Submitted:
03 September 2024
Posted:
04 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
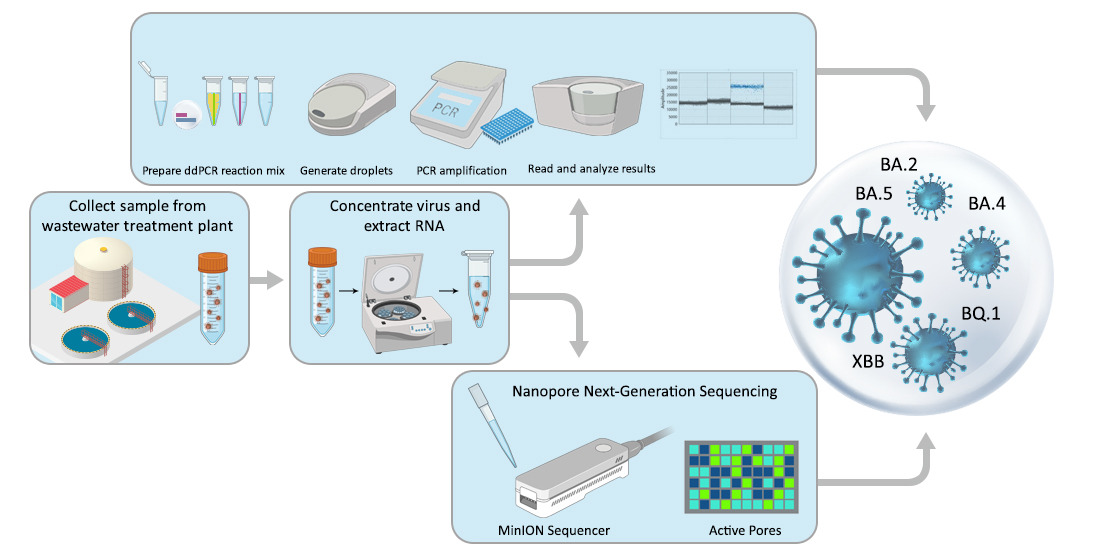
2. Materials and Methods
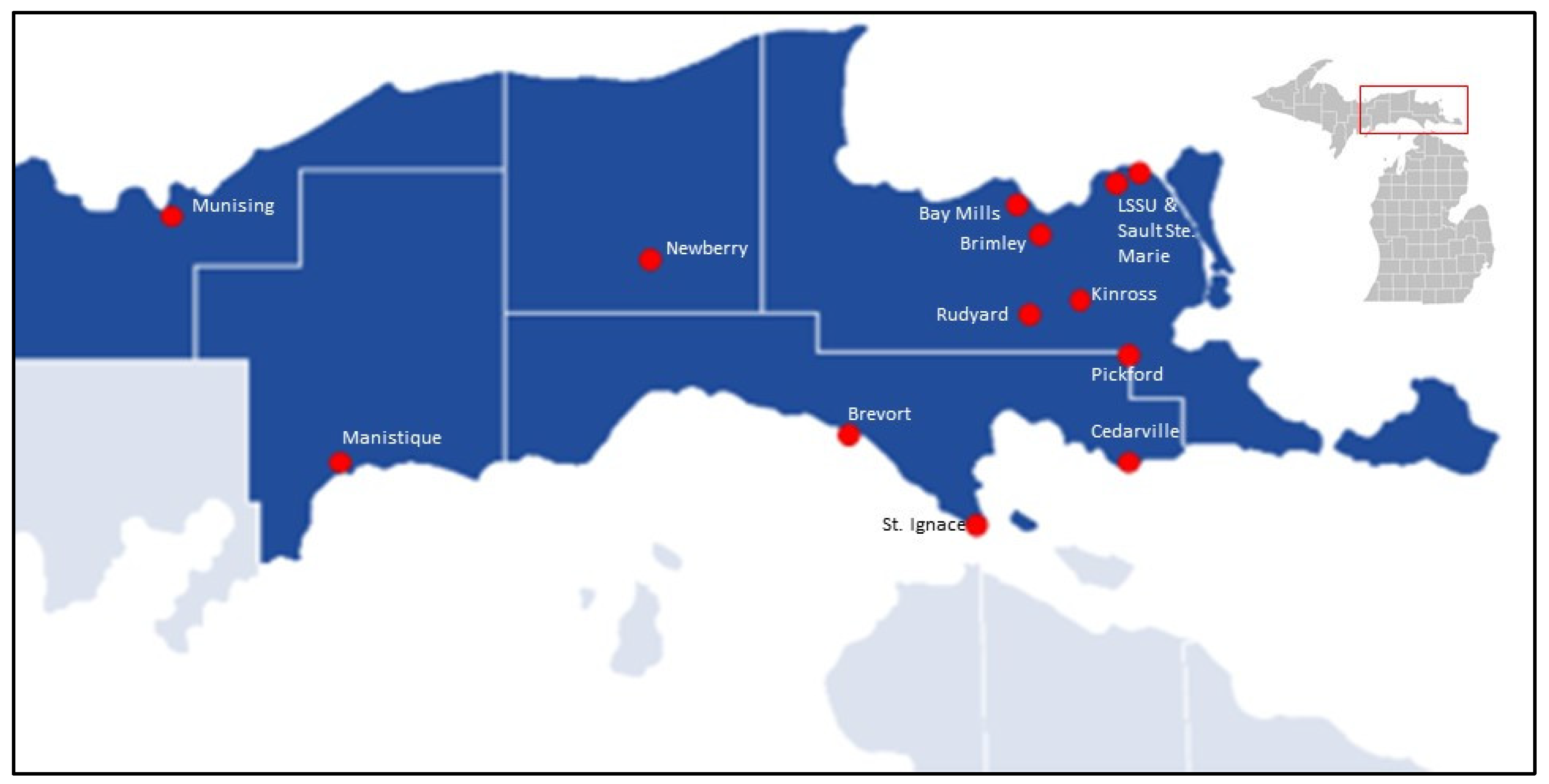
2.1. Study Location
2.2. Wastewater Sampling
2.3. Viral Concentration
2.4. RNA Extraction
2.5. Initial Virus Detection and Quantification
2.6. Variant Determination using ddPCR
2.7. Variant Determination using Genome Sequencing
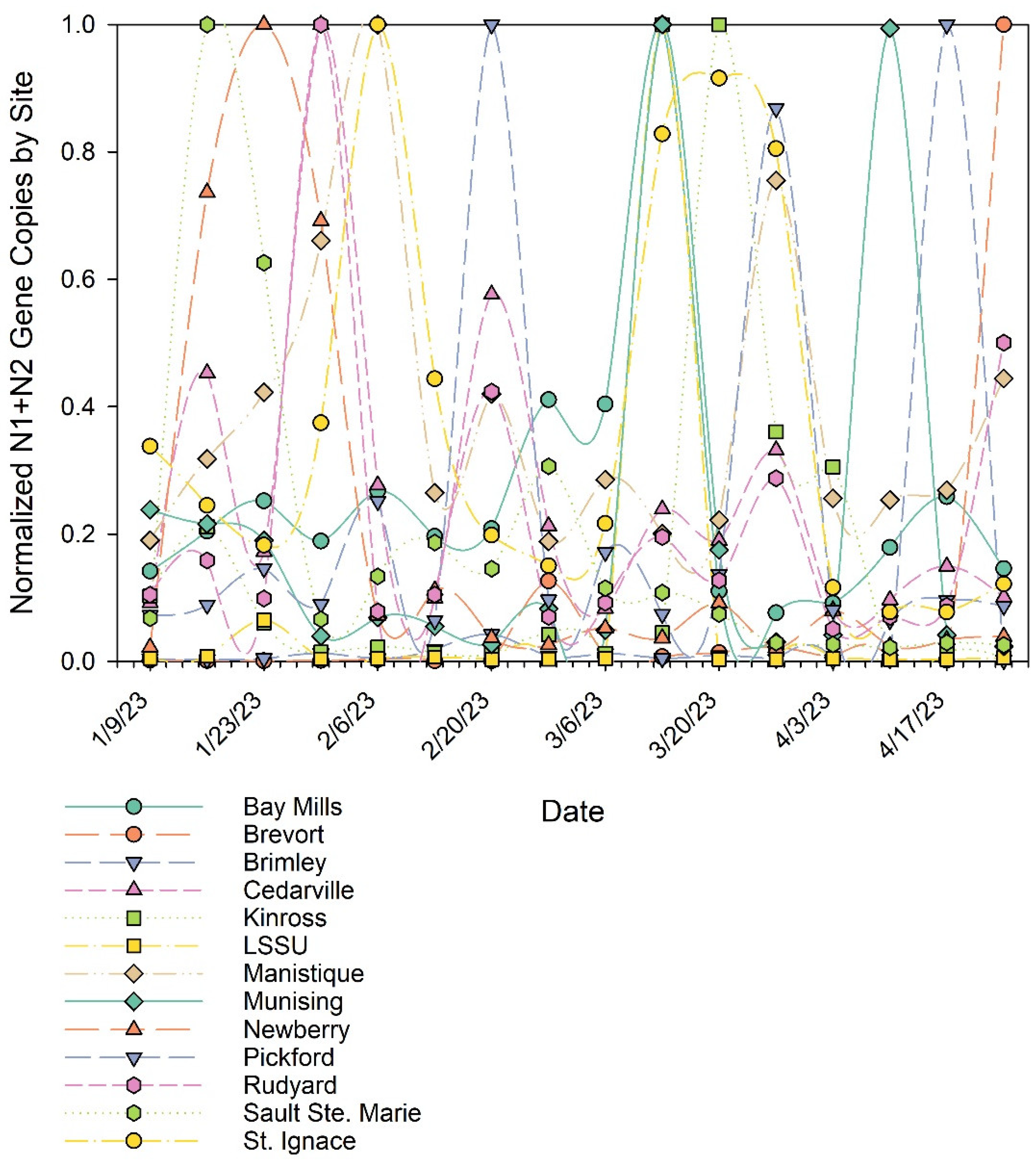
3. Results and Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kaku, Y.; Okumura, K.; Padilla-Blanco, M.; Kosugi, Y.; Uriu, K.; A Hinay, A.; Chen, L.; Plianchaisuk, A.; Kobiyama, K.; Ishii, K.J.; et al. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect. Dis. 2024, 24, e82. [Google Scholar] [CrossRef] [PubMed]
- Kunal, S. , Ish, P., Aditi, & Gupta, K. (2022). Emergence of COVID-19 variants and its global impact. In S. Adibi, P. Griffin, M. Sanicas, M. Rashidi, & F. Lanfranchi (Eds.), Frontiers of COVID-19: Scientific and Clinical Aspects of the Novel Coronavirus 2019 (pp. 183–201). Springer International Publishing. [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Wang R, Chen J, Gao K, Hozumi Y, Yin C, Wei GW. Analysis of SARS-CoV-2 mutations in the United States suggests presence of four substrains and novel variants. Commun Biol. 2021 Feb 15;4(1):228. 10.1038/s42003-021-01754-6. Erratum in: Commun Biol. 2021 Mar 3;4(1):311. 10.1038/s42003-021-01867-y.
