1. Introduction
The implementation of molecular Human Papilloma Virus (HPV) assays has accelerated programs based on HPV-primary screening for the prevention of HPV-related Cervical Cancer (CC). The improved clinical sensitivity of molecular HPV assays over cervical cytology in screening programs has helped to reduce the incidence and mortality arising from this cancer [
1,
2,
3,
4,
5]. Moreover, the introduction of molecular HPV assays has allowed the possible transition to the use of HPV-primary screening on self-collected samples, thereby broadening women’s participation to cervical cancer screening programs.
Self-collected sampling has been widely accepted as a method of cervical screening, especially in low-income and middle-income countries [
6,
7,
8,
9,
10]. Its acceptance is largely due to its convenience and flexibility in implementation, as it can be performed in various settings such as
health care facilities, homes or workplaces facilitated by community health outreach teams. Additionally, HPV molecular assays can be used on self-collected samples as point-of-care cervical cancer screening [
7,
11,
12,
13,
14,
15,
16,
17,
18,
19,
20,
21,
22]. Validation of HPV assays on self-samples is therefore becoming increasingly important and essential to provide assurance for their use in routine clinical settings.
The internationally recognized criteria for the validation of HPV tests to be used in cervical cancer screening programs have focused on the importance of assuring clinical sensitivity and specificity of the assays for the detection of women with precancerous lesions but less attention has been placed on sample adequacy assessment [
5].
Currently, there are several commercially available HPV kits in the global market, but not all of them have been validated according to the international validation criteria, and a recent review by
Poljak et al. showed that only about 21% of these kits meet the validation criteria [
23]. Furthermore, a systemic review by Arbyn et al. reported that not all validated HPV assays contain an Internal Control (IC) [
24]. In the majority of assays which include an IC, this is associated with the qualitative detection of a
Beta-globin gene target, used to assess both the presence of human cells in the sample as well as any potential PCR inhibition in the same reaction well used for the detection of HPV DNA targets.
As previously reported, the inclusion of a sample adequacy control (SAC) in a separate reaction well from the molecular diagnostic target allows a more reliable assessment to calculate HPV viral copies/cell, avoiding potential competition for the reaction reagents[
25,
26,
27]. Moreover, SAC would represent an important quality assurance tool in HPV molecular testing, boosting confidence in negative results [
25,
28,
29]. The inclusion of a reliable SAC assessment would help to reduce the potential risk of false-negative results due to inadequate sample collection, particularly in cervical screening programs based on self-collected samples, as clearly pointed out in a recent report [
5,
26].
The importance of evaluating cell counts in other diagnostic laboratory investigations by means of quantitative tools has previously been established and adopted for diagnostic and prognostic analysis in illness such as leukemia and lymphoma [
30,
31].
C-C Motif Chemokine Receptor 5 (CCR5) gene has been employed in the OncoPredict HPV assays (Hiantis Srl, Milan, Italy) as an independent Quality Control (QC) reaction to assess sample adequacy, by the quantitative determination of the single-copy human
CCR5 gene, allowing also a more reliable HPV viral load determination by normalization according to sample cellularity [
32,
33]. Moreover, the use of
CCR5 gene target has also been previously described for the normalization of HIV-1 pro-viral loads in clinical samples [
34,
35].
This study aimed to evaluate the performance of the molecular target CCR5 included in the OncoPredict QC module for the reliable quantitation of cell numbers by comparing results with cellularity assessment based on Thoma cell-counting chamber and Fluorescence-activated Cell Sorting (FACS).
2. Materials and Methods
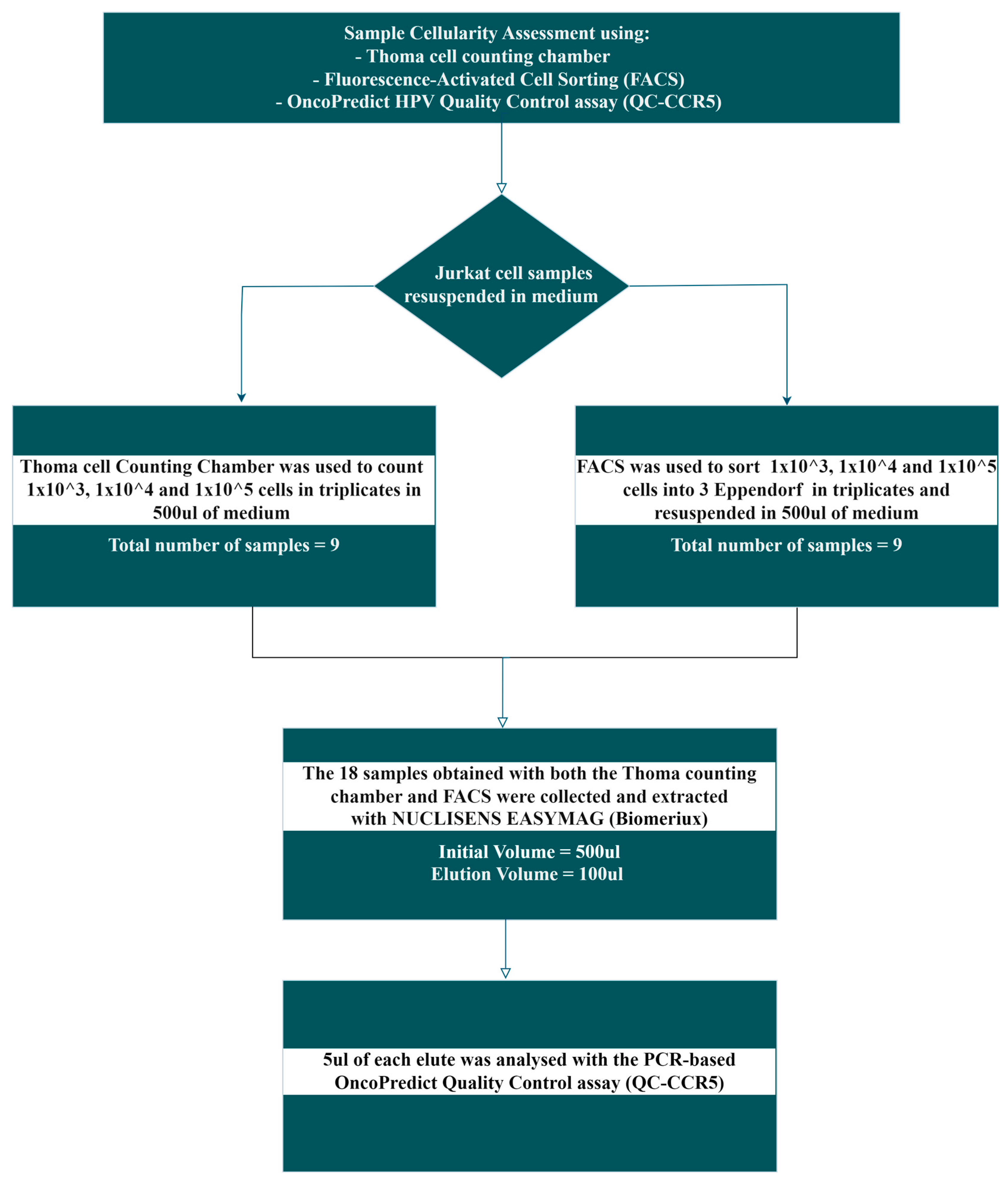
Human Jurkat cell lines were used to evaluate the accuracy of molecular quantitative cellularity assessment using
CCR5, a single copy human gene located on chromosome 3, included as an independent quantitative Real Time PCR (qRT-PCR) reaction of the OncoPredict HPV kit (Hiantis Srl, Milan, Italy), by comparing the results with those obtained using two other cell-counting methods based on microscopy cell-counting using a Thoma chamber and Fluorescence-activated Cell Sorting (FACS), as described in the workflow reported in
Figure 1.
