Introduction
Duchenne Muscular Dystrophy (DMD) is a recessive X-linked disease that affects one in 3,500 to 5,000 boys (Mendell et al., 2012). Dystrophin plays a major role in the mechanical support of muscle fibers and its absence results in muscle degeneration, characterized by a progressive muscle wasting leading to loss of walking around teen age. The respiratory and heart muscles also gradually weaken, causing premature death (Ervasti & Campbell, 1993; Hoffman et al., 1987).
The dystrophin protein can be divided into four structural regions: the actin-binding domain (ABD) at the N-terminus, the central rod domain, the cysteine-rich domain (CR), and the C-terminal domain (C-term). The central rod domain is formed by 24 spectrin-like regions (R), intercalated by 4 intrinsically disordered regions called hinges (H). In skeletal muscle fiber, dystrophin interacts with several cytoskeletal proteins, particularly actin, microtubules and intermediate filaments, as well as with sarcolemma lipids (Amann et al., 1998; Bhosle et al., 2006; Prins et al., 2009; Renley et al., 1998; Zhao et al., 2016). Additionally, thanks to the CR domain, dystrophin is interacting with the dystroglycan complex (DGC), a transmembrane group of proteins which in turns bind to the laminin of the extracellular matrix (ECM) (Ervasti & Campbell, 1993; Jung et al., 1995). These interactions create a structural bridge between the cytoskeleton and the ECM, providing support to the myofibers and protecting the skeletal muscle cell from contraction-induced damage. Additionally, dystrophin interacts with several signaling proteins, such as neuronal nitric oxide (nNOS) (Brenman et al., 1995) and Par1b, a serine/threonine kinase that controls cell polarity and cell–cell interaction (Dumont et al., 2015). Moreover, the C-terminal region of dystrophin binds to syntrophin and dystrobrevin, thus ensuring the formation and maintenance of neuromuscular junctions, as well as stabilizing the binding with nNOS (Koo et al., 2011). All these interactions contribute to the maintenance of the homeostasis of the skeletal muscle fiber.
While DMD remains a fatal and incurable disease, a variety of promising gene therapy mediated by adeno-associated virus (AAV) have been recently translated into clinical trials. These approaches restore the sarcolemma stability of dystrophic myofibers, by the expression of shorter versions of dystrophin (µDys) which contain the domains necessary to connect the actin filaments and the DGC (Birch et al., 2023; Bostick et al., 2012; Le Guiner et al., 2017). The recent accelerated approval of Sarepta’s µDys by the US Food and Drug Administration (FDA) has brought hope to patients aged 4 and more (Hoy, 2023). Nevertheless, due to the lack of critical domains necessary for full dystrophin function, µDys gene therapies are de facto limited in their ability to fully restore the homeostasis of skeletal and cardiac tissues (Birch et al., 2023; Harper et al., 2002)(Bostick et al., 2012; Hart et al., 2024). In view of this limitation, we decided to generate larger versions of dystrophin including specific additional domains.
Several attempts have been made to deliver longer versions of dystrophin by harnessing dual- or triple-AAV approaches (Albini et al., 2023; Koo et al., 2011, 2014; Lostal et al., 2014; Odom et al., 2011; Tasfaout et al., 2024; Yan et al., 2000, 2007; Zhou et al., 2024). Various reconstitution mechanisms were employed, including cDNA trans-splicing, homologous recombination (HR) and mRNA and protein trans-splicing. The transfer of midi-dystrophins or full-length dystrophin by cDNA trans-splicing (Koo et al., 2014; Lai et al., 2005; Zhang et al., 2013) and overlapping HR (Albini et al., 2023; Lostal et al., 2014; Odom et al., 2011) successfully demonstrated therapeutic efficacy in dystrophin-null mice. However, these approaches showed around 25 to 50% efficacy of reconstitution (Albini et al., 2023; Duan et al., 2000; Koo et al., 2014; Lostal et al., 2014), thus requiring high doses of AAVs with the potential to be associated with immune response and toxicities (Ertl, 2022). Another method for creating larger proteins from smaller transcription units involves utilizing split-inteins to fuse two proteins. Split-inteins are small polypeptides that undergo a distinct post-translational process called protein trans-splicing (PTS). During PTS, the N- and C-terminal residues (exteins) flanking the inteins are naturally ligated, resulting in the formation of one complete protein while removing the reconstituted inteins (Saleh & Perler, 2006). Intein-mediated PTS has been widely explored in the context of retinal and liver diseases (Esposito et al., 2022; Padula et al., 2022; Tornabene et al., 2019) and recently in the context of DMD (Tasfaout et al., 2024; Zhou et al., 2024). Specifically, the delivery of intein-based dual-AAVs in dystrophin-null mice demonstrated nearly 100% PTS efficiency for midi-dystrophins (Tasfaout et al., 2024) and 60% efficiency for reconstituting full-length dystrophin (Zhou et al., 2024). These approaches led to significant improvement of the dystrophic phenotype and physiological correction in young and old mdx mice.
Here we generated three novel Midi-dystrophins by split-inteins dual-AAV approach based on Artificial Intelligence (AI) structural prediction to monitor the impact of the changes made during the design on protein structure. Because of the limited capacity of AAV, we strategically decided to reduce the size of the hinges to make room for additional functional domains. We then assessed the correct 3D conformation by AI tool upon these changes. Additionally, we digitally simulated the interactions between the two halves of the split Midi-Dys proteins and observed strong interactions between the N- and C-termini in Midi-Dys 1 and 2, with a weaker association in Midi-Dys 3. We demonstrated the high protein trans-splicing (PTS) efficiency using an in vitro GFP reporter system and in vivo studies in DBA2/mdx mice, with approximately 90% PTS efficiency for Midi-Dys1 and Midi-Dys2, while Midi-Dys 3 exhibited lower reconstitution efficiency. Consistent with that, we demonstrated a high therapeutic efficacy of Midi-Dys 1 and 2 in vivo in DBA2/mdx mice.
