Submitted:
03 September 2024
Posted:
04 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Patient Population
2.2. Image Protocol
2.3. Implementation
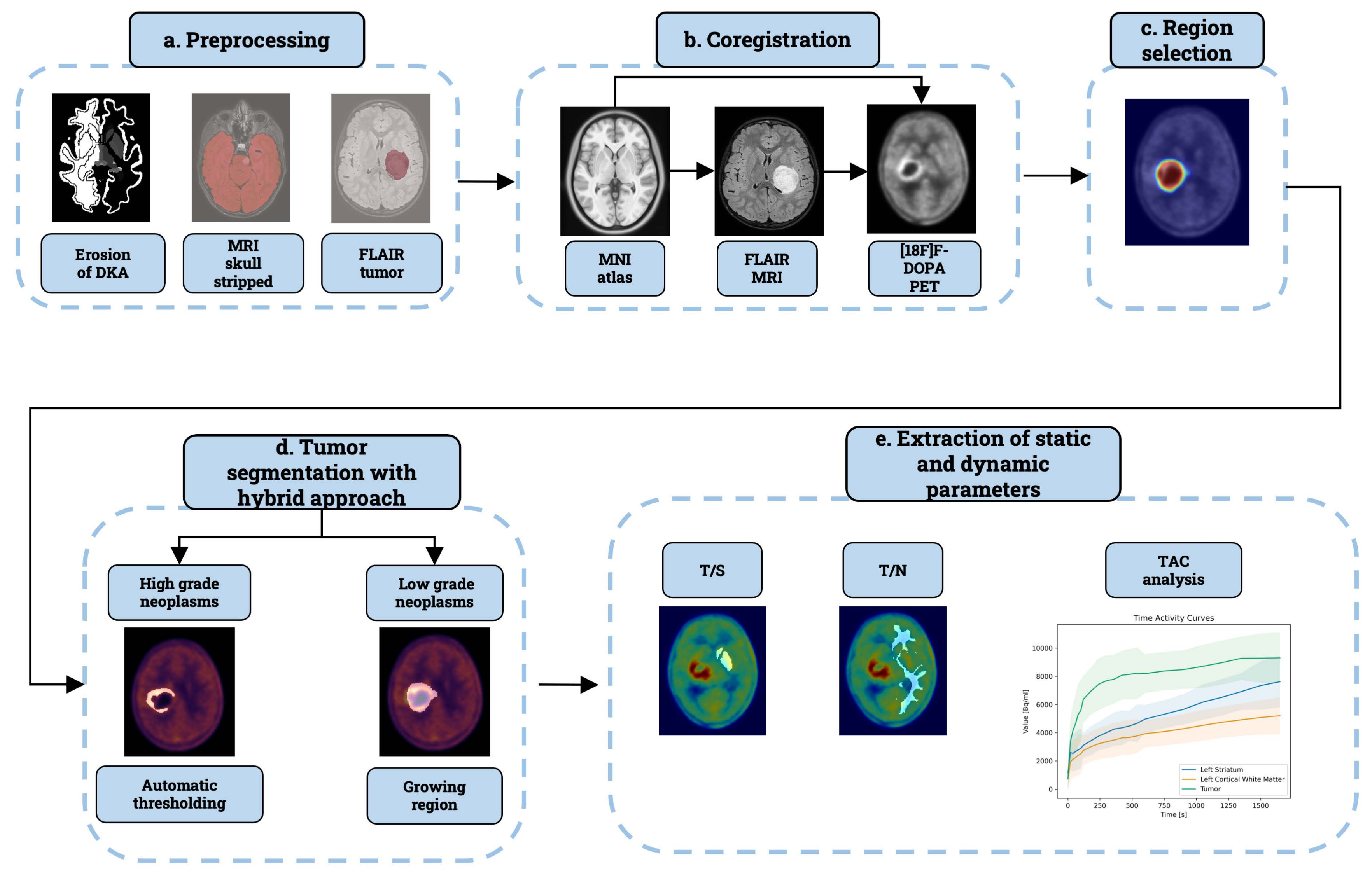
2.4. Preprocessing
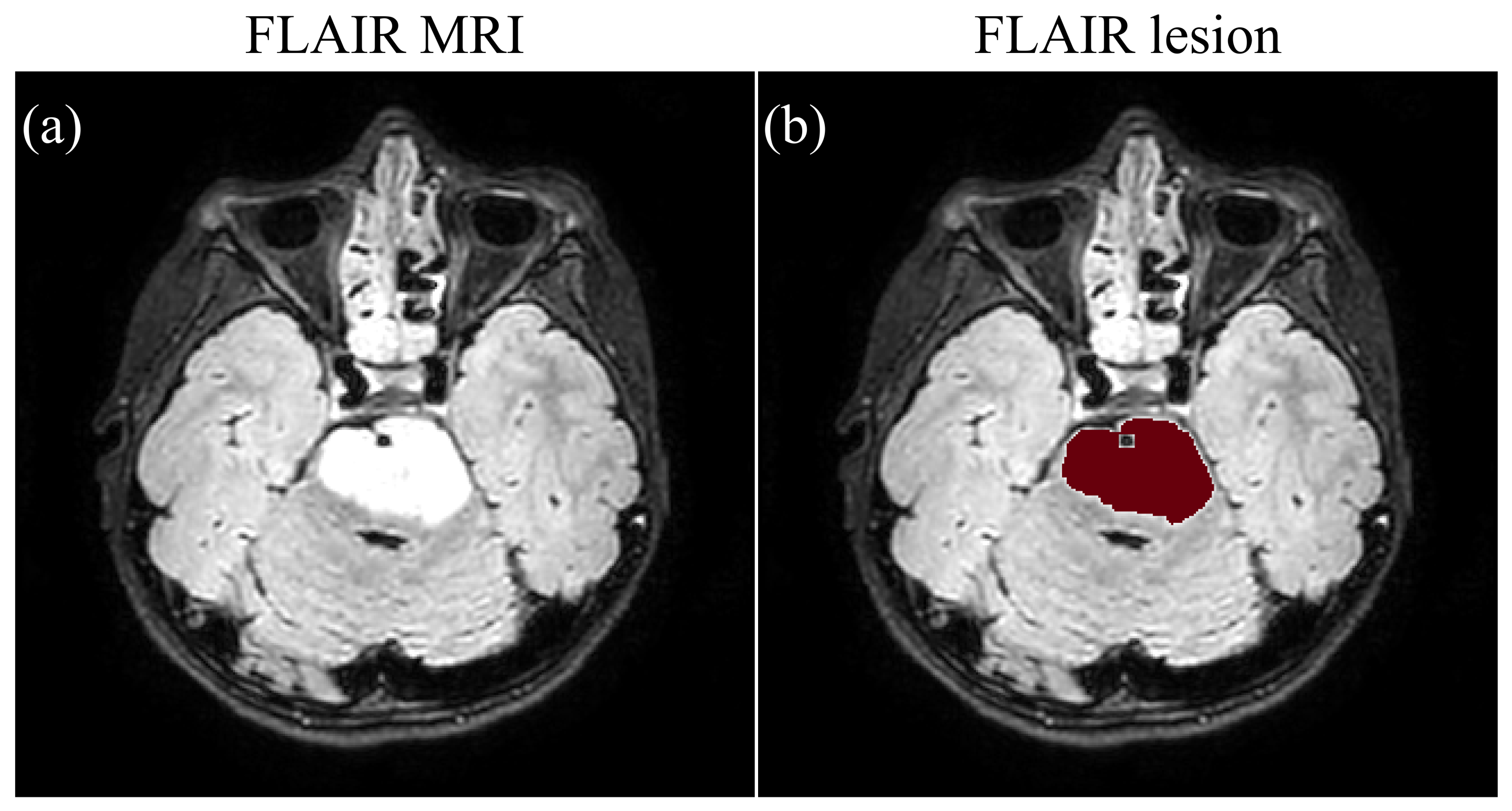
2.4.1. FLAIR MRI Segmentation
