveira, L. G. G. , Piona, M. N. M., Mousquer, C. J., Gomes, R. S., & Silveira, A. C. Sistemas de cria em áreas tropicais: desmama precoce. Revisão de Literatura. Ver. Bras. Hig. San. Anim. 2021,15(1), 1–14. [CrossRef]
- Murdoch, B. M., Murdoch, G. K., Greenwood, S., & McKay, S. Nutritional Influence on Epigenetic Marks and Effect on Livestock Production. Frontiers in Genetics. 2016, 7. [CrossRef]
- Ngo, V. , & Hein, L. Epigenetics concepts: An overview. In Epigenetics in Cardiovascular Disease (p. 19–40). Elsevier. 2021. [CrossRef]
- McKay, S. , Betancourt, F., Bhattarai, S., Buttolph, T., White, S., Lachance, H., Quijada, D., Friedman, S., Perlee, S., Cantrell, B., & Murdoch, B. 115 Profiling Conservation of DNA Methylation in Cattle. J. Anim. Sci. 2018, 96(suppl_3), 370–370. [CrossRef]
- Crouse, M. S., Caton, J. S., Claycombe-Larson, K. J., Diniz, W. J. S., Lindholm-Perry, A. K., Reynolds, L. P., Dahlen, C. R., Borowicz, P. P., & Ward, A. K. Epigenetic Modifier Supplementation Improves Mitochondrial Respiration and Growth Rates and Alters DNA Methylation of Bovine Embryonic Fibroblast Cells Cultured in Divergent Energy Supply. Frontiers in Genetics. 2022; 13, 812764. [CrossRef]
- Anderson, O. S. , Sant, K. E., & Dolinoy, D. C. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. The Journal of Nutritional Biochemistry. 2012, 23(8), 853–859. [CrossRef]
- Niculescu, M. D., & Zeisel, S. H. Diet, Methyl Donors and DNA Methylation: Interactions between Dietary Folate, Methionine and Choline. The Journal of Nutrition. 2012, 132(8), 2333S–2335S. [Google Scholar] [CrossRef]
- Lobley, G. E. Control of the metabolic fate of amino acids in ruminants: a review. J. Anim. Sci. 1992, 70(10), 3264–3275. [Google Scholar] [CrossRef] [PubMed]
- Amaral, C. L. D. , Bueno, R. D. B. E. L., Burim, R. V., Queiroz, R. H. C., Bianchi, M. D. L. P., & Antunes, L. M. G. The effects of dietary supplementation of methionine on genomic stability and p53 gene promoter methylation in rats. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 2011, 722(1), 78–83. [CrossRef]
- Mukherjee, N., Kumar, A. P., & Ghosh, R. DNA Methylation and Flavonoids in Genitourinary Cancers. Current Pharmacology Reports. 2015, 1(2), 112–120. [Google Scholar] [CrossRef]
- Ramos, P. M., Scheffler, T. L., Beline, M., Bodmer, J., Gerrard, D. E., Silva, S. L. Challenges and opportunities of using Bos indicus cattle to meet consumers’ demand for quality beef. Meat Science. 2024, 207, 109375. [CrossRef] [PubMed]
- Rogne, M. , & Taskén, K. Compartmentalization of cAMP Signaling in Adipogenesis, Lipogenesis, and Lipolysis. Hormone and Metabolic Research. 2014, 46(12), 833–840. [CrossRef]
- Tong, T. , Shen, Y., Lee, H.-W., Yu, R., & Park, T. Adenylyl cyclase 3 haploinsufficiency confers susceptibility to diet-induced obesity and insulin resistance in mice. Scientific Reports. 2016, 6(1), 34179. [CrossRef]
- Goni, L. , Riezu-Boj, J. I., Milagro, F. I., Corrales, F. J., Ortiz, L., Cuervo, M., & Martínez, J. A. Interaction between an ADCY3 Genetic Variant and Two Weight-Lowering Diets Affecting Body Fatness and Body Composition Outcomes Depending on Macronutrient Distribution: A Randomized Trial. Nutrients. 2018, 10(6), 789. [CrossRef]
- Wu, L. , Shen, C., Seed Ahmed, M., Östenson, C. -G., & Gu, H. F. Adenylate cyclase 3: a new target for anti-obesity drug development. Obesity Reviews. 2016, 17(9), 907–914. [CrossRef]
- London, E. , Bloyd, M., & Stratakis, C. A. PKA functions in metabolism and resistance to obesity: lessons from mouse and human studies. J. Endocri. 2020, 246(3), R51–R64. [CrossRef]
- Fitzpatrick, M. , & Solberg Woods, L. C. Adenylate cyclase 3: a potential genetic link between obesity and major depressive disorder. Physiological Genomics. 2024, 56(1), 1–8. [CrossRef]
- Hausman, G. J. , Dodson, M. V., Ajuwon, K., Azain, M., Barnes, K. M., Guan, L. L., Jiang, Z., Poulos, S. P., Sainz, R. D., Smith, S., Spurlock, M., Novakofski, J., Fernyhough, M. E., & Bergen, W. G. BOARD-INVITED REVIEW: The biology and regulation of preadipocytes and adipocytes in meat animals1,2. J. Anim. Sci. 2009, 87(4), 1218–1246. [CrossRef]
