Submitted:
04 September 2024
Posted:
05 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
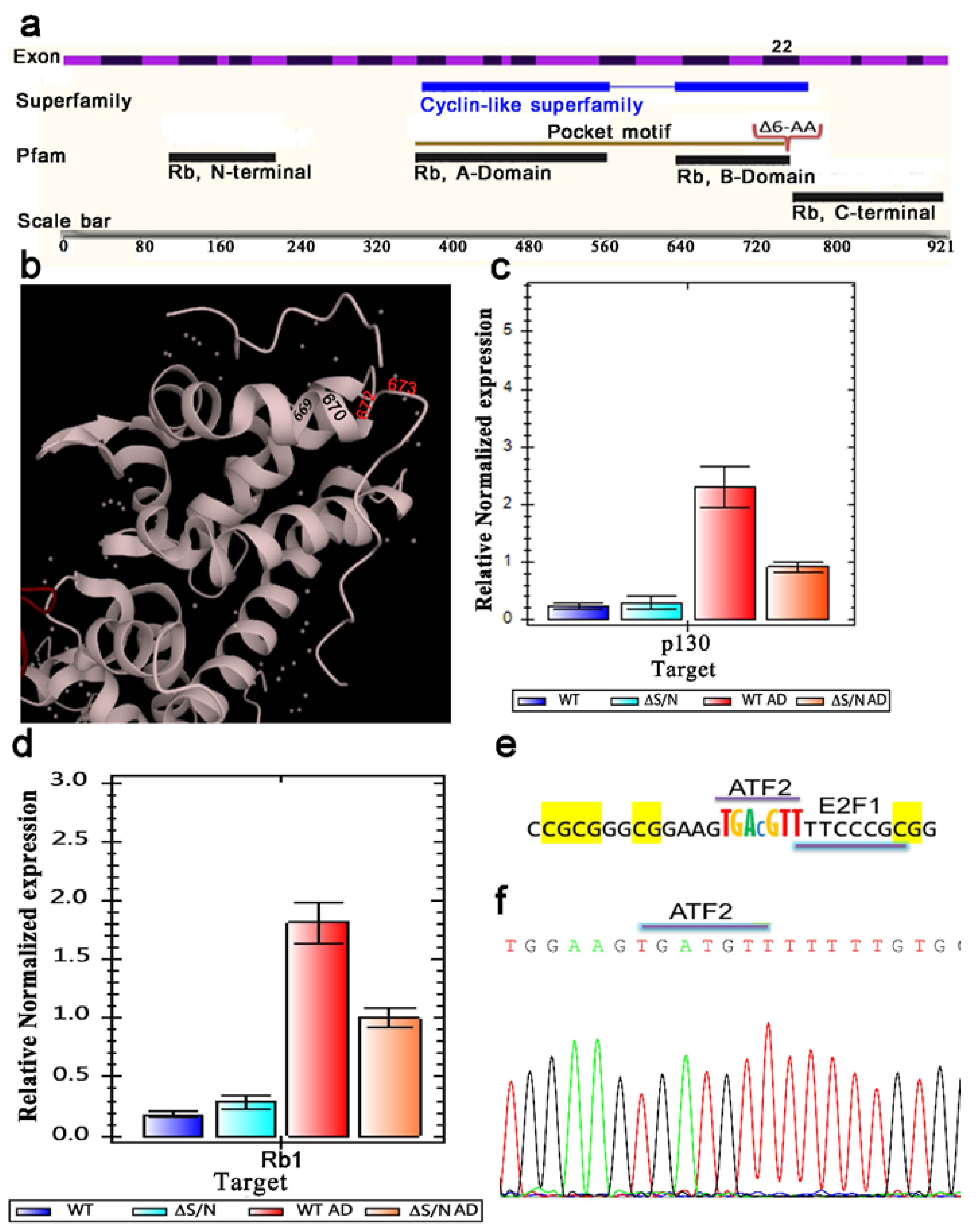
2.1. Alterations in Rb1 Expression in Cells That Temporarily Expressed ΔS/N Rb1
2.1.1. ΔS/N Cells Overexpress Rb1 in the Undifferentiated State
2.1.2. Rb1 and p130 Show Weak Induction during Adipogenic Differentiation in ΔS/N Cells
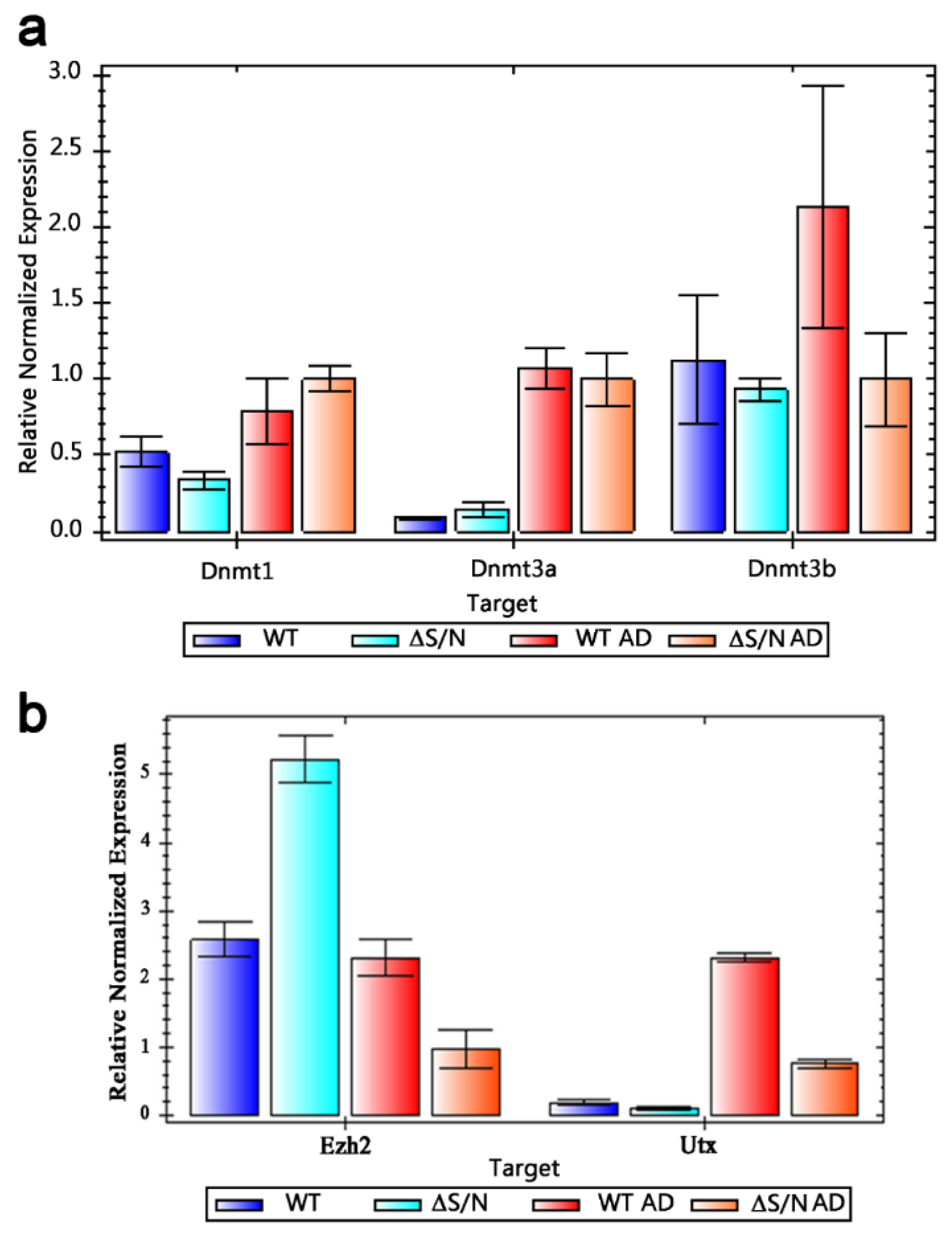
2.2. Dnmt3a Is Upregulated in Undifferentiated ΔS/N Cells
2.3. Ezh2 Is Highly Overexpressed in Undifferentiated ΔS/N Cells
2.4. Moderate UTX Induction Occurs in Differentiated ΔS/N Cells
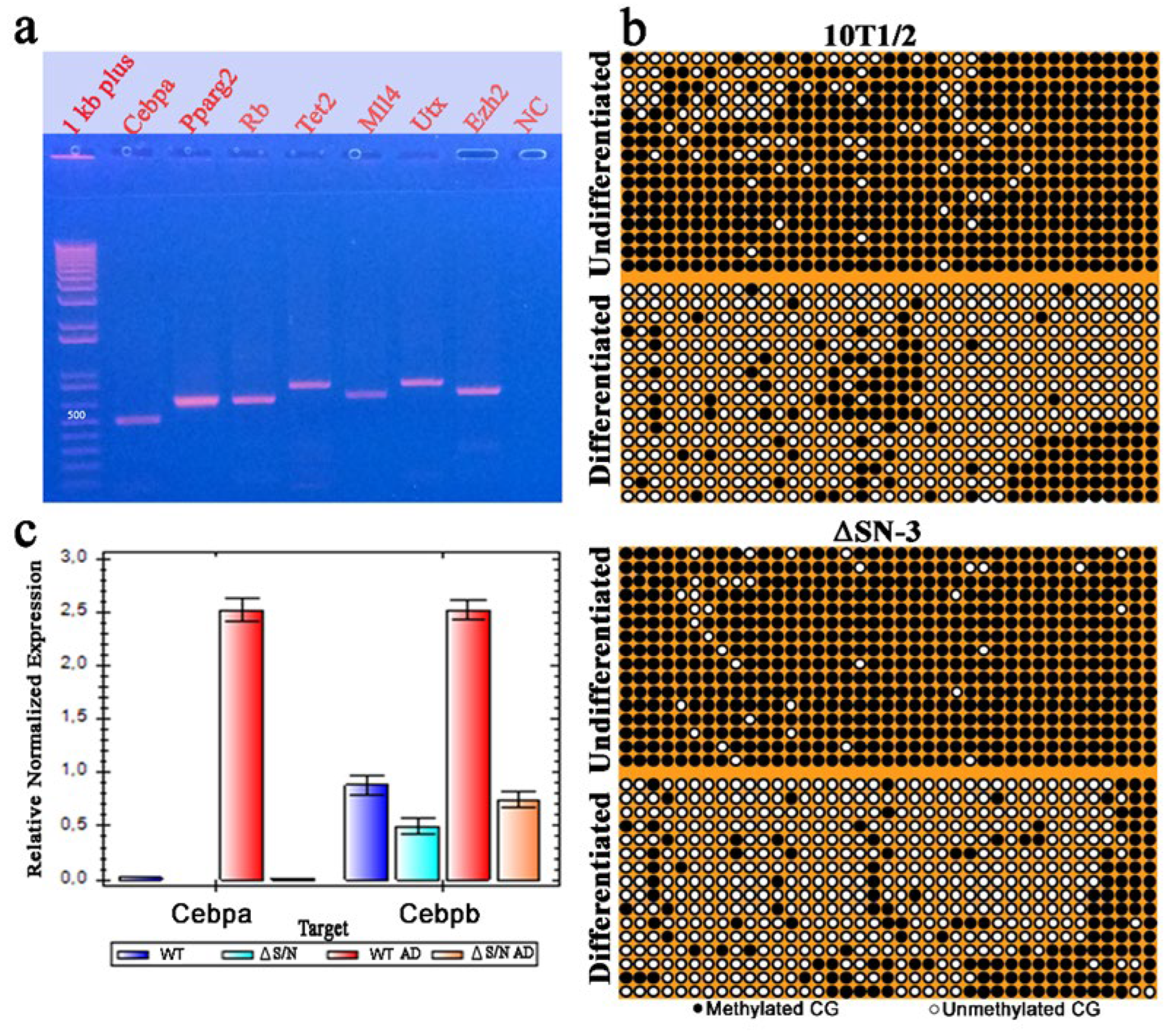
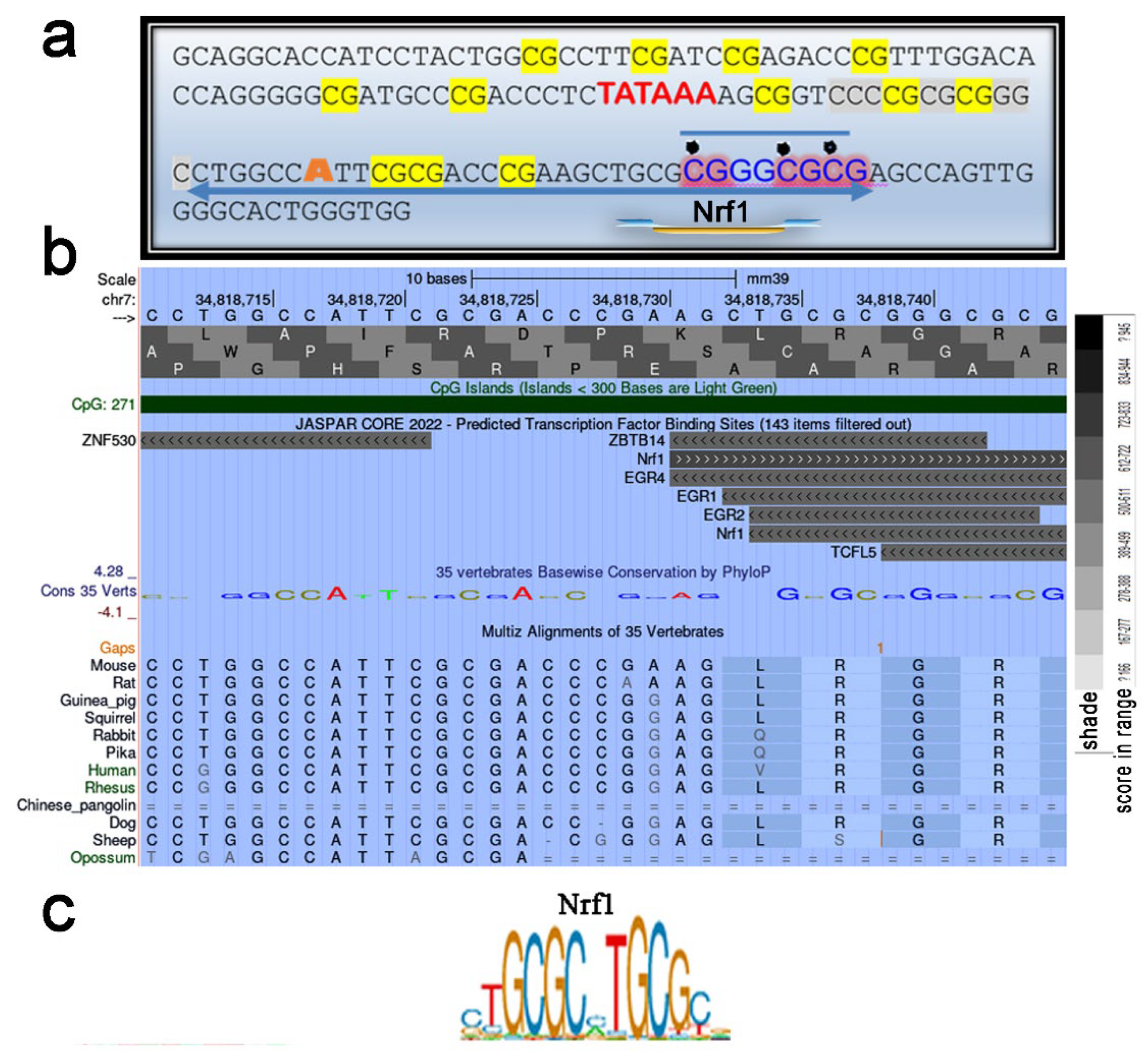
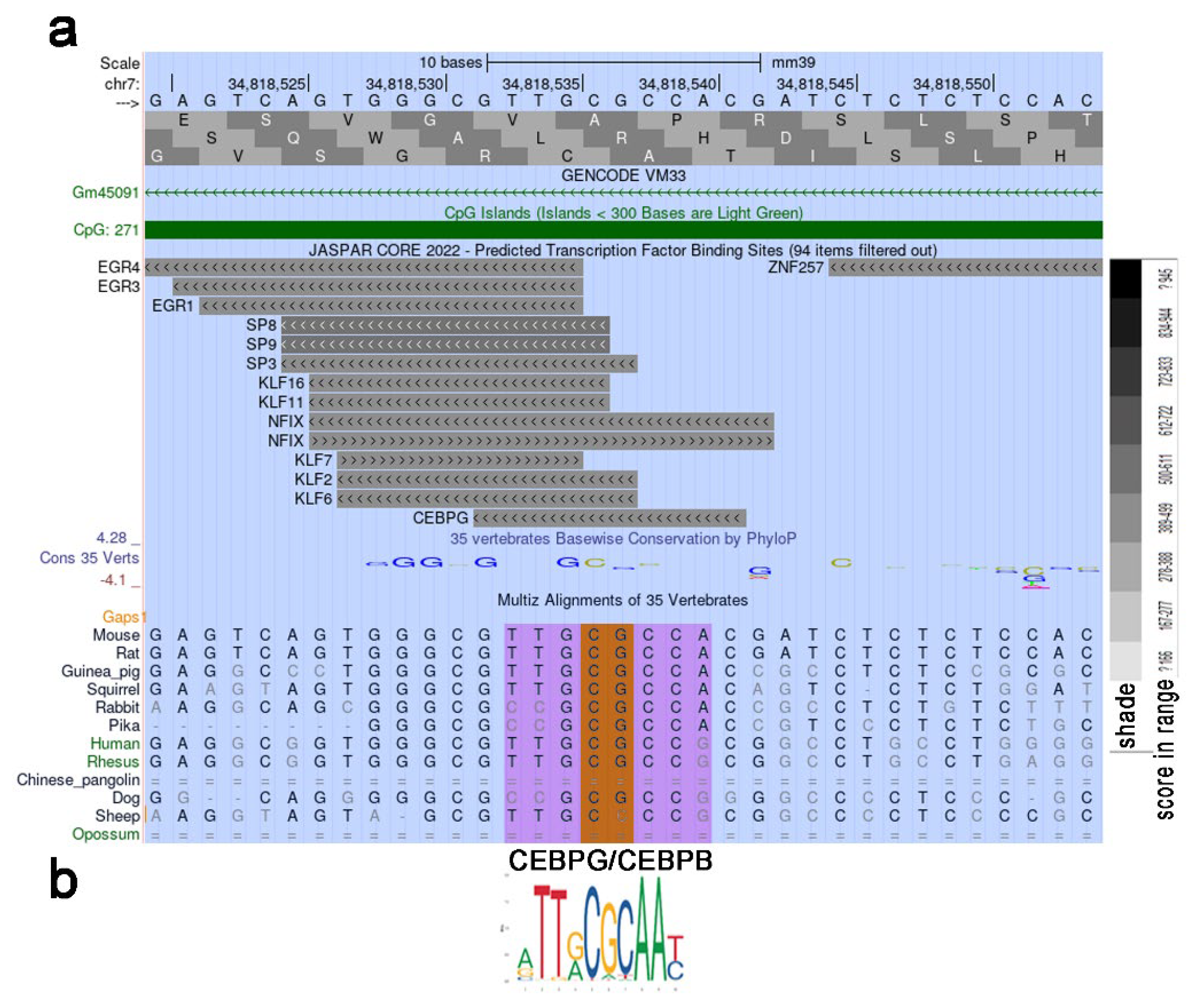
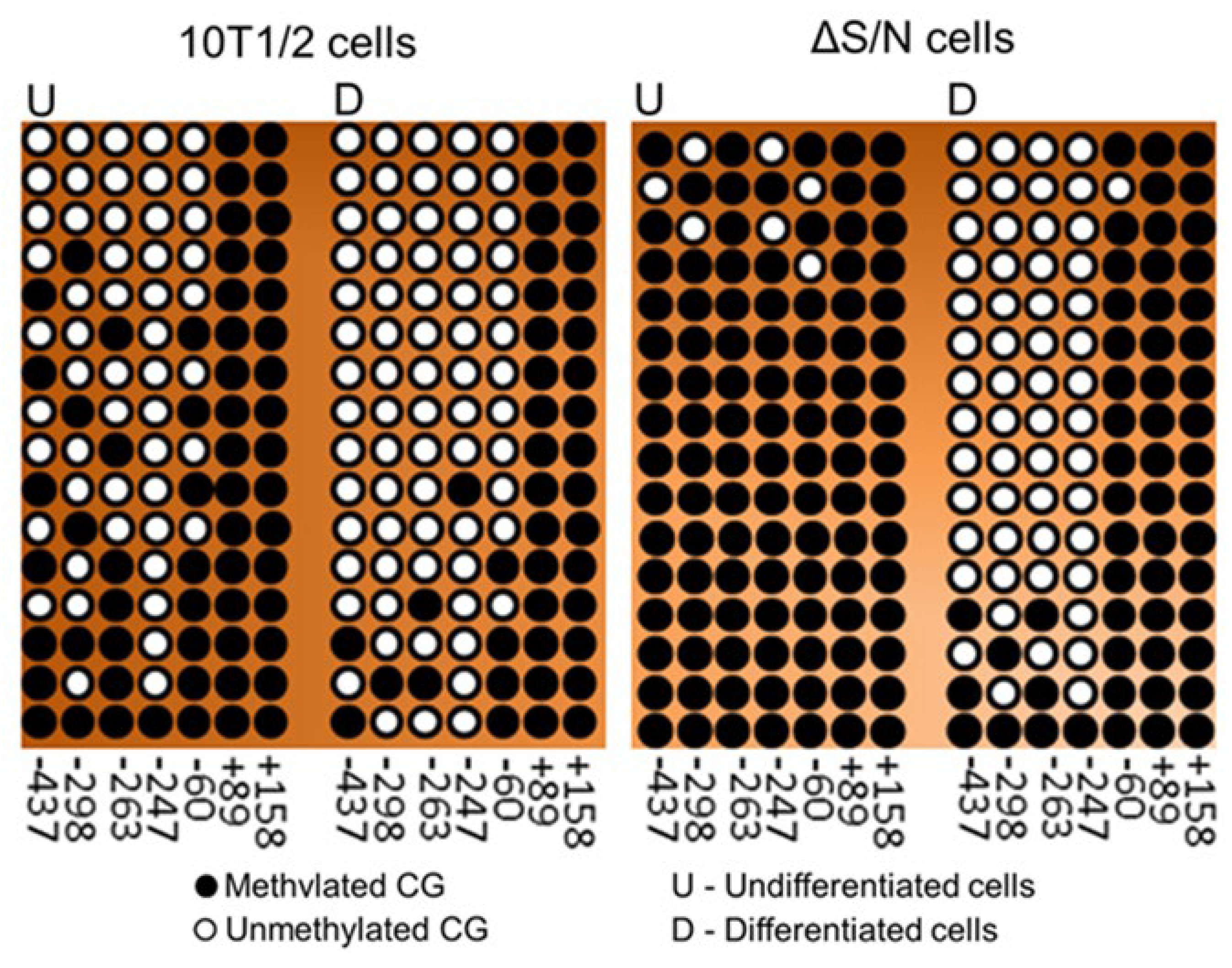
2.5. Heavily Methylated Cebpa in Wild-Type and ΔS/N Cells Loses CpG Methylation under AD, Maintaining an Unerased CpG Pattern near the TSS in ΔS/N Cells
2.5.1. Unerased CpG Methylation near the TSS May Interfere with the Formation of the Overall Transcription Complex
2.5.2. Unerased CpG Methylation Is Observed in the Cebpa Promoter Away from the TSS
2.6. CpG Methylation at Position 60 of the Pparγ2 Promoter Still Occurs in Postexpressing DN Rb1 Mutant ΔS/N Cells
2.7. The Promoters of the Adipogenesis-Related Genes Ezh2, Utx, Mll4, Rb1, and Tet2 Are Unmethylated Regardless of the Differentiation State
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation and Electrophoresis of RNA in Denaturing Gels and Its Quantitative Evaluation