The frequency of copy number variants and structural variant present in the human CCR5 gene were also investigated as part of this study to ensure that this gene target could represent a reliable marker in cellularity assessment.
2.1. Analysis of the Frequency of Copy Number Variants and Structural Variant in the Human CCR5 Gene
It has been shown that presence of copy number variants (CNVs) or structural variants (SVs) can affect the results of PCR analysis [
36]. We thus interrogated a catalog of genomic variants in the Jurkat cell line[
37]. We confirmed that
CCR5 is a single copy gene in these cells and that no copy number variants (CNVs) or structural variant (SVs) affect its coding sequence.
We also explored the frequency in the human population of CNVs and SVs that involve the
CCR5 gene. To this aim, we interrogated the Genome Aggregation Database (gnomAD, v4 release) [
38].
2.2. Cell Line: Cell Count with Thoma Chamber
Human Jurkat cell line sample obtained from ATCC (Manassas, VA, USA) was cultured in RPMI-1640 medium supplemented with 10% FBS, 1% Pen/Strep, Fungizone and Glutamine (Complete medium; Euroclone, Milan, Italy), at 37°C and 5% CO2.
After harvesting the Jurkat cells were centrifuged at 1200 rpm for 5 minutes at room temperature and the pellet was resuspended in 5 ml of complete medium. The cells were then filtered with 40 µm cell strainer (Euroclone) to eliminate cell aggregates.
Cell counting was performed by Trypan Blue solution (Sigma-Aldrich, ST Louis, MO, USA) to exclude non-viable cells, with living cells counted in a double grid Thoma chamber (Marienfeld Superior, Lauda-Konigshofen, Germany). The Thoma chamber slide was observed using an inverted microscope (Olympus) with a 10x objective. Subsequently, the following cell suspensions of 1000, 10000, and 100000 cells were prepared in triplicate in 500 µl (Total number = 9 Eppendorf tubes) of complete medium. The remaining cells were used for the cell sorting procedure.
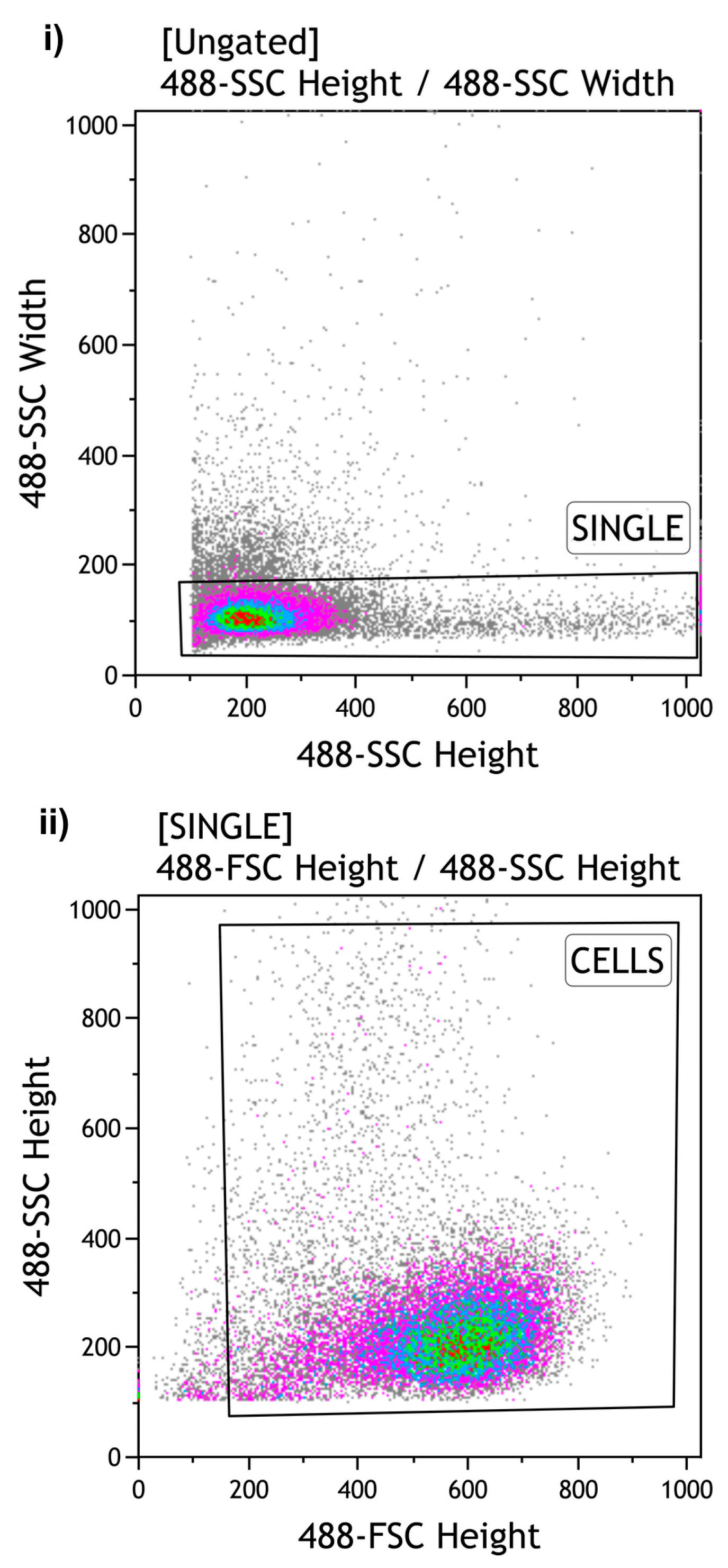
2.3. Cell Line: Fluorescent-Activated Cell Sorting (FACS)
The remaining Jurkat cell suspensions were sorted using a MoFlo Astrios Cell Sorter (Beckman Coulter, Brea, CA, USA). During the Set Sort Decision phase, the following criteria were used: “
Purify and 1-2 Drop”. Using the
“Limit Event function, 1000, 10000 and 100000 cells were sorted in triplicate (Total number = 9 Eppendorf tubes) according to the gate strategy shown in
Figure 2. An averaged sort rate of 200-2000 events per second and a sorting pressure of 25 PSI with a 100 µm nozzle were maintained throughout the process. Kaluza software version 1.2 was used for
.fcs file analysis.