- Tseng, Y.-H. , Cypess, A. M., Kahn, C. R. Cellular bioenergetics as a target for obesity therapy. Nature Reviews Drug Discovery. 2010, 9(6), 465–482. [CrossRef] [PubMed]
- Ladeira, M. , Schoonmaker, J., Gionbelli, M., Dias, J., Gionbelli, T., Carvalho, J., & Teixeira, P. Nutrigenomics and Beef Quality: A Review about Lipogenesis. Inter. J. Mol. Sci. 2016, 17(6), 918. [CrossRef]
- Dodson, M. V. , Jiang, Z., Chen, J., Hausman, G. J., Guan, L. L., Novakofski, J., Thompson, D. P., Lorenzen, C. L., Fernyhough, M. E., Mir, P. S., & Reecy, J. M. Allied Industry Approaches to Alter Intramuscular Fat Content and Composition in Beef Animals. J. Food Sci. 2010, 75(1). [CrossRef]
- Fernyhough, M. E., Okine, E., Hausman, G., Vierck, J. L., & Dodson, M. V. PPARγ and GLUT-4 expression as developmental regulators/markers for preadipocyte differentiation into an adipocyte. Domestic Animal Endocrinology. 2007, 33(4), 367–378. [Google Scholar] [CrossRef] [PubMed]
- Seale, P., Kajimura, S., Yang, W., Chin, S., Rohas, L. M., Uldry, M., Tavernier, G., Langin, D., & Spiegelman, B. M. Transcriptional Control of Brown Fat Determination by PRDM16. Cell Metabolism. 2007, 6(1), 38–54. [CrossRef]
- Frühbeck, G. , Sesma, P., & Burrell, M. A. PRDM16: the interconvertible adipo-myocyte switch. Trends in Cell Biology. 2009, 19(4), 141–146. [CrossRef]
- Pacini, F. , & Cantara, S. Molecular Diagnosis of Thyroid Cancer. In Genetic Diagnosis of Endocrine Disorders (p. 153–162). 2016, Elsevier. [CrossRef]
- Yokoyama, C., Wang, X., Briggs, M. R., Admon, A., Wu, J., Hua, X., Goldstein, J. L., & Brown, M. S. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993, 75(1), 187–197. [CrossRef]
- Li, Y. , Xu, S., Mihaylova, M. M., Zheng, B., Hou, X., Jiang, B., Park, O., Luo, Z., Lefai, E., Shyy, J. Y.-J., Gao, B., Wierzbicki, M., Verbeuren, T. J., Shaw, R. J., Cohen, R. A., & Zang, M. AMPK Phosphorylates and Inhibits SREBP Activity to Attenuate Hepatic Steatosis and Atherosclerosis in Diet-Induced Insulin-Resistant Mice. Cell Metabolism. 2011, 13(4), 376–388. [CrossRef]
- Shimano, H., & Sato, R. SREBP-regulated lipid metabolism: convergent physiology — divergent pathophysiology. Nature Reviews Endocrinology. 2017, 13(12), 710–730. [CrossRef]
- Pedro, A. E. , Torrecilhas, J. A., Torres, R. N. S., Ramírez-Zamudio, G. D., Baldassini, W. A., Chardulo, L. A. L., Curi, R. A., Russo, G. H., Napolitano, J. A., Bezerra Tinoco, G. L., Mariano, T. B., Caixeta, J. L., Moriel, P., & Pereira, G. L. Early Weaning Possibly Increases the Activity of Lipogenic and Adipogenic Pathways in Intramuscular Adipose Tissue of Nellore Calves. Metabolites. 2023, 13(9), 1028. [CrossRef]
- Shingfield, K. J., Bernard, L., Leroux, C., & Chilliard, Y. Role of trans fatty acids in the nutritional regulation of mammary lipogenesis in ruminants. Animal. 2010, 4(7), 1140–1166. [CrossRef]
- Fujino, T. , Kondo, J., Ishikawa, M., Morikawa, K., & Yamamoto, T. T. Acetyl-CoA Synthetase 2, a Mitochondrial Matrix Enzyme Involved in the Oxidation of Acetate. J. Bio. Chem. 2001, 276(14), 11420–11426. [CrossRef]
- Kaikaus, R. M., Bass, N. M., & Ockner, R. K. Functions of fatty acid binding proteins. Experientia. 1990, 46(6), 617–630. [CrossRef]
- Goszczynski, D. E. , Papaleo-Mazzucco, J., Ripoli, M. V., Villarreal, E. L., Rogberg-Muñoz, A., Mezzadra, C. A., Melucci, L. M., & Giovambattista, G. Genetic Variation in FABP4 and Evaluation of Its Effects on Beef Cattle Fat Content. Animal Biotechnology. 2017, 28(3), 211–219. [CrossRef]
- Barendse, W., Bunch, R. J., Thomas, M. B., & Harrison, B. E. A splice site single nucleotide polymorphism of the fatty acid binding protein 4 gene appears to be associated with intramuscular fat deposition in longissimus muscle in Australian cattle. Animal Genetics. 2009, 40(5), 770–773. [CrossRef]
- Maharani, D., Jung, Y., Jung, W. Y., Jo, C., Ryoo, S. H., Lee, S. H., Yeon, S. H., & Lee, J. H. Association of five candidate genes with fatty acid composition in Korean cattle. Molecular Biology Reports. 2012, 39(5), 6113–6121. [CrossRef]
- Silva A., Souza Filho E.E., & Cunha S.B. Padrões de canal do rio Paraguai na região de Cáceres (MT). Revista Brasileira de Geociências. 2008, volume 38 (1).