4.3. Real-Time PCR
4.4. DNA Extraction and Bisulfite Treatment
4.5. Bisulfite Sequencing Analysis
4.6. Primer Design
4.7. Bioinformatics
4.8. Statistical Methods and Associated Software
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeCaprio, J.A., Ludlow, J.W., Lynch, D., Furukawa, Y., Griffin, J., PiwnicaWorms, H., Huang, C.M., and Livingston, D.M. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. 1989, Cell 58, 1085–1095. [CrossRef]
- Goodrich, D.W., Wang, N.P., Qian, Y.W., Lee, E.Y., and Lee, W.H. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell, 1991, 67, 293–302. [CrossRef]
- Ishak, C.A., Marshall, A.E., Passos, D.T., White, C.R., Kim, S.J., Cecchini, Ferwati, S., MacDonald, W.A., Howlett, C.J., Welch, I.D., et al. An RB-EZH2 complex mediates silencing of repetitive DNA sequences. Mol. Cell, 2016, 64, 1074-1078. [CrossRef]
- W G Kaelin Jr., Functions of the retinoblastoma protein. BioEssays. 1999, 21:950-958. [CrossRef]
- Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. 2014, eLife. 3. doi:10.7554/eLife.02872. [CrossRef]
- Sun A, Bagella L, Tutton S, Romano G, Giordano A. From G0 to S phase: A view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway. J CellBiochem, 2007, 102:1400–1404. [CrossRef]
- Korenjak M, Brehm A. E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr Opin Genet Dev. 2005, 15:520–527. [CrossRef]
- Fernandez-Marcos PJ and Auwerx J. pRb, a switch between bone and brown fat. Dev Cell. 2010, Sep 14;19(3):360-2.