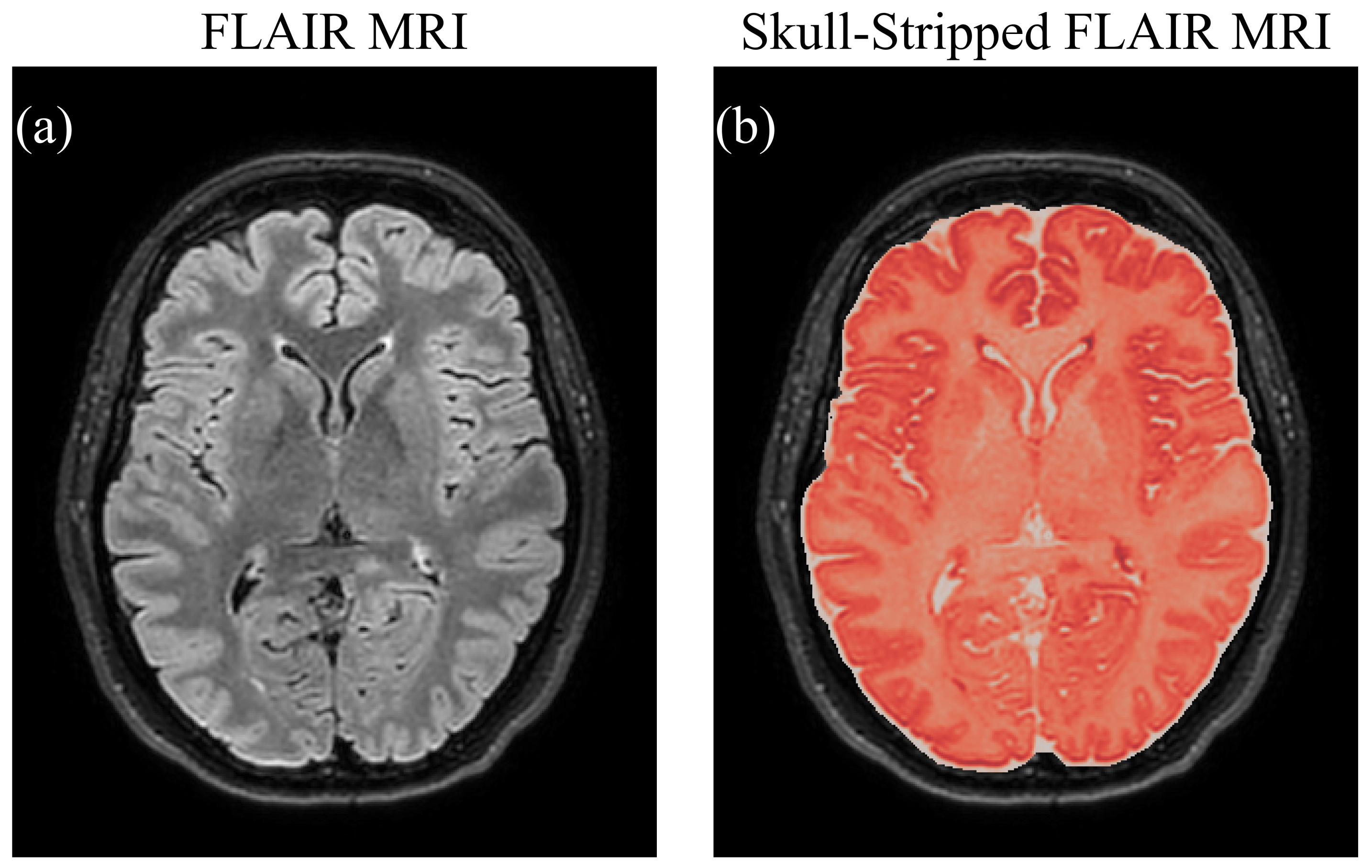
2.4.2. Skull Stripping
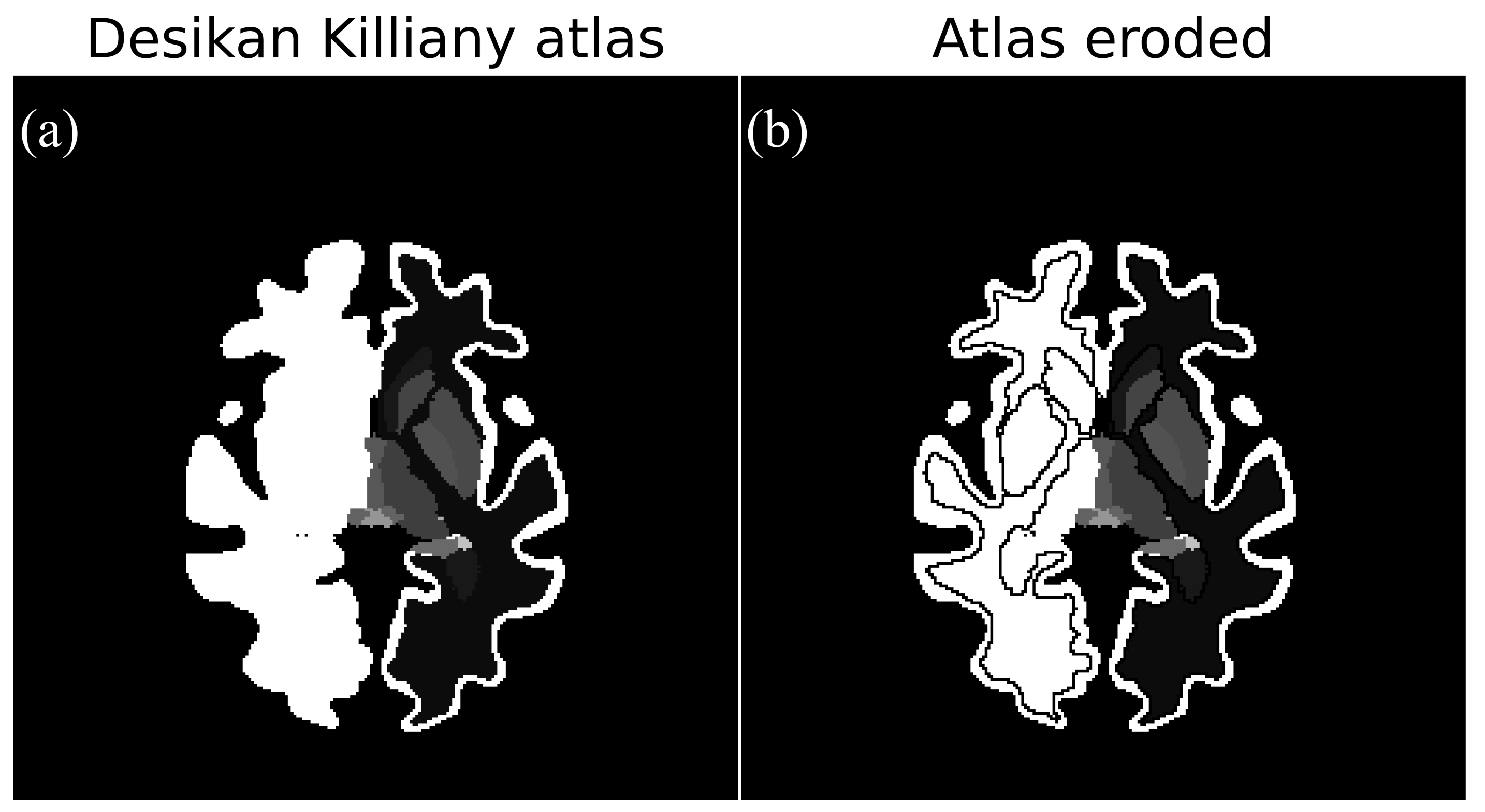
2.4.3. Erosion of Reference Region
2.5. Co-Registration
2.6. Region Selection
2.7. Tumour Segmentation
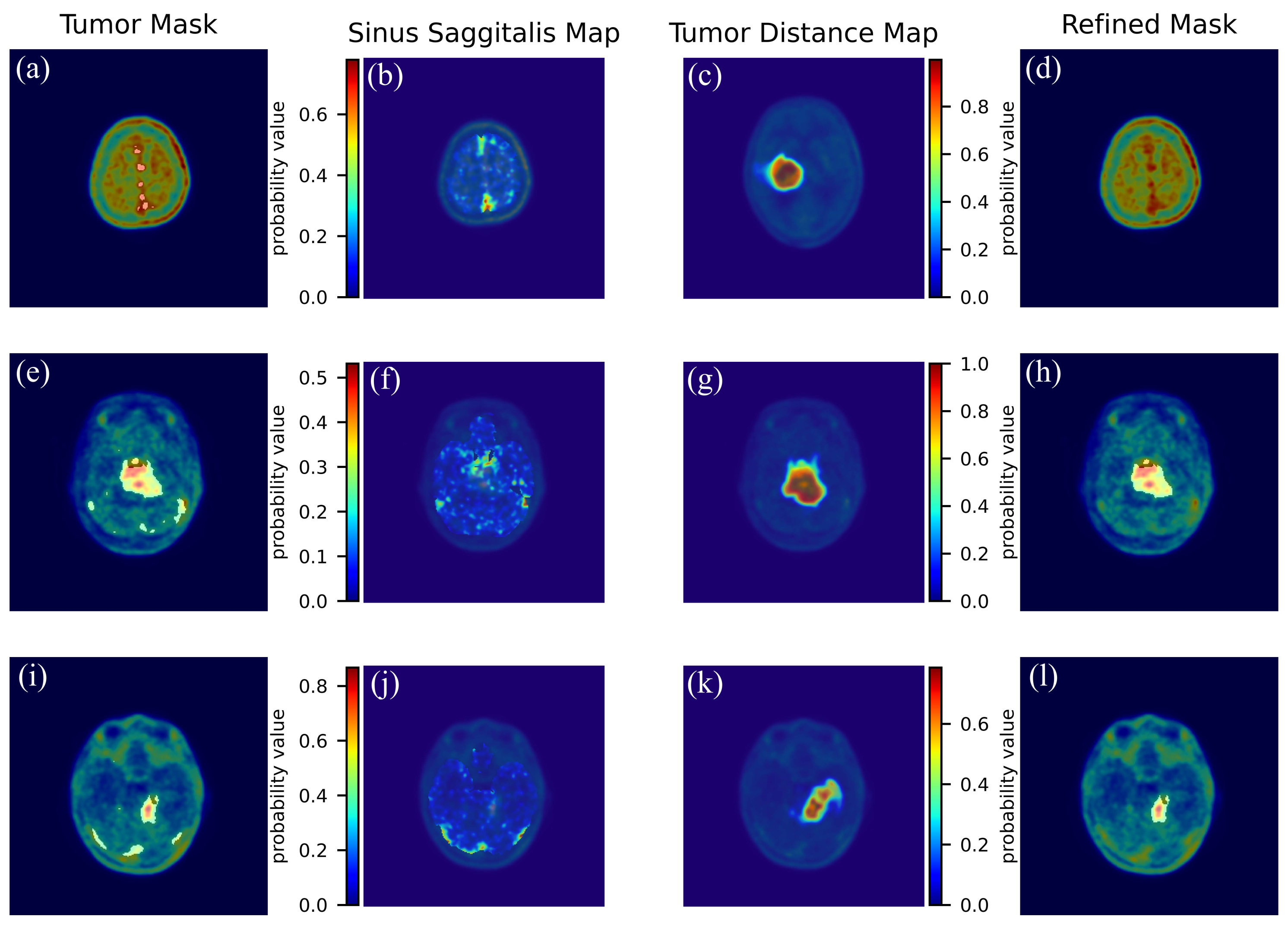
2.8. Enhancement of Tumour Segmentation
- We derived a probability map representing the sinus sagittalis by performing a weighted sum of the initial two frames of the 4D PET scan. We used the first two frames because the sagittal sinus signal decays in the following frames. Normalizing this volume to its maximum yields a pseudo-probability map.
- Next, we refined the tumor mask by removing small connected components and integrating it with the FLAIR tumor using logical OR operation. We then applied a Gaussian filter, with FWHM equal to 5, to generate a probability distance map.