Results
Design of Three Novel Midi-Dystrophins Proteins Based on Intein-Mediated Reconstitution Approach
The rationale behind the design of the new Midi-Dystrophins was based on evidence in the literature about the crucial domains for Dystrophin function (Amann et al., 1998; Bhosle et al., 2006; Dumont et al., 2015; Jung et al., 1995; Koo et al., 2011; Prins et al., 2009; Zhao et al., 2016, 2019). Because of the importance of the dystrophin signaling role, we decided to include the binding domains for Par1b (R8-9), nNOS (R16-17) and syntrophin/dystrobrevin (C-term) in all the three forms designed. However, we varied the inclusion of domains involved in dystrophin’s interaction with structural elements of the myofibers. Specifically, binding to membrane lipids (R1-3, R10-12) was fully included in Midi-Dys 1 and 3, but only partially in Midi-Dys 2. Interactions with microtubules (R4-15, R20-24) were included in Midi-Dys 2 and 3, and partially in Midi-Dys1, while the internal actin-binding site (R11-17) was partially included in Midi-Dys1 and 2, and not in Midi-Dys3. The ABD1 (N-term) and the binding with β-Dystroglycan are common in all our constructs (
Figure 1A–C). To make more space for key structural domains, we strategically sacrificed part of the hinges which link the several spectrin-like repeats in the central domain, and which are intrinsically disordered regions of dystrophin. In all our constructs, we shortened the hinges by 36 to 56%, where the most important reduction was applied to hinge 1, where we eliminated the first 54 amino acids (
Figure S1A). To determine if our modifications to the primary amino acid sequence caused abnormal protein conformation, we predicted the 3D structure of the designed Midi-Dystrophins. To do that, we employed AlphaFold3, an AI-based deep-learning algorithm that can predict protein structures with atomic accuracy, even in cases in which no similar structure is known (Abramson et al., 2024). Since the prediction with AlphaFold can only yield 2000 amino acids, we trimmed the 3 Midi-Dystrophins in three parts: the N-terminal (which includes the ABD1, the H1 and the R1), the central domain (which includes the R1 until the R24) and the C-terminal domain (which includes the R24, the H4, the Cystein rich domain and the C-terminal). Of note, the first and the last regions are in common in the three sequences. Firstly, these three parts were compared to the prediction obtained with the original untruncated hinge sequences. As depicted in
Figure S1B,C, no change in the structural conformation of the N-terminal and C-terminal regions was observed after the shortening of the hinges. Moreover, no significant alterations in the alpha-helix structure of the spectrin-like repeats and in the 3D conformation were detected with the inclusion of the shortened hinges (
Figure 1D–F). The predicted template modeling (pTM) score, which measures the accuracy of the entire protein structure (Xu & Zhang, 2010), is not significantly different between the structural prediction of the original and shortened hinges (
Figure 1G). This result indicates that the shortening of these regions is not affecting the 3D conformation of the proteins.
Once determined the aminoacidic sequence of the structures, we evaluated the possible site where inteins could be inserted. Based on a previously described intein library screening (Pinto et al., 2020), we selected GP41-1 as the most performant intein in terms of speed of reaction and efficiency of protein-trans splicing ability. Additionally, GP41-1 is one of the shortest intein known so far, thereby allowing more space to include functional domains in our constructs. Intein activity is context-dependent, particularly to the aminoacidic environment that surrounds their ligation junction (N- and C-exteins). For GP41-1, its native extein junction sequence is SGY/SSS, where the most crucial residue for the PTS reaction is the first amino acid in the C-extein, which is a Serine (Carvajal-Vallejos et al., 2012; Shah et al., 2013) (Figure 1H). Additionally, it was shown that the deletion of the native extein residues does not impact the cleavage reaction yields of GP41-1 (Carvajal-Vallejos et al., 2012), which makes it possible to insert the intein in the protein sequences, without including additional residues on the targeted proteins. Based on this evidence, we split the Midi-dystrophin protein sequences, maintaining a Serine in the first amino acid position the C-extein, and respecting the limited capacity of 4.7 kilobases of the rAAV, where possible. Specifically, the GP41-1 was inserted in the R11 and R17 in Midi-Dys1 and Midi-Dys2, respectively, while for Midi-Dys3 was inserted into the hinge 3.
In Silico Modeling of Intein Adjacent Sites Predicts Efficacy of Protein Reconstitution
The efficacy of split-intein protein trans-splicing is dependent on the association and folding of the split halves, influencing the ligation kinetics (Aranko et al., 2009). We therefore hypothesize that the residue interactions of the domains around intein subunits may influence the protein trans-splicing efficiency. To investigate these interactions, we performed a structure prediction of a GFP split-intein reporter by using AlphaFold3 (Abramson et al., 2024). We designed two plasmids each encoding either the N- and the C-terminal half of the reporter GFP fused to the N- and C- terminal halves of the GP41-1 intein under the control of CK8 promoter. Additionally, an HA tag was added to the C-terminal end of the GP41-1 N-terminal half (
Figure 2A). We initially aligned the GP41-1 structure predicted by AlphaFold3 for GFP with the structure obtained from the intein subunits alone (without any surrounding sequences). The two structures displayed identical conformations, with a Pymol root-mean-square deviation (RMSD) of 0.226. Furthermore, the intein-associated GFP structure demonstrated proper folding, with an RMSD of 0.154 compared to the intact, unsplit GFP. To evaluate the residue interaction within the intein subunits, we performed a structural prediction focusing on the GP41-1 intein. The GFP-associated intein showed high confidence of structure, with a predicted local distance difference test (pLDDT)
Intein-GFP of 89.09 (close to the 90 cut-off of high accuracy), and high confident residue relative positions, with a Predicted Aligned Error (PAE), a metric measuring interdomain structural error, of 2.06 Å, (
Figure 2B–D). The two GFP halves (colored in light cyan and light pink) in the predicted structure interact each other (
Figure 2C). Indeed, the PAE showed highly confident interaction of GFP N- and C-termini subunits with 77.92% of pairwise inter-domain PAE less than 10Å (%PAE
<10Å) (
Figure 2D). The prediction of GFP split-intein 3D structure indicated a correct incorporation of the intein, suggesting the possibility of high excision activity.
To actually evaluate intein protein reconstitution efficacy in vitro, we transfected HEK293T cells with either single or dual plasmids GFP-GP41-1 split intein, with, as a control, a plasmid encoding for the complete protein sequence of GFP under the control of the CK8 promoter (Figure 2E). Seventy-two hours after transfection, GFP fluorescence was observed in the dual plasmid condition albeit with a reduced intensity compared to the GFP plasmid) while no fluorescence was observed in the single-split controls. By fluorescence-activated cell sorting (FACS), we found that the GFP fluorescence intensity was 7.7E+05, in the dual GFP condition, while we obtained a GFP fluorescence of 1.1E+06 in the single plasmid GFP condition (Figure 2F). Based on these results we confirm the efficacy of reconstitution of the intein GP41-1 in the GFP construct.