- Calo, Eliezer, et al. “Rb regulates fate choice and lineage commitment in vivo.” Nature, 2010, 466, 7310, 1110-1114. [CrossRef]
- Puigserver P, Ribot J, Serra F et al. Involvement of the retinoblastoma protein in brown and white adipocyte cell differentiation: Functional and physical association with the adipogenic transcription factor C/EBPalpha. Eur J Cell Biol 1998, 77(2):117–123. [CrossRef]
- Kaelin, W. G., Jr., M. E. Ewen, and D. M. Livingston. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol. Cell. Biol. 1990, 10:3761–3769. [CrossRef]
- Kaye, F. J., R. A. Kratzke, J. L. Gerster, and J. M. Horowitz. A single amino acid substitution results in a retinoblastoma protein defective in phosphorylation and oncoprotein binding. PNAS USA, 1990, 87:6922–6926. [CrossRef]
- Aprile M, Cataldi S, Ambrosio MR, D’Esposito V, Lim K, Dietrich A, Blüher M, Savage DB, Formisano P, Ciccodicola A, Costa V. PPARγΔ5, a Naturally Occurring Dominant-Negative Splice Isoform, Impairs PPARγ Function and Adipocyte Differentiation. Cell Rep. 2018, Nov 6;25(6):1577-1592.e6. doi: 10.1016/j.celrep.2018.10.035. [CrossRef]
- Hamel PA, Cohen BL, Sorce LM, Gallie BL, Phillips RA. Hyperphosphorylation of the retinoblastoma gene product is determined by domains outside the simian virus 40 large-T-antigen-binding regions. Mol Cell Biol. 1990, 10:6586–6595. [CrossRef]