2.4. Extraction of Human Jurkat Cell Line Samples
A total of 18 Eppendorf tubes containing the human Jurkat cell line samples obtained following both cell-counting by Thoma chamber and sorting by FACS in a volume of about 500µl underwent nucleic acid extraction using the automated NucliSENS® easyMag® platform (bioMérieux, Marcy-l’Étoile, France) which exploits BOOM technology with magnetic silica particles diluted 1:2. Prior to extraction, 10 µl of the exogenous spike-in control gene target (synthetic gene - firefly luciferase DNA) included in the OncoPredict HPV Quantitative Typing (QT) kit, used to assess efficiency of nucleic acid extraction as part of the assay’s QC [
36], was added to the Jurkat cell line samples. The extraction protocol “Specific B Protocol 2.0.1”, characterized by an elution temperature of 70°C, was selected and performed according to manufacturer’s instructions using a final elution volume of 100 µl.
2.5. Testing of the Jurkat Cell Line Samples with OncoPredict HPV QC Module
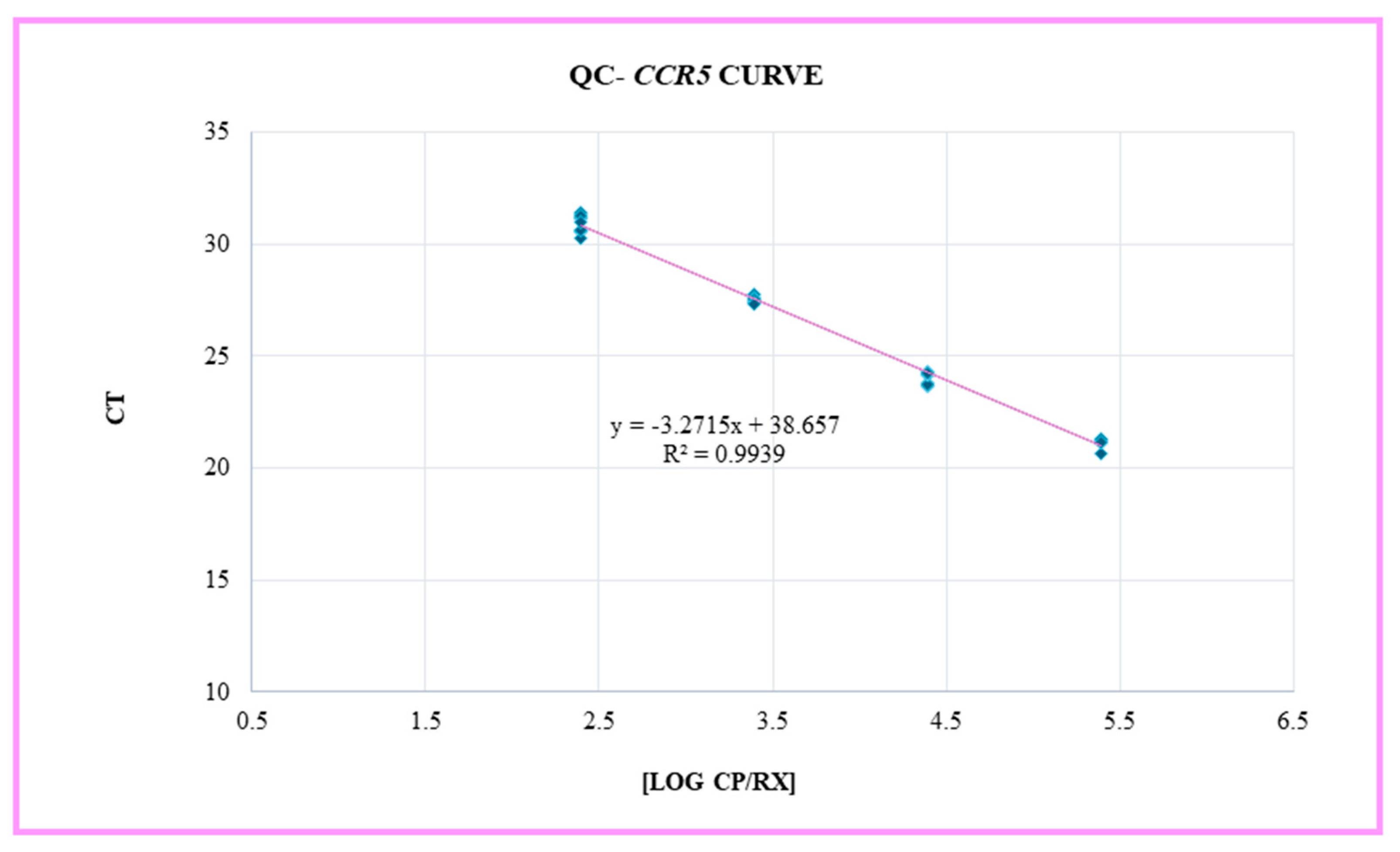
The real-time assay of the Quality Control (QC) module of OncoPredict HPV Quantitative Typing (QT) kit (Hiantis Srl, Milan, Italy) was performed on the CFX96™ Real-Time PCR Detection System (Bio-Rad) using 5 µl of extracted sample in a total reaction volume of 15 µl. The QC module includes an accurate evaluation of: the number of human cells present in the sample, by the quantitative determination of CCR5 gene; the efficiency of nucleic acid extraction, by the recovery of an exogenous control gene target added to the sample before preanalytical processing; and the evaluation of potential PCR inhibition, by the amplification of a control target (synthetic-custom designed gene/not human DNA) included in the OncoPredict HPV QT. Standard curves for the quantification of the CCR5 gene were constructed based on the cycle threshold (Ct) values of 4 quantitative calibrators run in triplicates.