2.9. Static Parameters Extraction
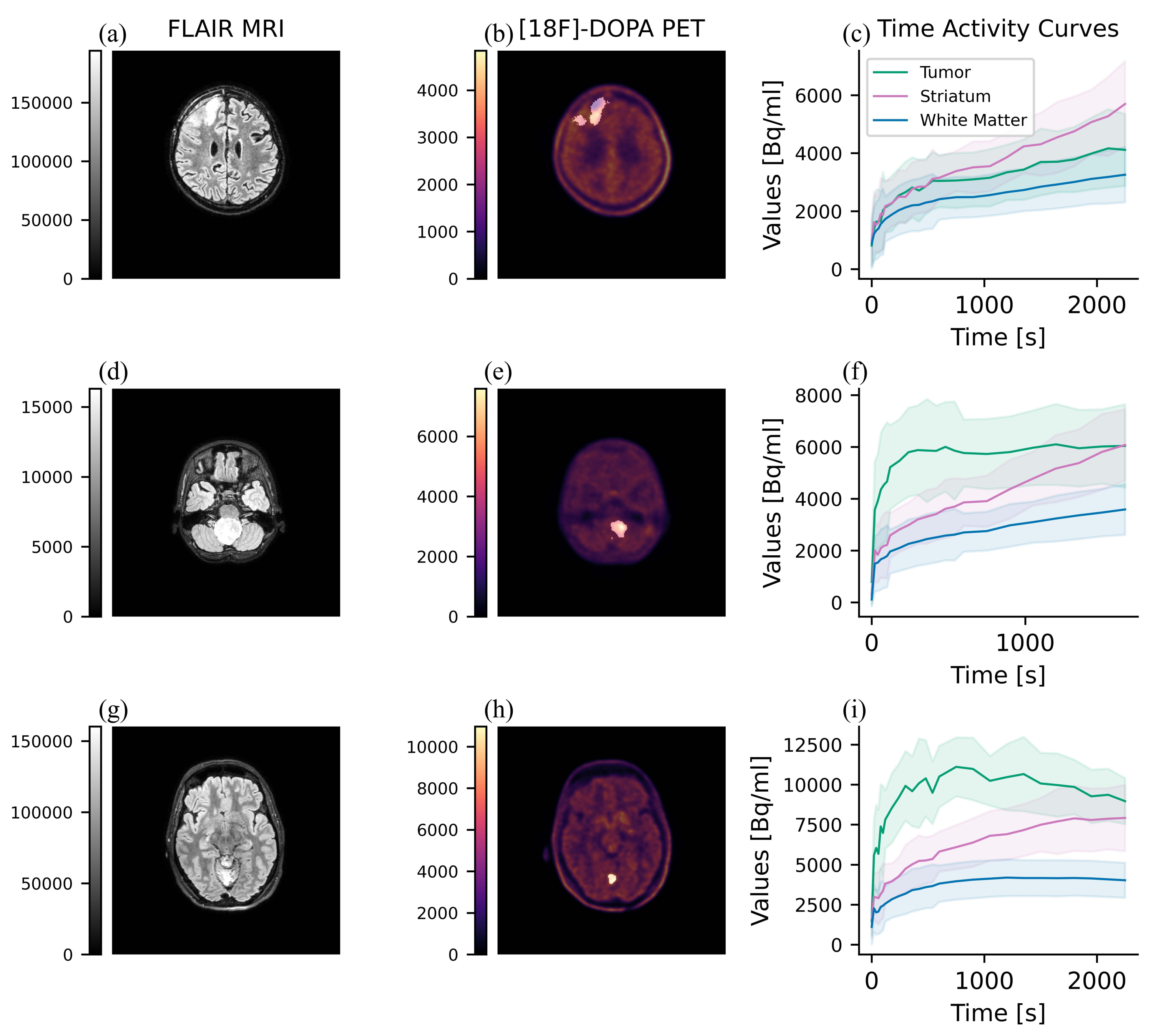
2.10. Dynamic Parameters Extraction
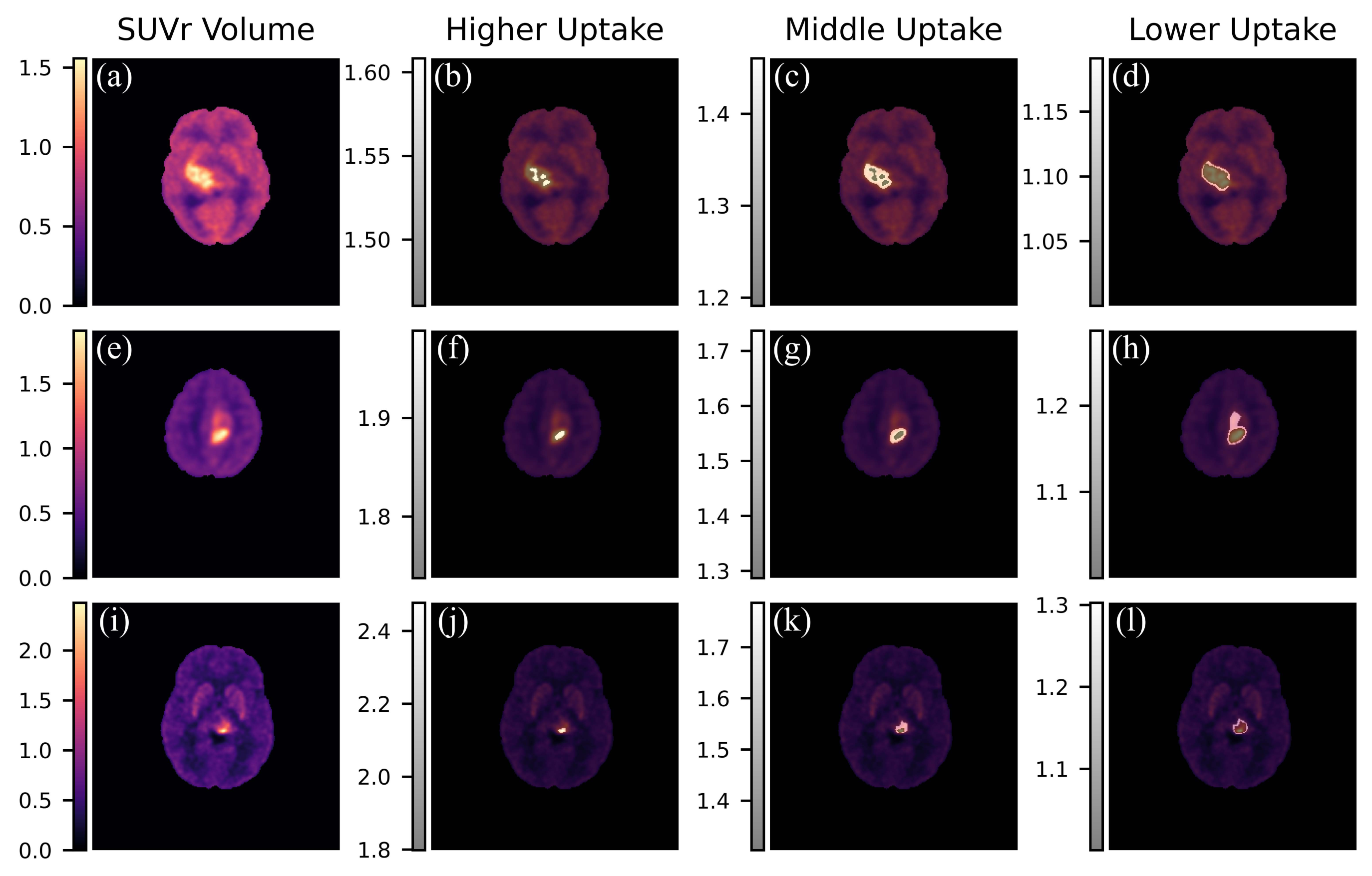
2.11. Intra-Tumor Heterogeneity Analysis
2.12. Manual Analysis
3. Results
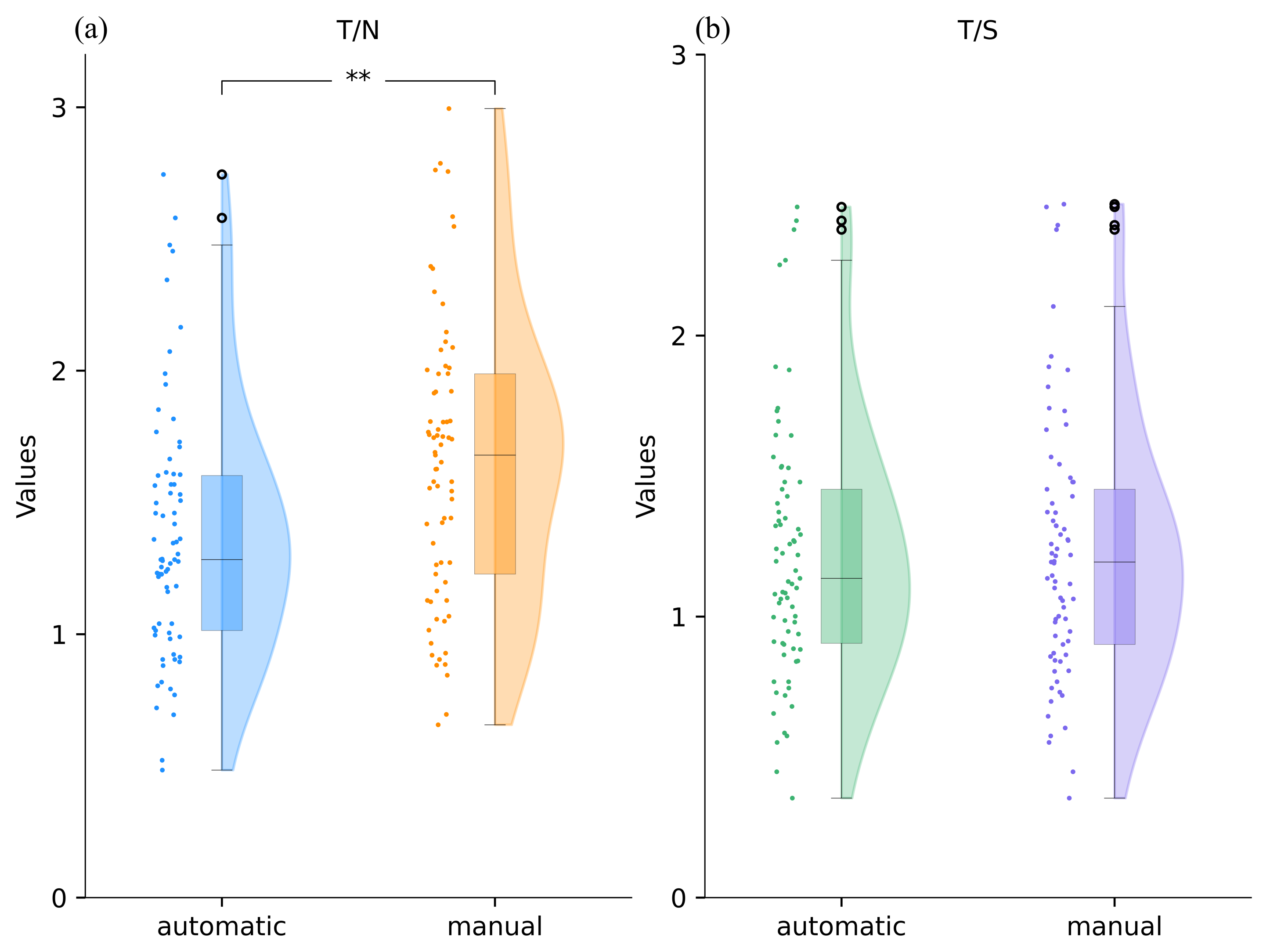
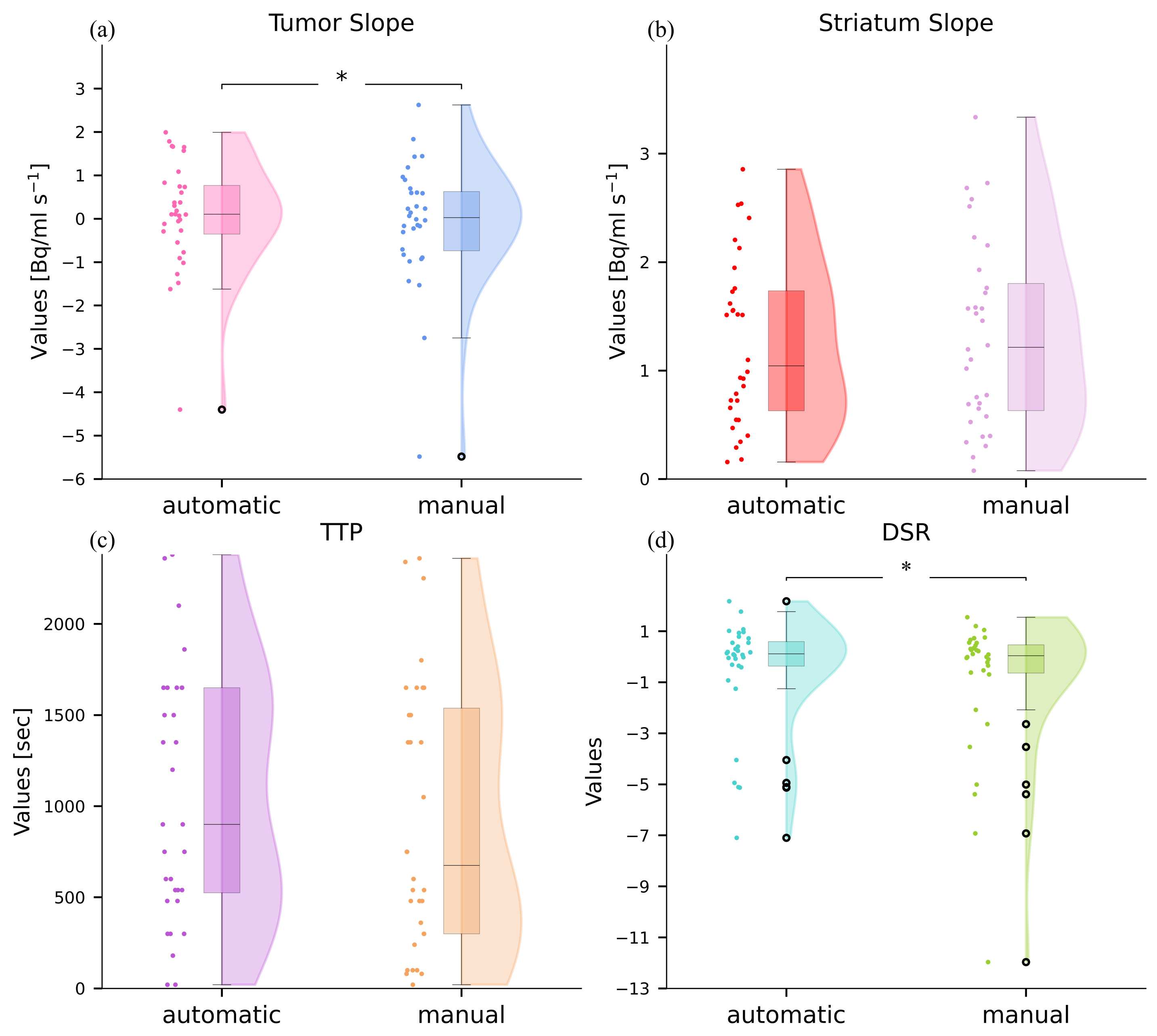
3.1. Comparison between Automatic and Manual Static Indexes
3.2. Comparison between Automatic and Manual Dynamic Indexes
3.3. Agreement between Ground Truth, Manual and Automatic Static Indexes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Hauser, P. Classification and treatment of pediatric gliomas in the molecular era. Children 2021, 8, 739. [Google Scholar] [CrossRef] [PubMed]
- Broniscer, A.; Gajjar, A. Supratentorial high-grade astrocytoma and diffuse brainstem glioma: two challenges for the pediatric oncologist. The Oncologist 2004, 9, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Langen, K.-J.; Galldiks, N. Update on amino acid PET of brain tumours. Curr. Opin. Neurol. 2018, 31, 354–361. [Google Scholar] [CrossRef]
- Masselli, G.; Casciani, E.; De Angelis, C.; Sollaku, S.; Gualdi, G. Clinical application of 18F-DOPA PET/CT in pediatric patients. Am. J. Nucl. Med. Mol. Imaging 2021, 11, 64. [Google Scholar] [PubMed]
- Carideo, L.; et al. 18F-DOPA uptake parameters in glioma: effects of patients’ characteristics and prior treatment history. Br. J. Radiol. 2018, 91, 20170847. [Google Scholar] [CrossRef]
- Janvier, L.; et al. Correlation of SUV-derived indices with tumoral aggressiveness of gliomas in static 18F-FDOPA PET: use in clinical practice. Clin. Nucl. Med. 2015, 40, e429–e435. [Google Scholar] [CrossRef]