- Baryshev M, Petrov N, Ryabov V, Popov B Transient expression of inactive RB in mesenchymal stem cells impairs their adipogenic potential and is associated with hypermethylation of the PPARγ2 promoter. Genes Dis, 2022, 9, 165-175. [CrossRef]
- Park, K., J. Choe, N. E. Osifchin, D. J. Templeton, P. D. Robbins, and S. J. Kim. The human retinoblastoma susceptibility gene promoter is positively autoregulated by its own product. J. Biol. Chem. 1994, 269:6083-6088. [CrossRef]
- Capasso S, Alessio N, Di Bernardo G, Cipollaro M, Melone MA, Peluso G, Giordano A and Galderisi U: Silencing of RB1 and RB2/P130 during adipogenesis of bone marrow stromal cells results in dysregulated differentiation. Cell Cycle, 2014, 13: 482-490. [CrossRef]
- Ayala Tovy, Jaime M Reyes, Linda Zhang, Yung-Hsin Huang, Carina Rosas, Alexes C Daquinag, Anna Guzman, Raghav Ramabadran, Chun-Wei Chen, Tianpeng Gu, Sinjini Gupta, Laura Ortinau, Dongsu Park, Aaron R Cox, Rachel E Rau, Sean M Hartig, Mikhail G Kolonin, Margaret A Goodell. Constitutive loss of DNMT3A causes morbid obesity through misregulation of adipogenesis. eLife, 2022, 11:e72359. [CrossRef]
- Li F, Jing J, Movahed M, et al. Epigenetic interaction between UTX and DNMT1 regulates diet-induced myogenic remodeling in brown fat. Nature Communications. 2021, Nov; 12(1):6838. DOI: 10.1038/s41467-021-27141-7. [CrossRef]
- Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene, 2009, 28:2492-501. [CrossRef]