Structural Predictions of GP41-1 Split Midi-Dys Revealed Efficiency of Protein Trans-Splicing
To determine if the residues interaction in the intein adjacent sites influences the protein trans-splicing efficiency of our Midi-Dystrophins, we performed a structure prediction of the complexes containing 200 amino-acids (aa) of exteins around the split intein protein by using AlphaFold3 (Abramson et al., 2024). Within all three Midi-Dys complexes, two split intein subunits (colored in cyan and pink) showed almost identical structures (RMSD
Intein-MD1/Intein= 0.189, RMSD
Intein-MD3/Intein=0.204, RMSD
Intein-MD3/Intein=0.170) of high confidence (pLDDT
Intein-MD1= 89.80, pLDDT
Intein-MD2= 91.89, pLDDT
Intein-MD3= 91.19) and highly confident interaction within the intein complexes (PAE
Intein-MD1= 3.02Å, PAE
Intein-MD2= 3.32Å, PAE
Intein-MD3=2.91Å) (
Figure 3A–I). On another hand, the N-ter and C-ter dystrophin exteins (colored in light pink and light cyan, respectively) in different Midi-Dys predicted structures interact differently (
Figure 3C,F,I). The Midi-Dys1 PAE showed highly confident interaction of Midi-Dys-5’ and Midi-Dys-3’ subunits with 40.74% of pairwise inter-domain PAE less than 10Å (%PAE
<10Å) (
Figure 3C) with an average contact probability of all highly confident PAE (contact_probs
PAE<10Å) of 0.00473. Fewer interactions were observed within the prediction of Midi-Dys2 (
Figure 3F, %PAE
<10Å=12.12%, contact_probs
PAE<10Å=0.00238) and almost no inter-dystrophin interactions within Midi-Dys3 prediction (
Figure 3I, %PAE
<10Å=1.12%, contact_probs
PAE<10Å=0.000625).
We next assessed in vitro the reconstitution efficacy of our dual Midi-Dystrophin constructs. To this end, we performed double transfection with our Midi-Dystrophin constructs (N-term + C-term) in HEK293T and checked after 48 hours the expression of reconstituted Midi-Dys (Figure 3J). We performed a capillary western blot by using two different antibodies for dystrophin, one recognizing the N-terminal part and one recognizing the C-terminal part. We observed the presence of a high protein band (around 240 kDa corresponding to the full size of the Midi-dystrophins) in cells transfected with Midi-Dys1 and Midi-Dys2, with both antibodies, while no band was detected at that position in cells transfected with Midi-Dys3 (Figure 3K). The lower bands detected correspond to the non-reconstituted Midi-Dys halves fused to the N- and C-inteins (124.8 kDa, and 136.3 kDa for the Midi-Dys1, 141.7 kDa, and 138 kDa for the Midi-Dys2, 138.3 kDa, and 131.8 kDa for the Midi-Dys3, respectively). The persistence of these non-reconstituted proteins is probably due to the balance between trans-splicing and protein translation, as shown in other short-term studies on inteins (Padula et al., 2022). These results demonstrate a high reconstitution efficiency for Midi-Dys1 and Midi-Dys2 while Midi-Dys3 fails to reconstitute. Importantly, these results indicate that the degree of interaction found within dystrophin domains surrounding intein splits positively correlated with the reconstitution levels (Figure 3J,K). This data revealed that the interactions between two dystrophin subunits adjacent to intein splits play an essential role in the protein splicing efficiency.
Restoration of Midi-Dystrophin 1 and Midi-Dystrophin 2 in DBA2-mdx Mice Following Systemic AAV9 Delivery of Split Midi-Dys
To assess the reconstitution of the Midi-Dystrophin constructs
in vivo, we injected intravenously one month-old DBA2-mdx mice, a severe DMD mouse model, with AAV9-Midi-Dystrophins at the dose of 2.0E+13 vg/kg for each vector (
Figure 4A). Given that the effectiveness of intein protein trans-splicing can vary between in vitro and in vivo contexts, we included all three Midi-Dystrophins in our studies, even though Midi-Dys3 did not show efficient reconstitution. In our constructs, the Midi-Dystrophins cDNA is under the activity of the muscle-specific CK8 promoter, which has been demonstrated to efficiently express transgene in skeletal and cardiac muscles (Himeda et al., 2011). At 7 weeks post-injection, the viral genome copy number (VGCN) assessment showed 0.29 viral copies per diploid genome for both Midi-Dys 1 and Midi-Dys 2, while Midi-Dys 3 had 0.12 copies per diploid genome in the gastrocnemius, tibialis anterior, and diaphragm, muscles. (
Figure S2A). This difference could be attributed to a lack of therapeutic efficacy in Midi-Dys 3, leading to the elimination of viral copies within the regenerative myofibers, as previously shown (Manini et al., 2021) Immunoblotting data were informative for assessing the ability of protein trans-splicing of Midi-Dystrophins, through the observation of the reconstituted form over the not reconstituted one. The capillary western blot revealed expression of the reconstituted Midi-Dys 1 and Midi-Dys 2 (band at 247.1 kDa and 265.6 kDa respectively), while no full-length Midi-Dys 3 was observed. Not-reconstituted isoforms of Midi-Dys 1 and Midi-Dys 3 were observed (bands at 124.8 kDa and 135 kDa respectively) (
Figure 4B). Quantification of reconstituted bands revealed that Midi-Dys 1 has higher expression levels compared to Midi-Dys 2 (
Figure 4C). Protein trans-splicing efficacy was evaluated by calculating the ratio between the full-length Midi-Dys and not-reconstituted form by using an antibody that can detect all the Midi-Dys constructs. The efficacy of reconstitution observed is 98% for Midi-Dys 1 and 2, while Midi-Dys 3 has around 13% efficacy in gastrocnemius, tibialis anterior and diaphragm (
Figure 4D). Additionally, we observed robust and uniform expression of Midi-Dys1 and Midi-Dys2 in the histological sections of the diaphragm, while discontinuous localization at the membrane was observed for Midi-Dys 3 (
Figure 4E). Quantification of dystrophin-positive fibers revealed higher expression of Midi-Dys 1 compared to Midi-Dys 2 in the diaphragm and in the tibialis anterior (50-100% of dystrophin positive fibers in mice injected with Midi-Dys 1 versus 40-90% in mice injected with Midi-Dys 2) (
Figure 4F), while no difference was observed in the gastrocnemius. Contrary to the Western blot results, the positive staining of Midi-Dys3 with the N-term antibody may indicate the presence of truncated forms of Midi-Dys3 in the membrane region, as they still contain sarcolemma-binding domain R1-3. All together these results demonstrated expression of Midi-Dys1 and Midi-Dys2, with a high reconstitution efficacy and a proper localization of these Midi-Dystrophins at the membrane.