- Davis, C.; Logan, N.; Tyson, G.; Orton, R.; Harvey, W.T.; Perkins, J.S.; Mollett, G.; Blacow, R.M.; Peacock, T.P.; Barclay, W.S.; et al. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLOS Pathog. 2021, 17, e1010022. [Google Scholar] [CrossRef] [PubMed]
- Pilapil, J.D.; Notarte, K.I.; Yeung, K.L. The dominance of co-circulating SARS-CoV-2 variants in wastewater. Int. J. Hyg. Environ. Heal. 2023, 253, 114224. [Google Scholar] [CrossRef] [PubMed]
- Rothman, J.A.; Saghir, A.; Zimmer-Faust, A.G.; Langlois, K.; Raygoza, K.; Steele, J.A.; Griffith, J.F.; Whiteson, K.L. Longitudinal Sequencing and Variant Detection of SARS-CoV-2 across Southern California Wastewater. Appl. Microbiol. 2024, 4, 635–649. [Google Scholar] [CrossRef]
- Ahmed, W.; Bivins, A.; Simpson, S.L.; Bertsch, P.M.; Ehret, J.; Hosegood, I.; Metcalfe, S.S.; Smith, W.J.; Thomas, K.V.; Tynan, J.; et al. Wastewater surveillance demonstrates high predictive value for COVID-19 infection on board repatriation flights to Australia. Environ. Int. 2022, 158, 106938–106938. [Google Scholar] [CrossRef]
- Barbé, L.; Schaeffer, J.; Besnard, A.; Jousse, S.; Wurtzer, S.; Moulin, L.; OBEPINE Consortium; Le Guyader, F. S.; Desdouits, M. SARS-CoV-2 Whole-Genome Sequencing Using Oxford Nanopore Technology for Variant Monitoring in Wastewaters. Front. Microbiol. 2022, 13, 889811. [Google Scholar] [CrossRef]
- Bar-Or, I.; Weil, M.; Indenbaum, V.; Bucris, E.; Bar-Ilan, D.; Elul, M.; Levi, N.; Aguvaev, I.; Cohen, Z.; Shirazi, R.; et al. Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci. Total. Environ. 2021, 789, 148002. [Google Scholar] [CrossRef]
- Cancela, F.; Ramos, N.; Smyth, D.S.; Etchebehere, C.; Berois, M.; Rodríguez, J.; Rufo, C.; Alemán, A.; Borzacconi, L.; López, J.; et al. Wastewater surveillance of SARS-CoV-2 genomic populations on a country-wide scale through targeted sequencing. PLOS ONE 2023, 18, e0284483. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A.; et al. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 2021, 12. [Google Scholar] [CrossRef]
- Tyson, J.R.; James, P.; Stoddart, D.; Sparks, N.; Wickenhagen, A.; Hall, G.; Choi, J.H.; Lapointe, H.; Kamelian, K.; Smith, A.D.; et al. Improvements to the ARTIC Multiplex PCR Method for SARS-CoV-2 Genome Sequencing Using Nanopore. Biorxiv Prepr. Serv. Biol. 2020. [Google Scholar] [CrossRef]
- Izquierdo-Lara, R.; Elsinga, G.; Heijnen, L.; Munnink, B.B.O.; Schapendonk, C.M.; Nieuwenhuijse, D.; Kon, M.; Lu, L.; Aarestrup, F.M.; Lycett, S.; et al. Monitoring SARS-CoV-2 Circulation and Diversity through Community Wastewater Sequencing, the Netherlands and Belgium. Emerg. Infect. Dis. 2021, 27, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Surya, K.; Buyukyoruk, M.; Cicha, C.; Vanderwood, K.K.; Wilkinson, R.; Wiedenheft, B. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. Cell Rep. Med. 2020, 1, 100098–100098. [Google Scholar] [CrossRef]
- Vigil, K.; D'Souza, N.; Bazner, J.; Cedraz, F.M.-A.; Fisch, S.; Rose, J.B.; Aw, T.G. Long-term monitoring of SARS-CoV-2 variants in wastewater using a coordinated workflow of droplet digital PCR and nanopore sequencing. Water Res. 2024, 254, 121338. [Google Scholar] [CrossRef] [PubMed]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- World Health Organization. (2022). Methods-for-the-detection-char-SARS-CoV-2-variants_2nd update_final.pdf (pp. 1–14) [Technical Report]. https://www.ecdc.europa.eu/sites/default/files/documents/Methods-for-the-detection-char-SARS-CoV-2-variants_2nd%20update_final.pdf.