- Zhang, T. Hosoya, A. Maruyama, K. Nishikawa, J.M. Maher, T. Ohta, H. Motohashi, A. Fukamizu, S. Shibahara, K. Itoh, M. Yamamoto. Nrf2 Neh5 domain is differentially utilized in the transactivation of cytoprotective genes. Biochem. J, 2007, 404 , pp. 459-466. [CrossRef]
- K. Itoh, K. Igarashi, N. Hayashi, M. Nishizawa, M. Yamamoto, Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell. Biol. 1995, 15, 4184–4193. [CrossRef]
- W. Xing, A. Singgih, A. Kapoor, C.M. Alarcon, D.J. Baylink, S. Mohan, Nuclear factor E2-related factor-1 mediates ascorbic acid induction of osterix expression via interaction with antioxidant-responsive element in bone cells. J. Biol. Chem. 2007, 282, 22052–22061. [CrossRef]
- K.S. Schneider, J.Y. Chan, Emerging role of Nrf2 in adipocytes and adipose biology. Adv. Nutr. 2013, 4, 62–66. [CrossRef]
- Kaluscha, S., Domcke, S., Wirbelauer, C. et al. Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation. Nat Genet, 2022, 54, 1895–1906. [CrossRef]
- Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009, 41:178–186. [CrossRef]
- Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet. 2016, Aug 1;17(9):551-65. doi: 10.1038/nrg.2016.83. PMID: 27479905; PMCID: PMC5559737. [CrossRef] [PubMed]
- Mann IK, et al. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 2013, 23:988–997. [CrossRef]
- Mori et al., Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, et al. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem, 2005, 280:12867–12875.