AAV9 Midi-Dys 1 and Midi-Dys 2 Systemic Delivery in DBA2 mdx Shows Therapeutic Efficacy
To assess the therapeutic efficacy of the Midi-Dystrophins, we injected intravenously one-month-old DBA2/mdx mice, with AAV9-Midi-Dystrophin1, 2, and 3 (2.0E+13 vg/kg each vector). After 7 weeks from the injection, we measured the biomarkers creatine kinase (CK) and Myomesin 3 (Myom3) in the serum of the mice. The damage to the muscle and the necrosis of the myofibers induce the release of these two molecules in the blood flow and for this reason, they are currently used as biomarkers for muscular dystrophies (Rouillon et al., 2015). The CK concentration in the serum decreased after gene transfer with Midi-Dys 1, while no differences with the mdx control were observed for Midi-Dys 2 and 3 (
Figure 5A). Consistent with these results, the Myom3 concentration in mice injected with Midi-Dys 1 decreases at the same level of the WT, while for Midi-Dys2 and Midi-Dys3 no differences between injected and not injected have been observed (
Figure 5B). Then, we assessed the dystrophic pathophysiology in histological sections of the diaphragm (the most affected muscle in mdx mice) (
Figure 5C), gastrocnemius and tibialis anterior. Fibrosis decreased in mice treated with Midi-Dys1 and of a lower degree with Midi-Dys2, while no decrease was observed in mice injected with Midi-Dys3, in all the three muscles analyzed (
Figure 5D). Moreover, the presence of calcific deposits, another hallmark of the dystrophic phenotype, was significantly decreased in mice treated with Midi-Dys1 and Midi-Dys 2 (
Figure 5E,F). Gene expression analysis of
Tmem8c, TGF-β, and
Pdgfr-α—genes linked to the progression of DMD—showed a significant decrease in mice treated with Midi-Dys1 and Midi-Dys2 in both the Tibialis Anterior and gastrocnemius muscles (
Figure 5G). However, treatment with Midi-Dys3 did not reduce the expression of these genes, indicating that it lacks therapeutic effectiveness (
Figure 5G). Additionally, a decrease in
Tmem8c, TGF-β, and
Pdgfr-α expression was observed in the diaphragm of mice injected with Midi-Dys1 (
Figure S3B). In contrast, no changes in gene expression were seen in mice treated with Midi-Dys2 or Midi-Dys3, which aligns with our previous findings. These results confirm the high therapeutic efficacy of Midi-Dys1 and Midi-Dys2 in restoring key dystrophic features while highlighting the therapeutic inefficacy of Midi-Dys3. Overall, these data show the potential of our strategy and underscore the importance of including functional domains of dystrophin.
Discussion
In this study, we developed three novel Midi-dystrophins using a split-intein dual-AAV strategy, guided by deep-learning-based structural predictions to evaluate how design modifications impact protein structure. Due to the limited capacity of AAV vectors, we strategically reduced hinge regions to prioritize the inclusion of functional domains, and we demonstrated that these modifications do not impact the structural stability of the protein. Additionally, we simulated the residue interactions between the two halves of the split Midi-Dys proteins, observing strong associations between the N- and C-termini exteins in Midi-Dys 1 and 2, with a weaker interaction in Midi-Dys 3. The strong residue interaction in the adjacent sites of the intein correlates with efficient protein trans-splicing (PTS) both in vitro and in vivo in DBA2-mdx mice. Consistent with these findings, we demonstrated the high therapeutic efficacy of Midi-Dys 1 and 2 in DBA2-mdx mice, underscoring their potential therapeutic applications.
Gene replacement methods using AAV vectors have posed significant challenges for DMD, primarily due to the limited cargo capacity of these vectors. The most promising approach involves delivering microdystrophin (µDys) to restore the mechanical properties of myofibers by including membrane and cytoskeleton binding domains. However, limited clinical trial improvements suggest µDys may be suboptimal in humans, potentially due to missing key protein domains. This suggests that expressing larger dystrophin proteins may be necessary for more effective disease correction. In our approach, we incorporated additional functional domains that are absent in micro-dystrophin. Specifically, we added interaction regions with the membrane, the cytoskeleton, Par1b, and nNOS. The connection between dystrophin and nNOS, through the R16-R17 region, is well-known for its importance. In fact, delivery of µDys including the R16-R17 has been shown to improve muscle strength and performance compared to those without the nNOS binding ability (Hakim et al., 2017; Lai et al., 2009). Additionally, the binding of dystrophin with Par1b was shown to be crucial for the proper asymmetrical division of satellite cells, which is vital for effective regeneration in dystrophic conditions (Dumont et al., 2015). In view of these added functional domains, it may be necessary to exacerbate the pathological condition to fully assess their impact, such as through cardiotoxin-induced muscle damage or long-term experiments, where µDys alone is ineffective (Hart et al., 2024).
In our study, in order to accommodate these additional domains, we strategically removed a great part of the hinge 1, by reducing it by 56.25%, maintaining a good therapeutic efficacy at low dose. These results are consistent with the recent work of Duan’s lab, where they demonstrated that hinge 1 is not necessary for the activity of micro-dystrophin in ameliorating the DMD phenotype, whereas hinge 4 is crucial for improving the pathophysiology(Wasala et al., 2023). Interestingly, the region that encodes for hinge 1 corresponds to an epitope linked with an immune response against dystrophin’s exon 8-11 in µDys injected patients. Indeed, a recent study reported that five DMD patients across four different clinical trials by Sarepta, Roche (using Sarepta’s vector), Pfizer, and Genethon—receiving three different gene therapy products varying in AAV serotype, promoter, and dose experienced strikingly similar severe adverse events suggestive of a cytotoxic T-cell immune response against micro-dystrophin proteins (Bönnemann et al., 2023). To the best of our knowledge, no other study has demonstrated the therapeutic efficacy of hinge-reduced Midi- or Micro-dystrophin, proving the impact of our study for the development of less-immunogenic DMD gene therapies. Further efforts will focus on investigating potential immune responses in comparison to micro-dystrophin, to understand the impact of the hinge 1 reduction in developing an immune response.
To accommodate additional functional domains in our Midi-Dys constructs, we harnessed the split-intein technology to deliver dual-AAVs. Intein activity is known to be context-dependent, particularly at the level of the amino-acid environment surrounding their ligation junctions (N- and C-exteins). For GP41-1, the deletion of the native extein residues was shown not to impact the cleavage reaction yields (Carvajal-Vallejos et al., 2012), allowing the insertion of the intein into sequences without adding extra residues to the targeted proteins. Since trans-splicing occurs at the protein level, the design of the split proteins (N- and C-terminal) must be precise to avoid incorrect polypeptide folding and to ensure efficient PTS. Some half-peptides may exhibit limited stability or incorrect targeting within the cell, resulting in low recombination efficiency between the two proteins. To evaluate the structural stability of the Midi-Dys half-peptides and the residue interactions involving the intein, we used AlphaFold3, a deep-learning algorithm that predicts protein structures from amino acid sequences. A major advantage of AlphaFold is its high accuracy in predicting protein conformations, including engineered forms that do not exist in nature, such as new therapeutic proteins. Our analysis confirmed the correct folding of our constructs and demonstrated the stability of the intein within the protein sequence. Additionally, AlphaFold3’s capability to predict protein-protein interactions within biomolecular complexes allowed us to observe residue interactions between intein-adjacent sites in some constructs, correlating with efficient PTS in vitro.
In conclusion, we validated the efficacy of our in-silico approach which predicts inter-domains interactions, and we confirmed our structural hypothesis on the importance of extein interactions for the occurrence of protein trans-splicing. Overall, our findings underscore the remarkable potential of deep-learning-based tools in the design of novel gene therapy products, paving the way for even more advanced approaches in the treatment of genetic diseases.