- Child, H.T.; Airey, G.; Maloney, D.M.; Parker, A.; Wild, J.; McGinley, S.; Evens, N.; Porter, J.; Templeton, K.; Paterson, S.; et al. Comparison of metagenomic and targeted methods for sequencing human pathogenic viruses from wastewater. mBio 2023, 14, e0146823. [Google Scholar] [CrossRef]
- Joshi, M.; Kumar, M.; Srivastava, V.; Kumar, D.; Rathore, D.S.; Pandit, R.; Graham, D.W.; Joshi, C.G. Genetic sequencing detected the SARS-CoV-2 delta variant in wastewater a month prior to the first COVID-19 case in Ahmedabad (India). Environ. Pollut. 2022, 310, 119757. [Google Scholar] [CrossRef]
- Tiwari, A.; Adhikari, S.; Zhang, S.; Solomon, T.B.; Lipponen, A.; Islam, A.; Thakali, O.; Sangkham, S.; Shaheen, M.N.F.; Jiang, G.; et al. Tracing COVID-19 Trails in Wastewater: A Systematic Review of SARS-CoV-2 Surveillance with Viral Variants. Water 2023, 15, 1018. [Google Scholar] [CrossRef]
- Lind, A.; Barlinn, R.; Landaas, E.T.; Andresen, L.L.; Jakobsen, K.; Fladeby, C.; Nilsen, M.; Bjørnstad, P.M.; Sundaram, A.Y.; Ribarska, T.; et al. Rapid SARS-CoV-2 variant monitoring using PCR confirmed by whole genome sequencing in a high-volume diagnostic laboratory. J. Clin. Virol. 2021, 141, 104906–104906. [Google Scholar] [CrossRef]
- Umunnakwe, C. , Makatini, Z., Maphanga, M., Mdunyelwa, A., Mlambo, K. M., Manyaka, P., Nijhuis, M., Wensing, A., & Tempelman, H. (2022). Evaluation of a commercial SARS-CoV-2 multiplex PCR genotyping assay for variant identification in resource-scarce settings. PLOS ONE, 17, e0269071. https://doi.org/10.1371/journal.pone.0269071Lind, A., Barlinn, R., Landaas, E. T., Andresen, L. L., Jakobsen, K., Fladeby, C., Nilsen, M., Bjørnstad, P. M., Sundaram, A. Y. M., Ribarska, T., Müller, F., Gilfillan, G. D., & Holberg-Petersen, M. (2021). Rapid SARS-CoV-2 variant monitoring using PCR confirmed by whole genome sequencing in a high-volume diagnostic laboratory. Journal of Clinical Virology, 141, 104906. https://doi.org/10.1016/j.jcv.2021.104906.
- D'Agostino, Y.; Rocco, T.; Ferravante, C.; Porta, A.; Tosco, A.; Cappa, V.M.; Lamberti, J.; Alexandrova, E.; Memoli, D.; Terenzi, I.; et al. Rapid and sensitive detection of SARS-CoV-2 variants in nasopharyngeal swabs and wastewaters. Diagn. Microbiol. Infect. Dis. 2022, 102, 115632–115632. [Google Scholar] [CrossRef]
- Tangwangvivat, R.; Wacharapluesadee, S.; Pinyopornpanish, P.; Petcharat, S.; Hearn, S.M.; Thippamom, N.; Phiancharoen, C.; Hirunpatrawong, P.; Duangkaewkart, P.; Supataragul, A.; et al. SARS-CoV-2 Variants Detection Strategies in Wastewater Samples Collected in the Bangkok Metropolitan Region. Viruses 2023, 15, 876. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.; Sashittal, P.; Zhou, A.; Wang, L.; El-Kebir, M.; Nguyen, T.H. Design of SARS-CoV-2 Variant-Specific PCR Assays Considering Regional and Temporal Characteristics. Appl. Environ. Microbiol. 2022, 88, e0228921. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, L.D.; Richter, S.R.; Midgley, S.E.; Franck, K.T. Detecting SARS-CoV-2 Omicron B.1.1.529 Variant in Wastewater Samples by Using Nanopore Sequencing. Emerg. Infect. Dis. 2022, 28, 1296–1298. [Google Scholar] [CrossRef]
- Gregory, D.A.; Wieberg, C.G.; Wenzel, J.; Lin, C.-H.; Johnson, M.C. Monitoring SARS-CoV-2 Populations in Wastewater by Amplicon Sequencing and Using the Novel Program SAM Refiner. Viruses 2021, 13, 1647. [Google Scholar] [CrossRef]
- Focosi, D.; Quiroga, R.; McConnell, S.; Johnson, M.C.; Casadevall, A. Convergent Evolution in SARS-CoV-2 Spike Creates a Variant Soup from Which New COVID-19 Waves Emerge. Int. J. Mol. Sci. 2023, 24, 2264. [Google Scholar] [CrossRef] [PubMed]
- Pastorio, C.; Noettger, S.; Nchioua, R.; Zech, F.; Sparrer, K.M.; Kirchhoff, F. Impact of mutations defining SARS-CoV-2 Omicron subvariants BA.2.12.1 and BA.4/5 on Spike function and neutralization. iScience 2023, 26, 108299. [Google Scholar] [CrossRef]
- Biobot Analytics. (n.d.). COVID-19, Influenza, and RSV wastewater monitoring in the U.S. Retrieved May 1, 2023, from https://biobot.io/data/.