- Banerjee SS, Feinberg MW, Watanabe M, Gray S, Haspel RL, Denkinger DJ, Kawahara R, Hauner H, Jain MK. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem, 2003, 278:2581–2584. [PubMed: 12426306].
- Y. Liu, T. He, Z. Li, Z. Sun, S. Wang, H. Shen, L. Hou, S. Li, Y. Wei, B. Zhuo, S. Li, C. Zhou, H. Guo, R. Zhang, B. Li. TET2 is recruited by CREB to promote Cebpb, Cebpa, and Pparg transcription by facilitating hydroxymethylation during adipocyte differentiation. iScience, 2023, 26 (11). [CrossRef]
- Lee et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife, 2013, 2:e01503. [CrossRef]
- Herskowitz, I. dominant negative mutations CS-G-CS. Nature, 1987, 329, 1–4.
- Bergendahl, L. T. et al. The role of protein complexes in human genetic disease. Protein Sci. 2019, 28, 1400–1411. [CrossRef]
- Veitia RA. Exploring the molecular etiology of dominant-negative mutations. Plant Cell, 2007, 19:3843–51. [CrossRef]
- Aprelikova ON, Fang BS, Meissner EG, Cotter S, Campbell M, Kuthiala A, Bessho M, Jensen RA, Liu ET. BRCA1-associated growth arrest is RB dependent. PNAS USA, 1999, 96:11866–11871. [CrossRef]
- Yarden and Brody, BRCA1 Interacts with Components of the Histone Deacetylase Complex. PNAS USA, 1999, 96: 4983-4988. [CrossRef]
- Friedman J, Cho WK, Chu CK, Keedy KS, Archin NM, Margolis DM, et al. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J Virol, 2011, 85(17):9078–89. [CrossRef]
- Viré, C. Brenner, R. Deplus, L. Blanchon, M. Fraga, C. Didelot, et al. The Polycomb group protein EZH2 directly controls DNA methylation Nature, 2006, 439, pp. 871-874. [CrossRef]