2.6. Statistical Analysis
The cellularity was extrapolated from the CCR5 standard curve and a general estimate of the cellularity value for samples was calculated using the equation below. The p-value was calculated using t-Test for independent samples.
The cellularity value for samples was calculated using:
Equation (1) used to determine the sample cellularity value
3. Results
3.1. Frequency of CNVs and SVs in the Human CCR5 Gene
Upon the interrogation of the Genome Aggregation Database (gnomAD, v4 release) [
38], we detected 6 CNVs and 9 SVs that involve the human
CCR5 gene. However, the frequency of individual CNVs and SVs was found to be less than 0.0033, suggesting that they do not pose a problem in the use of this gene as a Sample Adequacy Control.
3.2. Performance of CCR5
After extracting and performing real-time PCR analysis on the 18 Jurkat cell line samples obtained from both the Thoma counting chamber and FACS, the Ct values obtained from each tested sample were used to calculate the number of cells from the CCR5 standard curve (
Figure 3). These cell numbers were reported as
CCR5-Cellularity (cells/sample) and Log10
CCR5-cellularity (cells/sample).
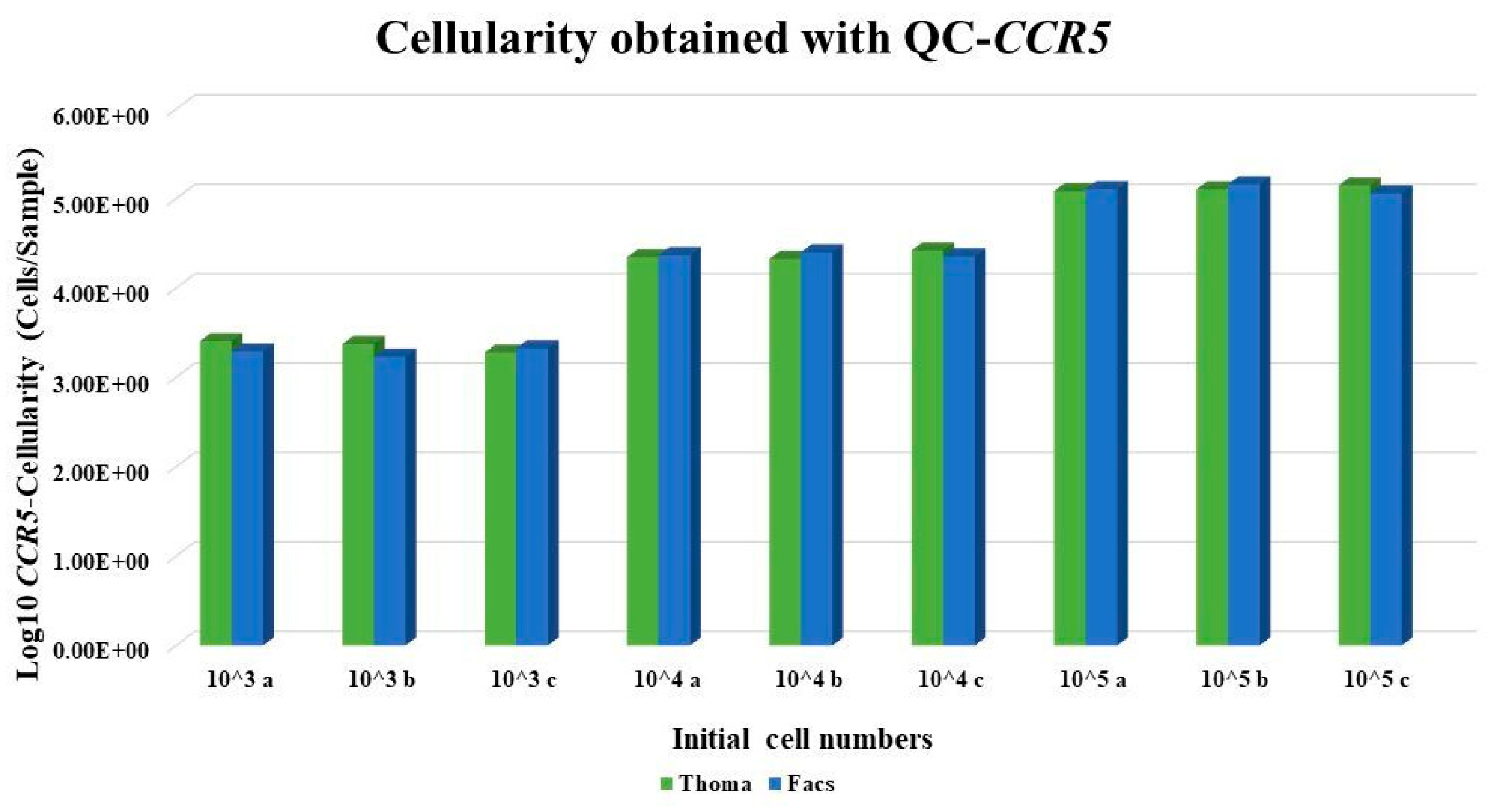
From
Table 1 and
Figure 4, we observed that the number of cells obtained by the use of the QC-
CCR5 assay correlates to the actual number of cells counted using the Thoma chamber and FACS.