- Gupta, S.; Kumar, A.; Gupta, N.; Bharti, D.R.; Aggarwal, N.; Ravi, V. A two-step process for in silico screening to assess the performance of qRTPCR kits against variant strains of SARS-CoV-2. BMC Genom. 2022, 23, 755. [Google Scholar] [CrossRef]
- Sharma, S.; Shrivastava, S.; Kausley, S.B.; Rai, B.; Pandit, A.B. Coronavirus: a comparative analysis of detection technologies in the wake of emerging variants. Infection 2022, 51, 1–19. [Google Scholar] [CrossRef]
- Dip, S.D.; Sarkar, S.L.; Setu, A.A.; Das, P.K.; Pramanik, H.A.; Alam, A.S.M.R.U.; Al-Emran, H.M.; Hossain, M.A.; Jahid, I.K. Evaluation of RT-PCR assays for detection of SARS-CoV-2 variants of concern. Sci. Rep. 2023, 13, 2342. [Google Scholar] [CrossRef]
- Nextstrain. (2024). Nextstrain Annual Update March 2024. https://nextstrain.org/blog/2024-03-27-annual-update-march-2024.
- Fontenele, R.S.; Kraberger, S.; Hadfield, J.; Driver, E.M.; Bowes, D.; Holland, L.A.; Faleye, T.O.; Adhikari, S.; Kumar, R.; Inchausti, R.; et al. High-throughput sequencing of SARS-CoV-2 in wastewater provides insights into circulating variants. Water Res. 2021, 205, 117710. [Google Scholar] [CrossRef]
- Rader, B.; Gertz, A.; Iuliano, A.D.; Gilmer, M.; Wronski, L.; Astley, C.M.; Sewalk, K.; Varrelman, T.J.; Cohen, J.; Parikh, R.; et al. Use of At-Home COVID-19 Tests — United States, August 23, 2021–March 12, 2022. Mmwr. Morb. Mortal. Wkly. Rep. 2022, 71, 489–494. [Google Scholar] [CrossRef]