- Li F, Jing J, Movahed M, et al. Epigenetic interaction between UTX and DNMT1 regulates diet-induced myogenic remodeling in brown fat. Nature Communications. 2021, Nov; 12(1):6838. [CrossRef]
- Wang JK, Tsai MC, Poulin G, Adler AS, Chen S, Liu H, Shi Y, Chang HY. The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev. 2010, 24:327–32. [CrossRef]
- Chen X, Zhou W, Song RH, Liu S, Wang S, Chen Y, et al. Tumor suppressor CEBPA interacts with and inhibits DNMT3A activity. Sci Adv. 2022, 8:eabl5220. [CrossRef]
- Davey, K., Pennings, S. and Allan, J. “CpG methylation remodels chromatin structure in vitro”, J. Mol. Biol.1997, pp. 276–288. [CrossRef]
- Miglė Tomkuvienė, Markus Meier, Diana Ikasalaitė, Julia Wildenauer, Visvaldas Kairys, Saulius Klimašauskas, Laura Manelytė, Enhanced nucleosome assembly at CpG sites containing an extended 5-methylcytosine analog. Nucleic Acids Research, 2022, Volume 50, Issue 11, 24 June , Pages 6549–6561. [CrossRef]
- Trouche D., Le Chalony,C., Muchardt,C., Yaniv,M. and Kouzarides,T. (1997) RB and hbrm cooperate to repress the activation functions of E2F1. PNAS USA, 1997, 94, 11268–11273. [CrossRef]
- Lee, J.; Inoue, K.; Ono, R.; Ogonuki, N.; Kohda, T.; Kaneko-Ishino, T.; Ogura, A.; Ishino, F. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 2002, 129, 1807–1817. [CrossRef]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 2006; 126:663-76.