Finally, no significant difference was observed between samples obtained from Thoma chamber and FACS analysis (p= 0.96) using t-Test for independent samples.
4. Discussion
The diagnostic accuracy of HPV molecular assays has received an important focus in the scientific community, which have led to a consensus on international validation guidelines, known as “Meijer criteria”, which establish minimum requirements for novel HPV tests in terms of clinical sensitivity, specificity, and reproducibility [
39]. Additionally, two complementary international initiatives - VALGENT (VALidation of HPV GENotyping Tests) and VALHUDES (VALidation of HUman papillomavirus assays and collection DEvices for HPV testing on Self-samples and urine samples
) - have been introduced to evaluate the clinical performance validation of HPV assays on cervical and self-collected samples [
40,
41]. Notwithstanding these validation criteria, only a limited number of studies have focused on evaluating the importance of sample adequacy assessment in molecular HPV-based primary testing in cervical cancer screening [
5,
25,
42]. Moreover, the causes of “invalid results” in molecular HPV testing, which could result from multiple pre-analytical and analytical issues, such as inadequate sampling, sample processing and/or PCR inhibition, have not been fully investigated. Furthermore, presently commercially available HPV assays which include an internal human gene control (IC) often apply arbitrarily chosen IC cut-offs, in terms of cycle threshold (Ct) values, by assays’ manufactures, which have not undergone more stringent validation, such as those that are presently required for determining HPV cut-offs in terms of clinical sensitivity and specificity. To our knowledge there are no previously reported studies evaluating what should be considered an appropriate number of human cells in relation to sample adequacy for molecular HPV testing.
The results of our study, comparing the performances of molecular CCR5 detection to microscopy and FACS cell-counting, showed that the cellularity assessment was comparable between the different methods.
Our results for using
CCR5 to assess cell counts align with the study conducted by Malnati and his group. In their study,
CCR5 was used to determine the cellularity of human cells from clinical samples to achieve the absolute quantification of proviral HIV-1 DNA [
34]. Similarly, a study carried out by Hong F, et. al, used
CCR5 copy numbers to calculate HIV-1 DNA copies per 1 million cells [
35]. Additionally, Cocuzza et al. reported the use of
CCR5 to determine the number of human cells present in clinical samples, thereby enabling the normalization of viral copy numbers per cell for accurate quantification of HPV viral loads. This highlights an additional advantage of using
CCR5 [
33].
The presence of a sample adequacy control (SAC) in HPV assays is expected to ensure the reliability of negative results by confirming the quality of the clinical sample to be tested and of the sample-collection procedure. In cervical cancer screening molecular HPV testing, a ”fixed” volume of resuspended cervical or self-sample is used to perform the analysis, based on manufacturers indications, irrespective of the abundance of sample collection. The latter is influenced by the fact that both clinician-collected cervical samples and self-collected vaginal samples originate from brushing or swabbing of a mucosal surface which is influenced by the sampling procedure and by the person who is performing the collection, as also previously reported for mucosal sampling used for SARS-CoV2 molecular testing[
43]. Moreover, negative results from molecular tests may not only be due to the absence of cells but also to errors in extraction or amplification processes[
26]. The OncoPredict HPV QC test addresses these issues by including an exogenous control for both nucleic acid extraction and PCR amplification, which allow to separately monitor both the extraction efficiency and potential PCR inhibition for each sample.
A SAC might be external, as in the case of the OncoPredict HPV assay, where it is amplified in a separate reaction well, or as an internal control (IC) where it is co-amplified with HPV nucleic acid [
25]. Nevertheless, appropriate IC molecular assays’ cut-off needs to be established in order to appropriately assess sample adequacy.
The reliability of negative results in HPV screening is critical for patient health, as it influences clinical decisions. HPV testing, with its strong negative predictive value has allowed the extension of screening intervals from approximately three years with cytology-based primary screening protocols to five years for those who test negative [
29]. This could mean that an undetected infection or early-stage precancerous lesions could progress into more advanced disease, potentially resulting in cervical cancer due to untimely follow-up care.
The risk of “false negatives” is expected to increase with the more recent move, promoted by the World Health Organization (WHO), towards the use of self-collection, such as vaginal samples collected by the woman herself, for cervical cancer prevention. Additionally, inadequate samples may also represent a problem in clinician-collected cervical samples particularly in relation to women’s age. It has been shown for example that in women of post-menopause, the transformation zone retreats further into the cervical canal making it more difficult to obtain adequate samples. All these factors can impact on the quality assurance of molecular HPV testing of clinical sample[
44,
45,
46].
One of the major issues with the use of molecular HPV assays is the lack of standardized criteria regarding the minimum cellularity required to define sample adequacy. On the contrary this parameter has been well defined for both traditional Pap smears and for cervical liquid-based cytology, with a required minimum cellularity count of 8000 cells and 5000 cells respectively, according to the Bethesda system [
47]. This requires a better understanding and further studies to assess quality assurance in the more recently introduced HPV-based primary cervical cancer screening programs. In order to evaluate the minimum cellularity required to define molecular cut-offs for sample adequacy it is necessary to have an accurate quantitative molecular assessment of sample cellularity with an appropriate human gene target with a reliable number of copies per cell.
To the best of our knowledge, this is the first study comparing the performance of
CCR5, a single copy human gene target, to other laboratory reference cell-counting methods, for the accurate molecular assessment of human cell numbers. Limitations of this study however include the small numbers of cell-line samples tested and that these were not true clinical samples, including human vaginal or cervical cells. Future studies will need to include a larger number of clinical samples allowing to evaluate the range of cellularity required for quality assurance in molecular HPV testing [
42].
5. Conclusions
The results obtained from this study show that CCR5 represents a promising molecular marker to assess sample cellularity in molecular diagnostics, with particular reference to molecular HPV testing used in cervical cancer screening. It will be important in future to define specific cellularity cut-off values for specific sample types and for the quality assurance in molecular HPV-based primary screening through the use of samples collected from human mucosal surfaces.