- Usher, A.D. FIND documents dramatic reduction in COVID-19 testing. Lancet Infect. Dis. 2022, 22, 949. [Google Scholar] [CrossRef] [PubMed]
- Flood, M.T.; Sharp, J.; Bruggink, J.; Cormier, M.; Gomes, B.; Oldani, I.; Zimmy, L.; Rose, J.B. Understanding the efficacy of wastewater surveillance for SARS-CoV-2 in two diverse communities. PLOS ONE 2023, 18, e0289343. [Google Scholar] [CrossRef] [PubMed]
- Rabe, A.; Ravuri, S.; Burnor, E.; Steele, J.A.; Kantor, R.S.; Choi, S.; Forman, S.; Batjiaka, R.; Jain, S.; León, T.M.; et al. Correlation between wastewater and COVID-19 case incidence rates in major California sewersheds across three variant periods. J. Water Heal. 2023, 21, 1303–1317. [Google Scholar] [CrossRef] [PubMed]
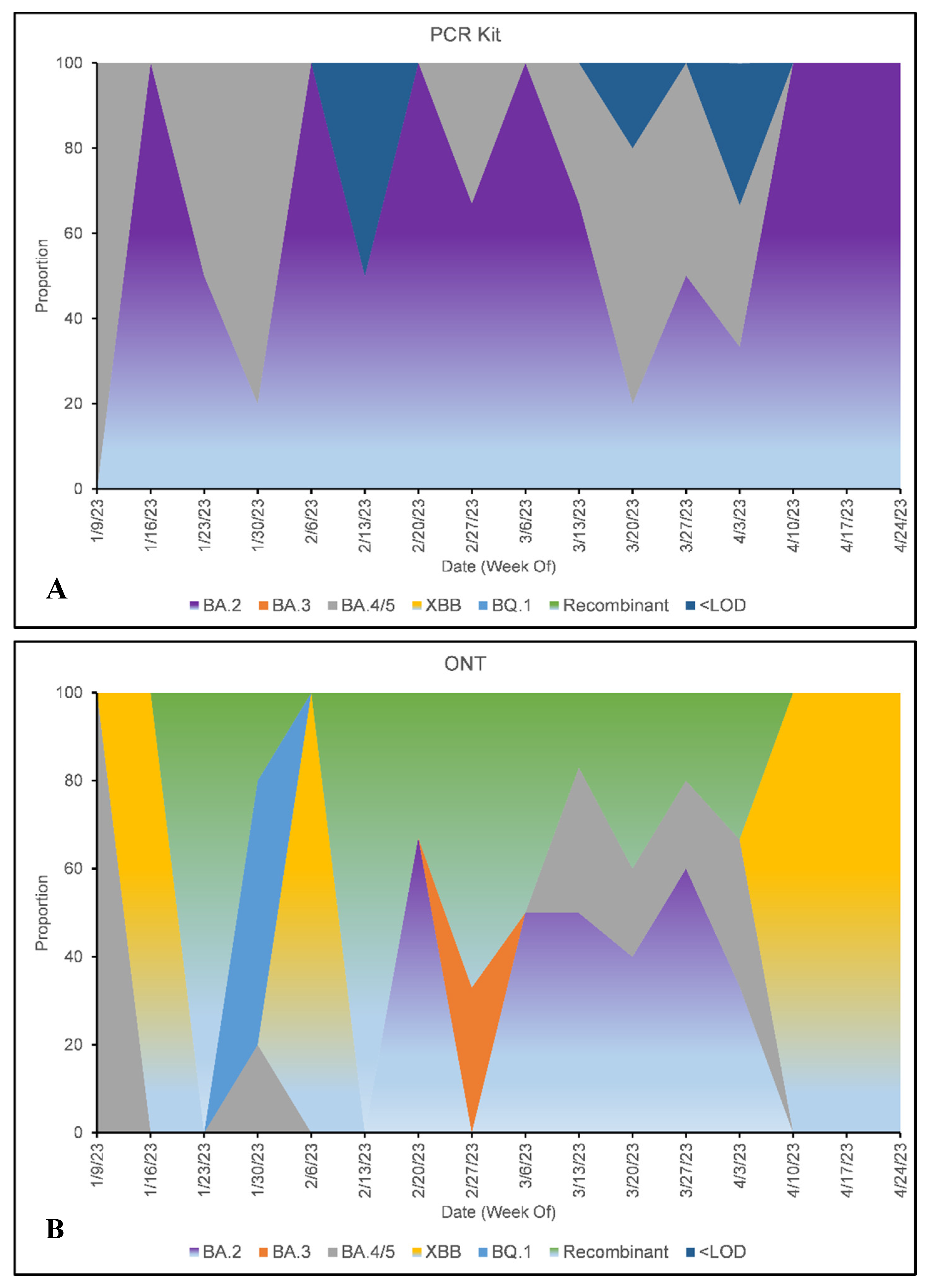
| DATE | SITE | ONT | PCR KIT |
| 3/14/2023 | Bay Mills Community | BA.5 | BA.1 and BA.2 (4/5) |
| 3/1/2023 | Brevort | BA.3 | BA.2 |
| 3/8/2023 | Brevort | BA.2 | BA.2 |
| 3/15/2023 | Brevort | BA.5 | BA.1 and BA.2 (4/5) |
| 3/22/2023 | Brevort | BA.5 | BA.1 and BA.2 (4/5) |
| 3/29/2023 | Brevort | BA.2 | BA.1 and BA.2 (4/5) |
| 4/5/2023 | Brevort | BA.5 | BA.1 and BA.2 (4/5) |
| 4/26/2023 | Brevort | XBB | BA.2 |
| 1/31/2023 | Brimley | Recombinant | BA.2 |
| 3/7/2023 | Brimley | Recombinant | BA.2 |
| 4/18/2023 | Brimley | XBB | BA.2 |
| 2/2/2023 | Cedarville | BQ.1 | BA.1 and BA.2 (4/5) |
| 2/23/2023 | Cedarville | BA.2.10 | BA.2 |
| 1/10/2023 | Kinross | BA.5 | BA.1 and BA.2 (4/5) |
| 2/28/2023 | Kinross | Recombinant | BA.1 and BA.2 (4/5) |
| 3/14/2023 | Kinross | Recombinant | BA.2 |