- Baryshev M.; Merculov, D.; Mironov, I. A new device-mediated miniprep method. AMB Express. 2022, 12, 21. [CrossRef]
| Gene | Sequence (5’⟶ 3’) | NCBI RefSeq | Tm | Size |
|---|---|---|---|---|
| Symbol | Forward/Reverse | Locus | (°C) | (bp) |
| Cebpa | GTAACCTTGTGCCTTGGATACT GGAAGCAGGAATCCTCCAAATA |
NM_007678 | 60 | 100 |
| Cebpb | CTTGATGCAATCCGGATCAAAC CCCGCAGGAACATCTTTAAGT |
NM_009883.4 | 60 | 113 |
| Dnmt1 | GAAGGCTACCTGGCTAAAGTCAAG ACTGAAAGGGTGTCACTGTCCGA |
NM_001199431.2 | 64 | 216 |
| Dnmt3a |
TGGAGAATGGCTGCTGTGTGAC CACTCATCCCGTTTCCGTTTGC |
NM_001271753.2 | 64 | 223 |
| Dnmt3b | AGTGACCAGTCCTCAGACACGA ATCAGAGCCATTCCCATCATCTAC |
NM_001003961.5 | 64 | 209 |
| Rb1 | ACAGTATGCCTCCACCAGGC AATCCGTAAGGGTGAACTAGAAAAC |
NM_009029 | 60 | 91 |
| P130 | TTTACTACTTCAGCAACAGCCC GAATCCCTCTCTTTTTAGTTGGAG | NM_001282000 | 60 | 93 |
| Ezh2 | ACTTACTGCTGGCACCGTCT GTTGAACAGAAAGCTGCACA | NM_007971 | 60 | 167 |
| Utx | GGGTTGGATTATGTATTTTTAGATTT CCAACAAAAATTCTCTACCTCAAA |
NM_009483 | 60 | 172 |
| Gene | Sequence (5’⟶ 3’) | Chromosomal | Tm | Size | No. of |
|---|---|---|---|---|---|
| Symbol | Forward/Reverse | Location | (°C) | (bp) | CpGs |
| Rb | GAAGGTTATTAATGGTTTTATTTTGG CTCCTCACCTAACCAAAAACAA |
Chr14:73430298- 73563446 |
670 | 110 | |
| Cebpa | GGGAGATAGGTTTAGTTTTAGT CACCCAATACCCCAACTAA |
Chr7:34818718-34821353 | 510 | 64 | |
| Pparg | TTTTAGATGTGTGATTAGGAGTTT ACAATTTCACCCACACATAAATA |
Chr6:115337912-115467360 | 685 | 7 | |
| Ezh2 | TTTGTTTATGGTTTTTTTGAGAGG CAAAACCAAACTCCAAAACAAAAAC | Chr6:47507208-47572309 | 722 | 133 | |
| Utx | GAAGGGATATAGTTTGGATTTTTTTA TCCTCCACTATCAAACTAAAAAAC | ChrX:18028814-18146175 | 829 | 67 | |
| Tet2 | GTTAAAGTAAATAGAAGGTGGGTT CCTTTCTAACAAATCCTACAAAACAA |
Chr3:133169438-133250882 | 812 | 99 | |
| Mll4 | GGGTTGGATTATGTATTTTTAGATTT CCAACAAAAATTCTCTACCTCAAA |
Chr15:98729550-98769085 | 696 | 84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