- Verger, A.; et al. Static and dynamic 18F–FET PET for the characterization of gliomas defined by IDH and 1p/19q status. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 443–451. [Google Scholar] [CrossRef]
- Piccardo, A.; et al. Advanced MR imaging and 18F-DOPA PET characteristics of H3K27M-mutant and wild-type pediatric diffuse midline gliomas. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 1685–1694. [Google Scholar] [CrossRef]
- Buczkowicz, P.; Bartels, C.; Bouffet, E.; Becher, O.; Hawkins, C. Histopathological spectrum of paediatric diffuse intrinsic pontine glioma: diagnostic and therapeutic implications. Acta Neuropathol. 2014, 128, 573–581. [Google Scholar] [CrossRef]
- Piccardo, A.; et al. Joint EANM/SIOPE/RAPNO practice guidelines/SNMMI procedure standards for imaging of paediatric gliomas using PET with radiolabelled amino acids and [18F] FDG: version 1.0. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 3852–3869. [Google Scholar] [CrossRef]
- Verger, A.; Imbert, L.; Zaragori, T. Dynamic amino-acid PET in neuro-oncology: a prognostic tool becomes essential. Eur. J. Nucl. Med. Mol. Imaging 2021, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Fiz, F.; et al. Role of dynamic parameters of 18F-DOPA PET/CT in pediatric gliomas. Clin. Nucl. Med. 2022, 47, 517–524. [Google Scholar] [CrossRef]
- Law, I.; et al. Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and [18F] FDG: version 1.0. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 540–557. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; et al. 18F-FDOPA PET imaging of brain tumors: comparison study with 18F-FDG PET and evaluation of diagnostic accuracy. J. Nucl. Med. 2006, 47, 904–911. [Google Scholar] [PubMed]