| 3/21/2023 | Kinross | BA.2.10 | BA.2 |
| 3/28/2023 | Kinross | BA.2.10 | BA.2 |
| 4/4/2023 | Kinross | BA.2 | BA.2 |
| 3/15/2023 | LSSU | BA.2.10 | BA.2 |
| 1/30/2023 | Manistique | BQ.1 | BA.1 and BA.2 (4/5) |
| 3/20/2023 | Manistique | BA.2 | <LOD |
| 1/23/2023 | Munising | Recombinant | BA.2 |
| 3/13/2023 | Munising | BA.2 | BA.2 |
| 3/20/2023 | Munising | Recombinant | BA.1 and BA.2 (4/5) |
| 4/10/2023 | Munising | XBB | BA.2 |
| 1/31/2023 | Newberry | BA.5 | BA.1 and BA.2 (4/5) |
| 2/14/2023 | Newberry | Recombinant | <LOD |
| 4/4/2023 | Newberry | Recombinant | <LOD |
| 2/22/2023 | Pickford | BA.2 | BA.2 |
| 3/29/2023 | Pickford | BA.5 | BA.1 and BA.2 (4/5) |
| 2/2/2023 | Rudyard | BQ.1 | BA.1 and BA.2 (4/5) |
| 2/23/2023 | Rudyard | Recombinant | BA.2 |
| 3/30/2023 | Rudyard | Recombinant | BA.2 |
| 4/27/2023 | Rudyard | XBB | BA.2 |
| 1/18/2023 | Sault Ste. Marie | XBB | BA.2 |
| 1/24/2023 | Sault Ste. Marie | Recombinant | BA.1 and BA.2 (4/5) |
| 2/14/2023 | Sault Ste. Marie | Recombinant | BA.2 |
| 2/28/2023 | Sault Ste. Marie | Recombinant | BA.2 |
| 2/8/2023 | St. Ignace | XBB | BA.2 |
| 3/15/2023 | St. Ignace | BA.2 | BA.2 |
| 3/22/2023 | St. Ignace | Recombinant | BA.1 and BA.2 (4/5) |
| 3/29/2023 | St. Ignace | BA.2.10 | BA.2 |
| SUBVARIANT | ONT | PCR KIT |
|---|---|---|
| BA.2 | 28% | 58% |
| BA.3 | 2% | 0% |
| BA.4 or BA.5 | 14% | 25% |
| XBB | 14% | 0% |
| BQ.1 | 7% | 0% |
| Recombinant | 35% | 0% |
| <LOD | N/A | 7% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