Author Contributions
Conceptualization: C.E.C.; methodology: C.E.C, R.C.N, M.M., C.G., S.D.M., B.T., and C.B; formal analysis, R.C.N., M.M., C.G.; investigation, C.E.C.; data curation, R.C.N.,M.M.,C.G, S.D.M., and B.T; writing—original draft preparation, R.C.N., C.E.C; writing, review and editing, R.C.N, M.M., C.G., S.D.M., B.T., M.D.A., M.S., C.B and C.E.C.; supervision, C.E.C.; project administration, C.E.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no funding.
Data Availability Statement
Study datasets generated by the study are stored locally and securely at the University of Milano-Bicocca.
Acknowledgments
The study group received free Oncopredict HPV Quality Control assay from Hiantis (Milan, Italy).
Conflicts of Interest
CEC received research funding or gratis consumables to support research from the following commercial entities in the last 3 years: BD, Seegene, Copan, Novosanis and Hiantis; CEC is a minority shareholder of Hiantis Srl. R.C.N., M.M., C.G., S.D.M., B.T., M.D.A., M.S., C.B have no conflicts of interest to declare.
References
- Human Papillomavirus (HPV) Nucleic Acid Amplification Tests (NAATs) to Screen for Cervical Pre-Cancer Lesions and Prevent Cervical Cancer. Available online: https://www.who.int/publications/i/item/9789240045248 (accessed on 30 January 2024).
- Arbyn, M.; De Sanjosé, S.; Saraiya, M.; Sideri, M.; Palefsky, J.; Lacey, C.; Gillison, M.; Bruni, L.; Ronco, G.; Wentzensen, N.; et al. EUROGIN 2011 Roadmap on Prevention and Treatment of HPV-Related Disease. Int J Cancer 2012, 131, 1969–1982. [Google Scholar] [CrossRef]
- Rijkaart, D.C.; Berkhof, J.; Rozendaal, L.; van Kemenade, F.J.; Bulkmans, N.W.J.; Heideman, D.A.M.; Kenter, G.G.; Cuzick, J.; Snijders, P.J.F.; Meijer, C.J.L.M. Human Papillomavirus Testing for the Detection of High-Grade Cervical Intraepithelial Neoplasia and Cancer: Final Results of the POBASCAM Randomised Controlled Trial. Lancet Oncol 2012, 13, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Ronco, G.; Dillner, J.; Elfström, K.M.; Tunesi, S.; Snijders, P.J.F.; Arbyn, M.; Kitchener, H.; Segnan, N.; Gilham, C.; Giorgi-Rossi, P.; et al. Efficacy of HPV-Based Screening for Prevention of Invasive Cervical Cancer: Follow-up of Four European Randomised Controlled Trials. The Lancet 2014, 383, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Cuschieri, K.; Fellner, M.D.; Sara, L.; Mühr, A.; Padalko, E.; Correa, R.M.; Dillner, J.; Gultekin, M.; Picconi, M.A. Quality Assurance in Human Papillomavirus Testing for Primary Cervical Screening. Int J Gynecol Cancer 2023, 0, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, H.; Yeh, P.T.; Oguntade, H.; Kennedy, C.E.; Narasimhan, M.; Topp, S.M. HPV Self-Sampling for Cervical Cancer Screening: A Systematic Review of Values and Preferences. BMJ Glob Health 2021, 6, 3743. [Google Scholar] [CrossRef]
- Arbyn, M.; Smith, S.B.; Temin, S.; Sultana, F.; Castle, P. Detecting Cervical Precancer and Reaching Underscreened Women by Using HPV Testing on Self Samples: Updated Meta-Analyses. BMJ 2018, 363, 4823. [Google Scholar] [CrossRef]
- Kamath Mulki, A.; Withers, M. Human Papilloma Virus Self-Sampling Performance in Low- and Middle-Income Countries. BMC Womens Health 2021, 21, 12. [Google Scholar] [CrossRef]
- Serrano, B.; Ibáñez, R.; Robles, C.; Peremiquel-Trillas, P.; de Sanjosé, S.; Bruni, L. Worldwide Use of HPV Self-Sampling for Cervical Cancer Screening. Prev Med (Baltim) 2022, 154, 106900. [Google Scholar] [CrossRef]
- Le, A.; Rohweder, C.; Wheeler, S.B.; Lafata, J.E.; Teal, R.; Giannone, K.; Zaffino, M.S.; Smith, J.S. Self-Collection for Primary HPV Testing: Perspectives on Implementation From Federally Qualified Health Centers. Prev Chronic Dis 2023, 20, 1–15. [Google Scholar] [CrossRef]
- Ketelaars, P.J.W.; Bosgraaf, R.P.; Siebers, A.G.; Massuger, L.F.A.G.; van der Linden, J.C.; Wauters, C.A.P.; Rahamat-Langendoen, J.C.; van den Brule, A.J.C.; IntHout, J.; Melchers, W.J.G.; et al. High-Risk Human Papillomavirus Detection in Self-Sampling Compared to Physician-Taken Smear in a Responder Population of the Dutch Cervical Screening: Results of the VERA Study. Prev Med (Baltim) 2017, 101, 96–101. [Google Scholar] [CrossRef]
- Tranberg, M.; Jensen, J.S.; Bech, B.H.; Blaakær, J.; Svanholm, H.; Andersen, B. Good Concordance of HPV Detection between Cervico-Vaginal Self-Samples and General Practitioner-Collected Samples Using the Cobas 4800 HPV DNA Test. BMC Infect Dis 2018, 18, 1–7. [Google Scholar] [CrossRef]