Material and Methods
Midi-Dystrophins Design and Viral Particle Preparation
Midi-Dystrophin 1, Midi-Dystrophin 2 and Midi-Dystrophin 3 protein sequences was derived by dystrophin muscular isoform (Dp427m; RefSeq: NM_004006.3) and optimized to remove the rare human codons, CpG and possible alternative ORFs by GeneArt service (Life Technologies). Gene fragments were produced by GeneArt gene synthesis (Life Technologies) and cloned into a plasmid containing CK8 promoter, and poly A SV40. Plasmids were amplified by endotoxin-free methods. For recombinant AAV production, HEK293-T cells (obtained from Stanford University School of Medicine), cultured in suspension, were transfected with the three plasmids coding for the adenovirus helper proteins, the AAV Rep and Cap proteins, and the ITR-flanked transgene expression cassette. Three days after transfection, cells were harvested, chemically lysed and treated with benzonase (Merck-Millipore, Darmstadt, Germany). After filtration, viral capsids were purified by affinity chromatography, formulated in sterile PBS, and the vector stocks were stored at −80 °C. Titers of AAV vector were determined by using digital droplet polymerase chain reaction (ddPCR). Viral particles were treated with DNAse I for 30 minutes at 37°C (Invitrogen, MA, USA) and then viral DNA was amplified by using polyA SV40 specific primers with ddPCR Supermix for probes (Biorad, CA, USA).
Structural Prediction Using AlphaFold3
The predicted 3D prediction of the Midi-Dystrophins structure was obtained with AlphaFold3 server (AlphaFold Server). The Midi-Dystrophin proteins were trimmed in three parts: the N-terminal region (including the ABD1, the hinge 1 and R1), central region of the rod domain, and the C-terminal region (R24, hinge 4, the CR and the C-term). The first and the last parts are in common in the three Midi-Dys, while the central rod domain was predicted separately for all the proteins. For protein subunits interaction, prediction was obtained by AlphaFold3 and root square mean deviation (RMSD) was defined by Pymol. (Molecular Graphics System, Version 1.3, Schrödinger, LLC.). Predicted local distance difference test (pLDDT) and predicted aligned error (PAE) was calculated by a custom Python code.
GFP-GP41-1 Split Cassette Design and Protein-Trans Splicing Assessment
GFP protein was split in two parts, whereas Serine was present in position +1 in C-terminal extein. The coding sequence is under the control of a CK8 promoter, and the mRNA stabilized by a polyA SV40 sequence. Additionally, a sequence HA tag was added at the end of the N-term vector. HEK293T were transfected at 70% of confluence by Lipofectamine 3000 kit (Invitrogen, MA, USA) accordingly to manufacturer’s instructions. After 72 hours, cells were harvested and washed twice with PBS. Flow cytometry experiment was performed using the CytoFLEX S (Beckman-Coulter) and FloJO software (TreeStar, Ashland, OR).
Capillary Western Blot and Protein Trans-Splicing Efficacy Assessment
Capillary western blot of microdystrophin was performed on a Simple Western™ Jess system (ProteinSimple, Bio-Techne) according to the manufacturer’s instructions, using a 60-440 kDa separation module (ProteinSimple SMW004) and the anti-mouse detection module (Protein simple DM-002) or anti-rabbit-detection module. In brief, protein samples consisting of either green fluorescent protein (GFP) or midi-dystrophin proteins obtained from lysed HEK 293-transfected cells were loaded at a concentration of 0.5 µg/µL. For capillary western blot on muscle lysates, proteins were extracted in RIPA buffer and 2.5 ng were used for Dystrophin detection in mdx mice injected with vehicle, AAV-Midi-Dystrophins or AAV-Micro-Dystrophin. Primary antibodies used for recognizing the N-term and the C-term part of dystrophin are listed in
Table 1. PTS efficiency was calculated by doing the ratio between the not reconstituted and reconstituted protein fragments for each Midi-Dystrophin construct.
Animal Care and Use
DBA2 (B6;129S4-DBA2tm1Cpr/J, strain #000671) and DBA2-mdx (D2.B10-Dmdmdx/J) mice were supplied by the Jackson Laboratory. All animal procedures were approved by the National Ethical Committee, C2EA-51 (Evry, France), and the French Ministry of Research (MESRI) and received a national agreement number (APAFiS #38006). Four-week-old Dba2_mdx and Dba2_WT male mice were injected by retro-orbital injection with dual Midi-Dystrophin AAV9 vectors at the dose of 2 × 1013 vg/kg for each vector (total dose of 4 × 1013 vg/kg) and with Micro-Dystrophin AAV9 vector at the dose of 2 x 1013 vg/kg. AAV9 injected mice were compared to Dba2_mdx mice injected with PBS (negative control) as well as to the Dba2 WT mice (positive control). Seven weeks post-injection, an Escape test was performed (see below), and muscles and serum collected for molecular and histological analysis.
Genomic DNA Extraction and Viral Copy Number Analysis
Genomic DNA was extracted from muscles by NucleoMag Pathogen kit (Macherey Nagel) according to the manufacturer’s instructions and purified using the KingFisher Flex purification system (Thermo-Fisher-Scientific). Droplet digital PCR was performed for the detection of vector genome number by using N-terminal and C-terminal specific primers. For detection of the N-terminal Midi-Dystrophins and Micro-Dys (at the 5’ end) R1-specific primers were used. For the detection of the C-terminal Midi-Dystrophin and Micro-Dys (at the 3’ end), R24-specific primers were used in combination with ddPCR for probes technology (Biorad). Results are normalized for titin genomic region and showed as viral copy number (VCN) per diploid genome. Primers and probes details are shown in
Table 2.
Histological Staining
Skeletal muscles (tibialis anterior, diaphragm and gastrocnemius) were sampled and frozen in isopentane cooled in liquid nitrogen. Transverse cryosections (8-10 µm) were prepared from frozen muscles, air dried, and stored at -80°C. Muscle sections were processed for Sirius red and red Alizarin histological staining. Sections were visualized on an Axioscan Z1 automated slide scanner (Zeiss, Germany), using a Plan APO 10X / 0.45 NA objective. Sections were immunostained overnight at 4°C with primary antibodies listed in
Table 1. Following three washes with PBS, the muscle sections were incubated for 1 hour at room temperature with a goat secondary antibody conjugated with Alexa Fluor 488 or 594 dye (Molecular Probe, dilution 1:1000). The sections were then mounted using DAPI-Fluoromount-G (Southern Biotech) and visualized either on a LEICA TCS-SP8 confocal microscope (Leica) with a 63X APO CS2 1.4 NA objective or on an Axioscan Z1 automated slide scanner (Zeiss) with a Plan APO 10X / 0.45 NA objective.