- Patel, C.B.; et al. 18F-FDOPA PET and MRI characteristics correlate with degree of malignancy and predict survival in treatment-naïve gliomas: a cross-sectional study. J. Neuro-Oncol. 2018, 139, 399–409. [Google Scholar] [CrossRef]
- Funck, T.; Larcher, K.; Toussaint, P.-J.; Evans, A.C.; Thiel, A. APPIAN: automated pipeline for PET image analysis. Front. Neuroinformatics 2018, 12, 64. [Google Scholar] [CrossRef]
- Li, X.; Morgan, P.S.; Ashburner, J.; Smith, J.; Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 2016, 264, 47–56. [Google Scholar] [CrossRef]
- Gorgolewski, K.J.; et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 2016, 3, 1–9. [Google Scholar] [CrossRef]
- Jenkinson, M. BET2: MR-based estimation of brain, skull and scalp surfaces. In Eleventh Annual Meeting of the Organization for Human Brain Mapping, 2005.
- Desikan, R.S.; et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006, 31, 968–980. [Google Scholar] [CrossRef]
- Mazziotta, J.C.; et al. A probabilistic atlas of the human brain: theory and rationale for its development. Neuroimage 1995, 2, 89–101. [Google Scholar] [CrossRef]
- Avants, B.B.; et al. Advanced normalization tools (ANTS). Insight J. 2009, 2, 1–35. [Google Scholar]
- Hagos, Y.B.; Minh, V.H.; Khawaldeh, S.; Pervaiz, U.; Aleef, T.A. Fast PET scan tumor segmentation using superpixels, principal component analysis and K-means clustering. Methods and Protocols 2018, 1, 7. [Google Scholar] [CrossRef]