- Leinonen, M.K.; Schee, K.; Jonassen, C.M.; Lie, A.K.; Nystrand, C.F.; Rangberg, A.; Furre, I.E.; Johansson, M.J.; Tropé, A.; Sjøborg, K.D.; et al. Safety and Acceptability of Human Papillomavirus Testing of Self-Collected Specimens: A Methodologic Study of the Impact of Collection Devices and HPV Assays on Sensitivity for Cervical Cancer and High-Grade Lesions. Journal of Clinical Virology 2018, 99–100, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Jentschke, M.; Chen, K.; Arbyn, M.; Hertel, B.; Noskowicz, M.; Soergel, P.; Hillemanns, P. Direct Comparison of Two Vaginal Self-Sampling Devices for the Detection of Human Papillomavirus Infections. Journal of Clinical Virology 2016, 82, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.W.; Ouyang, Y.Q.; Hillemanns, P.; Jentschke, M. Excellent Analytical and Clinical Performance of a Dry Self-Sampling Device for Human Papillomavirus Detection in an Urban Chinese Referral Population. Journal of Obstetrics and Gynaecology Research 2016, 42, 1839–1845. [Google Scholar] [CrossRef] [PubMed]
- Ejegod, D.M.; Pedersen, H.; Alzua, G.P.; Pedersen, C.; Bonde, J. Time and Temperature Dependent Analytical Stability of Dry-Collected Evalyn HPV Self-Sampling Brush for Cervical Cancer Screening. Papillomavirus Research 2018, 5, 192–200. [Google Scholar] [CrossRef]
- Toliman, P.J.; Kaldor, J.M.; Badman, S.G.; Phillips, S.; Tan, G.; Brotherton, J.M.L.; Saville, M.; Vallely, A.J.; Tabrizi, S.N. Evaluation of Self-Collected Vaginal Specimens for the Detection of High-Risk Human Papillomavirus Infection and the Prediction of High-Grade Cervical Intraepithelial Lesions in a High-Burden, Low-Resource Setting. Clinical Microbiology and Infection 2019, 25, 496–503. [Google Scholar] [CrossRef]
- Woo, Y.L. The Feasibility and Acceptability of Self-Sampling and HPV Testing Using Cepheid Xpert® HPV in a Busy Primary Care Facility. J Virus Erad 2019, 5, 10–11. [Google Scholar] [CrossRef]
- Hawkes, D.; Keung, M.H.T.; Huang, Y.; McDermott, T.L.; Romano, J.; Saville, M.; Brotherton, J.M.L. Self-Collection for Cervical Screening Programs: From Research to Reality. Cancers 2020, Vol. 12, Page 1053 2020, 12, 1053. [Google Scholar] [CrossRef]
- Feltri, G.; Valenti, G.; Isidoro, E.; Kaur, J.; Treleani, M.; Bartelloni, A.; Mauro, C.; Spiga, F.; Ticich, G.; Di Napoli, M.; et al. Evaluation of Self-Sampling-Based Cervical Cancer Screening Strategy Using HPV Selfy CE-IVD Test Coupled with Home-Collection Kit: A Clinical Study in Italy. Eur J Med Res 2023, 28. [Google Scholar] [CrossRef]
- Fitzpatrick, M.B.; Dube Mandishora, R.S.; Katzenstein, D.A.; McCarty, K.; Weber, J.; Sahoo, M.K.; Manasa, J.; Chirenje, Z.M.; Pinsky, B.A. HrHPV Prevalence and Type Distribution in Rural Zimbabwe: A Community-Based Self-Collection Study Using near-Point-of-Care GeneXpert HPV Testing. Int J Infect Dis 2019, 82, 21–29. [Google Scholar] [CrossRef]
- Vallely, A.J.B.; Saville, M.; Badman, S.G.; Gabuzzi, J.; Bolnga, J.; Mola, G.D.L.; Kuk, J.; Wai, M.; Munnull, G.; Garland, S.M.; et al. Point-of-Care HPV DNA Testing of Self-Collected Specimens and Same-Day Thermal Ablation for the Early Detection and Treatment of Cervical Pre-Cancer in Women in Papua New Guinea: A Prospective, Single-Arm Intervention Trial (HPV-STAT). Lancet Glob Health 2022, 10, e1336–e1346. [Google Scholar] [CrossRef] [PubMed]
- Poljak, M.; Oštrbenk Valenčak, A.; Cuschieri, K.; Bohinc, K.B.; Arbyn, M. 2023 Global Inventory of Commercial Molecular Tests for Human Papillomaviruses (HPV). Journal of Clinical Virology 2024, 172, 105671. [Google Scholar] [CrossRef]
- Arbyn, M.; Simon, M.; Peeters, E.; Xu, L.; Meijer, C.J.L.M.; Berkhof, J.; Cuschieri, K.; Bonde, J.; Ostrbenk Vanlencak, A.; Zhao, F.H.; et al. 2020 List of Human Papillomavirus Assays Suitable for Primary Cervical Cancer Screening. Clinical Microbiology and Infection 2021, 27, 1083–1095. [Google Scholar] [CrossRef]
- Brukner, I.; Eintracht, S.; Papadakis, A.I.; Faucher, D.; Lamontagne, B.; Spatz, A.; Oughton, M. Maximizing Confidence in a Negative Result: Quantitative Sample Adequacy Control. J Infect Public Health 2020, 13, 991–993. [Google Scholar] [CrossRef]
- Pasquier, C.; Raymond, S.; Duchanois, D.; Sauné, K.; Oliveira-Mendes, K.; Vayssiere, C.; Izopet, J. Human Papillomavirus Testing Using HPV APTIMA® Assay and an External Cellularity Control in Self-Collected Samples. J Med Virol 2023, 95, e29283. [Google Scholar] [CrossRef] [PubMed]
- Brukner, I.; Resendes, A.; Eintracht, S.; Papadakis, A.I.; Oughton, M. Sample Adequacy Control (SAC) Lowers False Negatives and Increases the Quality of Screening: Introduction of “Non-Competitive” SAC for QPCR Assays. Diagnostics (Basel) 2021, 11. [Google Scholar] [CrossRef]
- Verberckmoes, B.; De Vos, T.; Maelegheer, K.; Ali-Risasi, C.; Sturtewagen, Y.; Praet, M.; Vanden Broeck, D.; Padalko, E. Evaluation of the Applicability of Internal Controls on Self-Collected Samples for High-Risk Human Papillomavirus Is Needed. BMC Womens Health 2023, 23. [Google Scholar] [CrossRef]