Fibrosis and Calcific Quantification
Sirius Red and Alizarin-stained transverse sections were used for fibrosis and calcification assessment using an open-source software for bioimages analysis QuPath (version 0.3.2). Two small artificial neural networks were trained on three representative muscle sections to classify positive and negative pixels. The first one is for delimiting the tissue to be considered and the second one is for identifying the region of collagen deposits or calcified fibers to be quantified. We subsequently used these networks for each muscle scan, to quantify fibrotic and calcified surface areas. Results were expressed in percentage (%) of areas in relation to the total muscle section surface area.
Dystrophin Positive Fibers Quantification
Muscle sections were stained with laminin or wheat germ agglutinin (WGA) to label cell membranes. The Cellpose2 cyto2 model 10 was fine-tuned using manually labeled images of myofibers based on laminin staining. This fine-tuning was performed with hyperparameters set to 200 epochs, a learning rate of 0.05, and a weight decay of 0.0001. The labeled dataset was carefully prepared to allow the model to simultaneously segment myofibers while ignoring areas with low-quality staining.
Once fine-tuned, these models were utilized to extract myofiber masks. The reconstruction of myofiber masks from whole-scan images was performed using the Cellpose package 10. These reconstructed masks were then converted into Regions of Interest (ROIs), with each ROI corresponding to an individual myofiber, using the Labels_To_Rois.py FIJI plugin 11.
Muscle sections were co-labeled with laminin for membrane labeling and dystrophin for further analysis. As with the initial labeling, ROIs were generated based on membrane labeling and used for the subsequent quantification of dystrophin signals using a FIJI macro.
Serum Biomarkers Quantification
Blood samples were collected by retro-orbital bleeding and quickly centrifuged for 10 min at 8000 rpm. Sera were harvested and stored at −80 °C until measurement. For serum MYOM3 quantification, a custom sandwich ELISA assay was employed. Initially, a polyclonal MYOM3 antibody (Proteintech, 17692-1-AP) diluted 1/10 was coated onto a 96-well plate and incubated overnight at 4°C. After this incubation, the plate was washed three times with PBST and then blocked with a saturation solution of 3% BSA in PBS. Dilutions of the serum samples were then added to the plate and incubated for 2 hours at room temperature. For detection, a custom monoclonal antibody (Proteogenix, REF: 51-H1-B4) coupled with SULFO-TAG (MSD) was used, and the plate was incubated for an additional 2 hours at room temperature. After incubation, the plate was washed three times with wash buffer (0.05% Tween-20 in PBS). The absorbance of the SULFO-TAG was measured using the MESO Quickplex SQ 120 (MSD). To quantify the MYOM3 concentration in the serum samples, a set of concentration-defined MYOM3 peptides (His-tagged, Proteogenix) was included in the same experiment and served as the standard for calculation. For creatin phosphor-kinase (PK) quantification, 10 μL of mouse serum was used to calorimetrically measure Creatine Phospho Kinase concentration by FUJI DRI-CHEM nx500 system (DMV Imaging).
RNA Extraction and Gene Expression Analysis
Total RNA was extracted from frozen muscle tissue using an IDEAL32 extraction robot. Any DNA contamination in the RNA samples was removed with the TURBO DNA-free kit (Thermo Fisher Scientific). For gene expression analysis, 1000 ng of total RNA was reverse-transcribed using a mix of random oligonucleotides and oligo-dT, along with the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The resulting cDNA was then quantified by qPCR on the LightCycler 480 system (Roche) using Taqman Gene Expression Assays (Thermo Fisher Scientific). Each PCR reaction was conducted in duplicate, and the data were normalized across samples using Rplp0 as a reference gene.
Statistical Analysis
All data were analyzed by GraphPad Prism 9.5.1 software. Parametric tests such as t-tests and ANOVA were used for statistical comparison. To compare two groups, initially, the F-test was used to compare variances. If there was no difference in variances, a statistical comparison was performed using an unpaired t-test. To compare multiple groups, we used one-way ANOVA with Tukey’s correction for multiple comparison tests. For grouped analysis, we performed two-way ANOVA statistic test with multiple comparison with main row effect. Results were considered significantly different at p < 0.05. Graphs were generated using Graphpad Prism v9 or R version 3.6.2, The figures display the mean ± standard deviation.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org.
Author Contributions
L.P. designed the Midi-Dystrophin constructs, designed and performed the majority of the experiments, and wrote the manuscript. M.F. performed all the in vivo experiments and in vitro reconstitution of Midi-Dys. AVH helped with AlphaFold3 predictions. SA conceived and helped with the experiments. IR and SA edited the manuscript.
Acknowledgments
This work was supported by the “Association Française contre les Myopathies” (AFM). The authors are Genopole’s members, first French biocluster dedicated to genetic, biotechnologies and biotherapies. We are grateful to the “Imaging and Cytometry Core Facility” and to the in vivo evaluation, services of Genethon for technical support, to Ile-de-France Region, to Conseil Départemental de l’Essonne (ASTRE), INSERM and GIP Genopole, Evry for the purchase of the equipment. We would like to acknowledge the technical help of Louna Pili and Nathalie Bourg-Alibert. We acknowledge Dr. G.E. Morris, who deposited the MANCHO11 (9F2) to the DSHB (DSHB Hybridoma Product MANCHO11(9F2)).
Conflicts of Interest
LP, SA and IR are inventors on PCT application EP2024/070501 for the generation of novel Midi-Dystrophins. I.R. is a part-time employee of Atamyo Therapeutics. The other authors declare that they have no competing interests.