- Groendahl, A.R.; et al. A comparison of methods for fully automatic segmentation of tumors and involved nodes in PET/CT of head and neck cancers. Phys. Med. Biol. 2021, 66, 065012. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Nomura, Y.; et al. Characteristics of time-activity curves obtained from dynamic 11C-methionine PET in common primary brain tumors. J. Neuro-Oncol. 2018, 138, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Dallessio, K.M. Volume Computer-Assisted Reading for PET/CT. Appl. Radiol. 2007, 36, 30. [Google Scholar] [CrossRef]
- Shrout, P.E.; Fleiss, J.L. Intraclass correlations: uses in assessing rater reliability. Psychol. Bull. 1979, 86, 420. [Google Scholar] [CrossRef] [PubMed]
- Nordio, G.; et al. An automatic analysis framework for FDOPA PET neuroimaging. J. Cereb. Blood Flow Metab. 2023, 43, 1285–1300. [Google Scholar] [CrossRef]
- Peira, E.; et al. Towards an Automated Approach to the Semi-Quantification of [18F] F-DOPA PET in Pediatric-Type Diffuse Gliomas. J. Clin. Med. 2023, 12, 2765. [Google Scholar] [CrossRef]
- Morana, G.; et al. Diagnostic and prognostic value of 18F-DOPA PET and 1H-MR spectroscopy in pediatric supratentorial infiltrative gliomas: a comparative study. Neuro-Oncol. 2015, 17, 1637–1647. [Google Scholar] [CrossRef]
| Variable | Categories | Subjects Included (%) |
|---|---|---|
| Sex | Female, n(%) | 37 (50.6) |
| Male, n(%) | 36 (49.3) | |
| Age on diagnosis, median | <12 | 35 (47.9) |
| >=12 | 38 (52) | |
| Classification of Tumors | Circumscribed astrocytic gliomas | 6 (8.2) |
| Glioneuronal and neuronal tumours | 4 (5.4) | |
| Pediatric type diffuse low-grade gliomas | 16 (21.9) | |
| Pediatric type diffuse high-grade gliomas | 47 (64.3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