- Cuschieri, K.; Wilson, A.; Palmer, T.; Stanczuk, G.; Bhatia, R.; Ejegod, D.; Bonde, J. The Challenges of Defining Sample Adequacy in an Era of HPV Based Cervical Screening. Journal of Clinical Virology 2021, 137, 104756. [Google Scholar] [CrossRef] [PubMed]
- Verso, M.L. Section of the History of Medicine, First Australian Medical Congress. [CrossRef]
- Maruccio, G.; Qasaimeh, M.A.; Vembadi, A.; Menachery, A. Cell Cytometry: Review and Perspective on Biotechnological Advances. Cell Cytometry: Review and Perspective on Biotechnological Advances. Front. Bioeng. Biotechnol 2019, 7, 147. [Google Scholar] [CrossRef]
- Dhillon, S.K.; Cocuzza, C.E.; Chung, P.Y.J.; Martinelli, M.; Giubbi, C.; Njoku, R.C.; Bhatia, R.; Cuschieri, K.; Arbyn, M. Evaluation of the Clinical Performance of OncoPredict HPV® SCR Assay within the VALGENT-2 Framework. J Med Virol 2023, 95. [Google Scholar] [CrossRef]
- Cocuzza, C.E.; Dhillon, S.K.; Martinelli, M.; Giubbi, C.; Njoku, R.C.; Bhatia, R.; Cuschieri, K.; Arbyn, M. Clinical Performance of the Novel Full-Genotyping OncoPredict HPV Quantitative Typing Assay Using the VALGENT Framework. Int J Cancer 2024, 154, 538–547. [Google Scholar] [CrossRef]
- Malnati, M.S.; Scarlatti, G.; Gatto, F.; Salvatori, F.; Cassina, G.; Rutigliano, T.; Volpi, R.; Lusso, P. A Universal Real-Time PCR Assay for the Quantification of Group-M HIV-1 Proviral Load. Nat Protoc 2008, 3, 1240–1248. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Aga, E.; Cillo, A.R.; Yates, A.L.; Besson, G.; Fyne, E.; Koontz, D.L.; Jennings, C.; Zheng, L.; Mellors, J.W. Novel Assays for Measurement of Total Cell-Associated HIV-1 DNA and RNA. J Clin Microbiol 2016, 54, 902–911. [Google Scholar] [CrossRef]
- Pös, O.; Radvanszky, J.; Styk, J.; Pös, Z.; Buglyó, G.; Kajsik, M.; Budis, J.; Nagy, B.; Szemes, T. Copy Number Variation: Methods and Clinical Applications. 2021. [Google Scholar] [CrossRef]
- Gioia, L.; Siddique, A.; Head, S.R.; Salomon, D.R.; Su, A.I. A Genome-Wide Survey of Mutations in the Jurkat Cell Line. BMC Genomics 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Francioli, L.C.; Goodrich, J.K.; Collins, R.L.; Kanai, M.; Wang, Q.; Alföldi, J.; Watts, N.A.; Vittal, C.; Gauthier, L.D.; et al. A Genomic Mutational Constraint Map Using Variation in 76,156 Human Genomes. Nature 2024, 625, 92–100. [Google Scholar] [CrossRef]
- Meijer, C.J.L.M.; Berkhof, J.; Castle, P.E.; Hesselink, A.T.; Franco, E.L.; Ronco, G.; Arbyn, M.; Bosch, F.X.; Cuzick, J.; Dillner, J.; et al. Guidelines for Human Papillomavirus DNA Test Requirements for Primary Cervical Cancer Screening in Women 30 Years and Older. Int J Cancer 2009, 124, 516–520. [Google Scholar] [CrossRef]
- Arbyn, M.; Depuydt, C.; Benoy, I.; Bogers, J.; Cuschieri, K.; Schmitt, M.; Pawlita, M.; Geraets, D.; Heard, I.; Gheit, T.; et al. VALGENT: A Protocol for Clinical Validation of Human Papillomavirus Assays. J Clin Virol 2016, 76 Suppl 1, S14–S21. [Google Scholar] [CrossRef]
- Arbyn, M.; Peeters, E.; Benoy, I.; Vanden Broeck, D.; Bogers, J.; De Sutter, P.; Donders, G.; Tjalma, W.; Weyers, S.; Cuschieri, K.; et al. VALHUDES: A Protocol for Validation of Human Papillomavirus Assays and Collection Devices for HPV Testing on Self-Samples and Urine Samples. Journal of Clinical Virology 2018, 107, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, L.; Napoli, Z.; Turrisi, S.; Donati, S.; Lari, R. Pre-Analytical and Post-Analytical Evaluation in the Era of Molecular Diagnosis of Sexually Transmitted Diseases: Cellularity Control and Internal Control. Microbiologia Medica 2014, 29. [Google Scholar] [CrossRef]
- Dimeglio, C.; Loubes, J.M.; Mansuy, J.M.; Izopet, J. Ct Values from SARS-CoV-2 Diagnostic PCR Assays Should Not Be Used as Direct Estimates of Viral Load. Journal of Infection 2021, 82, 426–427. [Google Scholar]
- Desai, K.T.; Hansen, N.; Rodriguez, A.C.; Befano, B.; Egemen, D.; Gage, J.C.; Wentzensen, N.; Lopez, C.; Jeronimo, J.; de Sanjose, S.; et al. Squamocolumnar Junction Visibility, Age, and Implications for Cervical Cancer Screening Programs. Prev Med (Baltim) 2024, 180, 107881. [Google Scholar] [CrossRef] [PubMed]
- Reich, O.; Regauer, S. Understanding the Age-Related Epithelial Shifts at the Uterine Cervix and the Area of Risk for Squamous Cell Cervical Cancer. Prev Med (Baltim) 2024, 184, 108006. [Google Scholar] [CrossRef] [PubMed]
- Castle, P.E.; Jeronimo, J.; Schiffman, M.; Herrero, R.; Rodríguez, A.C.; Bratti, M.C.; Hildesheim, A.; Wacholder, S.; Long, L.R.; Neve, L.; et al. Age-Related Changes of the Cervix Influence Human Papillomavirus Type Distribution. Cancer Res 2006, 66, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- ’Birdsong, G.G.; ’Davey, D.D. Specimen Adequacy. In The Bethesda System for Reporting Cervical Cytology: Definitions, Criteria, and Explanatory Notes; Ritu, N., David C. Wilbur, Eds.; Springer, 2015; pp. 1–28.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).