References
- Abramson, J., Adler, J., Dunger, J., Evans, R., Green, T., Pritzel, A., Ronneberger, O., Willmore, L., Ballard, A. J., Bambrick, J., Bodenstein, S. W., Evans, D. A., Hung, C.-C., O’Neill, M., Reiman, D., Tunyasuvunakool, K., Wu, Z., Žemgulytė, A., Arvaniti, E., … Jumper, J. M. (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature, 630(8016), 493–500. [CrossRef]
- Albini, S., Palmieri, L., Dubois, A., Bourg, N., Lostal, W., & Richard, I. (2023). Assessment of Therapeutic Potential of a Dual AAV Approach for Duchenne Muscular Dystrophy. International Journal of Molecular Sciences, 24(14), Article 14. [CrossRef]
- Amann, K. J., Renley, B. A., & Ervasti, J. M. (1998). A cluster of basic repeats in the dystrophin rod domain binds F-actin through an electrostatic interaction. The Journal of Biological Chemistry, 273(43), 28419–28423. [CrossRef]
- Aranko, A. S., Züger, S., Buchinger, E., & Iwaï, H. (2009). In vivo and in vitro protein ligation by naturally occurring and engineered split DnaE inteins. PloS One, 4(4), e5185. [CrossRef]
- Bhosle, R. C., Michele, D. E., Campbell, K. P., Li, Z., & Robson, R. M. (2006). Interactions of intermediate filament protein synemin with dystrophin and utrophin. Biochemical and Biophysical Research Communications, 346(3), 768–777. [CrossRef]
- Birch, S. M., Lawlor, M. W., Conlon, T. J., Guo, L.-J., Crudele, J. M., Hawkins, E. C., Nghiem, P. P., Ahn, M., Meng, H., Beatka, M. J., Fickau, B. A., Prieto, J. C., Styner, M. A., Struharik, M. J., Shanks, C., Brown, K. J., Golebiowski, D., Bettis, A. K., Balog-Alvarez, C. J., … Kornegay, J. N. (2023). Assessment of systemic AAV-microdystrophin gene therapy in the GRMD model of Duchenne muscular dystrophy. Science Translational Medicine, 15(677), eabo1815. [CrossRef]
- Bönnemann, C. G., Belluscio, B. A., Braun, S., Morris, C., Singh, T., & Muntoni, F. (2023). Dystrophin Immunity after Gene Therapy for Duchenne’s Muscular Dystrophy. The New England Journal of Medicine, 388(24), 2294–2296. [CrossRef]
- Bostick, B., Shin, J.-H., Yue, Y., Wasala, N. B., Lai, Y., & Duan, D. (2012). AAV micro-dystrophin gene therapy alleviates stress-induced cardiac death but not myocardial fibrosis in > 21-m-old mdx mice, an end-stage model of Duchenne muscular dystrophy cardiomyopathy. Journal of Molecular and Cellular Cardiology, 53(2), 217–222. [CrossRef]
- Brenman, J. E., Chao, D. S., Xia, H., Aldape, K., & Bredt, D. S. (1995). Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell, 82(5), 743–752. [CrossRef]
- Carvajal-Vallejos, P., Pallissé, R., Mootz, H. D., & Schmidt, S. R. (2012). Unprecedented rates and efficiencies revealed for new natural split inteins from metagenomic sources. The Journal of Biological Chemistry, 287(34), 28686–28696. [CrossRef]
- Duan, D., Yue, Y., Yan, Z., & Engelhardt, J. F. (2000). A new dual-vector approach to enhance recombinant adeno-associated virus-mediated gene expression through intermolecular cis activation. Nature Medicine, 6(5), 595–598. [CrossRef]
- Dumont, N. A., Wang, Y. X., von Maltzahn, J., Pasut, A., Bentzinger, C. F., Brun, C. E., & Rudnicki, M. A. (2015). Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nature Medicine, 21(12), 1455–1463. [CrossRef]
- Ertl, H. C. J. (2022). Immunogenicity and toxicity of AAV gene therapy. Frontiers in Immunology, 13. [CrossRef]
- Ervasti, J., & Campbell, K. (1993). A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. Journal of Cell Biology, 122(4), 809–823. [CrossRef]
- Esposito, F., Lyubenova, H., Tornabene, P., Auricchio, S., Iuliano, A., Nusco, E., Merlin, S., Olgasi, C., Manni, G., Gargaro, M., Fallarino, F., Follenzi, A., & Auricchio, A. (2022). Liver gene therapy with intein-mediated F8 trans-splicing corrects mouse haemophilia A. EMBO Molecular Medicine, 14(6), e15199. [CrossRef]
- Hakim, C. H., Wasala, N. B., Pan, X., Kodippili, K., Yue, Y., Zhang, K., Yao, G., Haffner, B., Duan, S. X., Ramos, J., Schneider, J. S., Yang, N. N., Chamberlain, J. S., & Duan, D. (2017). A Five-Repeat Micro-Dystrophin Gene Ameliorated Dystrophic Phenotype in the Severe DBA/2J-mdx Model of Duchenne Muscular Dystrophy. Molecular Therapy - Methods & Clinical Development, 6, 216–230. [CrossRef]
- Harper, S. Q., Hauser, M. A., DelloRusso, C., Duan, D., Crawford, R. W., Phelps, S. F., Harper, H. A., Robinson, A. S., Engelhardt, J. F., Brooks, S. V., & Chamberlain, J. S. (2002). Modular flexibility of dystrophin: Implications for gene therapy of Duchenne muscular dystrophy. Nature Medicine, 8(3), 253–261. [CrossRef]
- Hart, C. C., Lee, Y. il, Xie, J., Gao, G., Lin, B. L., Hammers, D. W., & Sweeney, H. L. (2024, May 7). Potential limitations of micro-dystrophin gene therapy for Duchenne muscular dystrophy. American Society for Clinical Investigation. [CrossRef]
- Himeda, C. L., Chen, X., & Hauschka, S. D. (2011). Design and testing of regulatory cassettes for optimal activity in skeletal and cardiac muscles. Methods in Molecular Biology (Clifton, N.J.), 709, 3–19. [CrossRef]
- Hoffman, E. P., Brown, R. H., & Kunkel, L. M. (1987). Dystrophin: The protein product of the duchenne muscular dystrophy locus. Cell, 51(6), 919–928. [CrossRef]
- Hoy, S. M. (2023). Delandistrogene Moxeparvovec: First Approval. Drugs, 83(14), 1323–1329. [CrossRef]
- Jung, D., Yang, B., Meyer, J., Chamberlain, J. S., & Campbell, K. P. (1995). Identification and characterization of the dystrophin anchoring site on beta-dystroglycan. The Journal of Biological Chemistry, 270(45), 27305–27310. [CrossRef]
- Koo, T., Malerba, A., Athanasopoulos, T., Trollet, C., Boldrin, L., Ferry, A., Popplewell, L., Foster, H., Foster, K., & Dickson, G. (2011). Delivery of AAV2/9-microdystrophin genes incorporating helix 1 of the coiled-coil motif in the C-terminal domain of dystrophin improves muscle pathology and restores the level of α1-syntrophin and α-dystrobrevin in skeletal muscles of mdx mice. Human Gene Therapy, 22(11), 1379–1388. [CrossRef]