- Andrews, S. "FastQC: a quality control analysis tool for high throughput sequencing data." *Github*. 2010.
- Krueger, F. , James, F., Ewels, P., Afyounian, E., Weinstein, M., Schuster-Boeckler, B., Hulselmans, G., & Sclamons. FelixKrueger/TrimGalore: v0.6.10 - add default decompression path (0.6.10). Zenodo. 2023. [Google Scholar] [CrossRef]
- Krueger, F. , & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011, 27(11), 1571–1572. [CrossRef]
- Akalin, A. , Kormaksson, M., Li, S., Garrett-Bakelman, F. E., Figueroa, M. E., Melnick, A., & Mason, C. E. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biology. 2012, 13(10), R87. [CrossRef]
- Akalin, A. , Franke, V., Vlahoviček, K., Mason, C. E., & Schübeler, D. genomation: a toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics. 2015, 31(7), 1127–1129. [CrossRef]
- Team, R. Core. "RA language and environment for statistical computing, R Foundation for Statistical." Computing. 2020.
- Chen, Y. , Chen, L., Lun, A. T. L., Baldoni, P. L., & Smyth, G. K. edgeR 4.0: powerful differential analysis of sequencing data with expanded functionality and improved support for small counts and larger datasets. 2024. [CrossRef]
- Chen, E. Y., Tan, C. M., Kou, Y., Duan, Q., Wang, Z., Meirelles, G. V., Clark, N. R., & Ma’ayan, A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013, 14(1), 128. [CrossRef]
| Early weaning | Conventional weaning | |
| Number of raw reads | 30,703,133 | 28,494,972 |
| Number of clean reads | 30,194,010 | 28,023,825 |
| Number of aligned reads | 23,084,052 | 21,452,788 |
| Percentage of aligned reads | 76.45% | 76.55% |
| Early weaning | Conventional weaning | |
| mCpG (%) | 3.84% | 3.79% |
| mCHG (%) | 0.54% | 0.51% |
| mCHH (%) | 0.47% | 0.44% |
| DMG | Description | LocChr | Start | End | DMR.p-value | Meth.diff | Category |
| ADCY3 | adenylate cyclase 3 | chr11 | 74335201 | 74335700 | 2,83E-24 | -20,7252 | TSSes |
| ADCY3 | chr11 | 74335301 | 74335800 | 1,92E-24 | -22,5866 | TSSes | |
| ADCY3 | chr11 | 74335201 | 74335700 | 2,83E-24 | -20,7252 | promoters | |
| ADCY3 | chr11 | 74335301 | 74335800 | 1,92E-24 | -22,5866 | promoters | |
| AKT1 | AKT serine/threonine kinase 1 | chr21 | 69246101 | 69246600 | 3,72E-15 | 31,18985 | promoters |
| ATP6 | mitochondrially encoded ATP synthase 6 | chrM | 6201 | 6700 | 5,75E-23 | 35,20596 | promoters |
| COX1 | cytochrome c oxidase subunit I | chrM | 6201 | 6700 | 5,75E-23 | 35,20596 | promoters |
| F2 | coagulation factor II, thrombin | chr15 | 76656801 | 76657300 | 2,77E-06 | 21,88917 | promoters |
| F2 | chr15 | 76656801 | 76657300 | 2,77E-06 | 21,88917 | TSSes | |
| MAP2K1 | mitogen-activated protein kinase 1 | chr10 | 13275401 | 13275900 | 5,80E-05 | 23,12619 | promoters |
| MAP2K1 | chr10 | 13275201 | 13275700 | 5,80E-05 | 23,12619 | promoters | |
| MAP2K1 | chr10 | 13275301 | 13275800 | 5,80E-05 | 23,12619 | promoters | |
| MRPL28 | mitochondrial ribosomal protein L28 | chr25 | 363101 | 363600 | 0,000182 | -20,7854 | promoters |
| RUVBL1 | RuvB like AAA ATPase 1 | chr22 | 59508001 | 59508500 | 2,67E-08 | -22,0339 | promoters |
| STUB1 | STIP1 homology and U-box containing protein 1 | chr25 | 585001 | 585500 | 1,74E-05 | -25 | promoters |
| STUB1 | chr25 | 584901 | 585400 | 1,74E-05 | -25 | promoters | |
| STUB1 | chr25 | 584801 | 585300 | 0,000112 | -22,4395 | promoters | |
| STUB1 | chr25 | 585101 | 585600 | 1,74E-05 | -25 | promoters | |
| ZNF557 | zinc finger protein 557 | chr7 | 15991901 | 15992400 | 2,77E-08 | -21,7254 | promoters |
| Term | Library | pvalue | qvalue | Genes |
| Growth hormone synthesis, secretion and action | KEGG_2021_Human | 0.00001 | 0.00162 | MAP2K1, ADCY3, ADCY5, MAP2K6, AKT1 |
| Rap1 signaling pathway | KEGG_2021_Human | 0.00001 | 0.00162 | ADCY3, MAP2K6, ADCY5, PGF, MAP2K1, AKT1 |
| Positive regulation of lipid metabolic process (GO:0045834) | GO_Biological_Process_2021 | 0.00003 | 0.02894 | F2, AKT1, PPARD |
| Phospholipase D signaling pathway | KEGG_2021_Human | 0.00004 | 0.00254 | AKT1, ADCY3, ADCY5, F2, MAP2K1 |
| Pathways in cancer | KEGG_2021_Human | 0.00007 | 0.00310 | MAP2K1, PGF, ADCY3, AKT1, ADCY5, F2, PPARD, HEYL |
| GnRH signaling pathway | KEGG_2021_Human | 0.00010 | 0.00323 | MAP2K1, ADCY3, MAP2K6, ADCY5 |
| Connective tissue development (GO:0061448) | GO_Biological_Process_2021 | 0.00013 | 0.70289 | COL11A2, SULF2, DYRK1B |
| Progesterone-mediated oocyte maturation | KEGG_2021_Human | 0.00014 | 0.00323 | MAP2K1, ADCY3, ADCY5, AKT1 |
| Surfactant homeostasis (GO:0043129) | GO_Biological_Process_2021 | 0.00014 | 0.02894 | CTSH, ABCA3 |
| Chemical homeostasis within a tissue (GO:0048875) | GO_Biological_Process_2021 | 0.00014 | 0.02894 | ABCA3, CTSH |
| Chemokine signaling pathway | KEGG_2021_Human | 0.00015 | 0.00323 | MAP2K1, FGR, ADCY3, AKT1, ADCY5 |
| Longevity regulating pathway | KEGG_2021_Human | 0.00015 | 0.00323 | ADCY3, AKT1, ADCY5, EHMT1 |
| Parathyroid hormone synthesis, secretion and action | KEGG_2021_Human | 0.00017 | 0.00323 | ADCY3, ADCY5, MAP2K1, PDE4A |
| Focal adhesion | KEGG_2021_Human | 0.00018 | 0.00323 | PGF, AKT1, ITGB5, TNXB, MAP2K1 |
| Cellular response to forskolin (GO:1904322) | GO_Biological_Process_2021 | 0.00024 | 0.03297 | ADCY3, ADCY5 |
| Response to forskolin (GO:1904321) | GO_Biological_Process_2021 | 0.00024 | 0.03297 | ADCY3, ADCY5 |
| cAMP biosynthetic process (GO:0006171) | GO_Biological_Process_2021 | 0.00030 | 0.03526 | ADCY3, ADCY5 |
| Regulation of proteolysis (GO:0030162) | GO_Biological_Process_2021 | 0.00039 | 0.03996 | F2, AKT1, STUB1 |
| Peptidyl-threonine phosphorylation (GO:0018107) | GO_Biological_Process_2021 | 0.00054 | 0.04846 | DYRK1B, AKT1, MAP2K1 |
| cAMP metabolic process (GO:0046058) | GO_Biological_Process_2021 | 0.00061 | 0.04958 | ADCY3, ADCY5 |
| Rearing phase | Early weaning | Conventional weaning |
| Birth to 120 days | Conventional suckling on Brachiaria pasture | |
| 120 to 205 days | Brachiaria decumbens pasture + concentrate-based diet (20% CP¹; 75% TDN²): 1 g of DM³/kg BW⁴ | Conventional suckling + Brachiaria decumbens |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