- Koo, T., Popplewell, L., Athanasopoulos, T., & Dickson, G. (2014). Triple trans-splicing adeno-associated virus vectors capable of transferring the coding sequence for full-length dystrophin protein into dystrophic mice. Human Gene Therapy, 25(2), 98–108. [CrossRef]
- Lai, Y., Thomas, G. D., Yue, Y., Yang, H. T., Li, D., Long, C., Judge, L., Bostick, B., Chamberlain, J. S., Terjung, R. L., & Duan, D. (2009, March 2). Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. American Society for Clinical Investigation. [CrossRef]
- Lai, Y., Yue, Y., Liu, M., Ghosh, A., Engelhardt, J. F., Chamberlain, J. S., & Duan, D. (2005). Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nature Biotechnology, 23(11), 1435–1439. [CrossRef]
- Le Guiner, C., Servais, L., Montus, M., Larcher, T., Fraysse, B., Moullec, S., Allais, M., François, V., Dutilleul, M., Malerba, A., Koo, T., Thibaut, J.-L., Matot, B., Devaux, M., Le Duff, J., Deschamps, J.-Y., Barthelemy, I., Blot, S., Testault, I., … Dickson, G. (2017). Long-term microdystrophin gene therapy is effective in a canine model of Duchenne muscular dystrophy. Nature Communications, 8, 16105. [CrossRef]
- Lostal, W., Kodippili, K., Yue, Y., & Duan, D. (2014). Full-Length Dystrophin Reconstitution with Adeno-Associated Viral Vectors. Human Gene Therapy, 25(6), 552–562. [CrossRef]
- Manini, A., Abati, E., Nuredini, A., Corti, S., & Comi, G. P. (2021). Adeno-Associated Virus (AAV)-Mediated Gene Therapy for Duchenne Muscular Dystrophy: The Issue of Transgene Persistence. Frontiers in Neurology, 12, 814174. [CrossRef]
- Mendell, J. R., Shilling, C., Leslie, N. D., Flanigan, K. M., al-Dahhak, R., Gastier-Foster, J., Kneile, K., Dunn, D. M., Duval, B., Aoyagi, A., Hamil, C., Mahmoud, M., Roush, K., Bird, L., Rankin, C., Lilly, H., Street, N., Chandrasekar, R., & Weiss, R. B. (2012). Evidence-based path to newborn screening for Duchenne muscular dystrophy. Annals of Neurology, 71(3), 304–313. [CrossRef]
- Odom, G. L., Gregorevic, P., Allen, J. M., & Chamberlain, J. S. (2011). Gene Therapy of mdx Mice With Large Truncated Dystrophins Generated by Recombination Using rAAV6. Molecular Therapy, 19(1), 36–45. [CrossRef]
- Padula, A., Petruzzelli, R., Philbert, S. A., Church, S. J., Esposito, F., Campione, S., Monti, M., Capolongo, F., Perna, C., Nusco, E., Schmidt, H. H., Auricchio, A., Cooper, G. J. S., Polishchuk, R., & Piccolo, P. (2022). Full-length ATP7B reconstituted through protein trans-splicing corrects Wilson disease in mice. Molecular Therapy. Methods & Clinical Development, 26, 495–504. [CrossRef]
- Prins, K. W., Humston, J. L., Mehta, A., Tate, V., Ralston, E., & Ervasti, J. M. (2009). Dystrophin is a microtubule-associated protein. Journal of Cell Biology, 186(3), 363–369. [CrossRef]
- Renley, B. A., Rybakova, I. N., Amann, K. J., & Ervasti, J. M. (1998). Dystrophin binding to nonmuscle actin. Cell Motility and the Cytoskeleton, 41(3), 264–270. [CrossRef]
- Rouillon, J., Poupiot, J., Zocevic, A., Amor, F., Léger, T., Garcia, C., Camadro, J.-M., Wong, B., Pinilla, R., Cosette, J., Coenen-Stass, A. M. L., Mcclorey, G., Roberts, T. C., Wood, M. J. A., Servais, L., Udd, B., Voit, T., Richard, I., & Svinartchouk, F. (2015). Serum proteomic profiling reveals fragments of MYOM3 as potential biomarkers for monitoring the outcome of therapeutic interventions in muscular dystrophies. Human Molecular Genetics, 24(17), 4916–4932. [CrossRef]
- Saleh, L., & Perler, F. B. (2006). Protein splicing in cis and in trans. Chemical Record (New York, N.Y.), 6(4), 183–193. [CrossRef]
- Shah, N. H., Eryilmaz, E., Cowburn, D., & Muir, T. W. (2013). Extein residues play an intimate role in the rate-limiting step of protein trans-splicing. Journal of the American Chemical Society, 135(15), 5839–5847. [CrossRef]
- Tasfaout, H., Halbert, C. L., McMillen, T. S., Allen, J. M., Reyes, T. R., Flint, G. V., Grimm, D., Hauschka, S. D., Regnier, M., & Chamberlain, J. S. (2024). Split intein-mediated protein trans-splicing to express large dystrophins. Nature, 632(8023), 192–200. [CrossRef]
- Tornabene, P., Trapani, I., Minopoli, R., Centrulo, M., Lupo, M., de Simone, S., Tiberi, P., Dell’Aquila, F., Marrocco, E., Iodice, C., Iuliano, A., Gesualdo, C., Rossi, S., Giaquinto, L., Albert, S., Hoyng, C. B., Polishchuk, E., Cremers, F. P. M., Surace, E. M., … Auricchio, A. (2019). Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Science Translational Medicine, 11(492), eaav4523. [CrossRef]
- Wasala, L. P., Watkins, T. B., Wasala, N. B., Burke, M. J., Yue, Y., Lai, Y., Yao, G., & Duan, D. (2023). The Implication of Hinge 1 and Hinge 4 in Micro-Dystrophin Gene Therapy for Duchenne Muscular Dystrophy. Human Gene Therapy, 34(9–10), 459–470. [CrossRef]
- Xu, J., & Zhang, Y. (2010). How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics, 26(7), 889–895. [CrossRef]
- Yan, Z., Lei-Butters, D. C. M., Zhang, Y., Zak, R., & Engelhardt, J. F. (2007). Hybrid adeno-associated virus bearing nonhomologous inverted terminal repeats enhances dual-vector reconstruction of minigenes in vivo. Human Gene Therapy, 18(1), 81–87. [CrossRef]
- Yan, Z., Zhang, Y., Duan, D., & Engelhardt, J. F. (2000). Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proceedings of the National Academy of Sciences, 97(12), 6716–6721. [CrossRef]
- Zhang, Y., Yue, Y., Li, L., Hakim, C. H., Zhang, K., Thomas, G. D., & Duan, D. (2013). Dual AAV therapy ameliorates exercise-induced muscle injury and functional ischemia in murine models of Duchenne muscular dystrophy. Human Molecular Genetics, 22(18), 3720–3729. [CrossRef]
- Zhao, J., Kodippili, K., Yue, Y., Hakim, C. H., Wasala, L., Pan, X., Zhang, K., Yang, N. N., Duan, D., & Lai, Y. (2016). Dystrophin contains multiple independent membrane-binding domains. Human Molecular Genetics, 25(17), 3647–3653. [CrossRef]
- Zhao, J., Yang, H. T., Wasala, L., Zhang, K., Yue, Y., Duan, D., & Lai, Y. (2019). Dystrophin R16/17 protein therapy restores sarcolemmal nNOS in trans and improves muscle perfusion and function. Molecular Medicine, 25(1), Article 1. [CrossRef]
- Zhou, Y., Zhang, C., Xiao, W., Herzog, R. W., & Han, R. (2024). Systemic delivery of full-length dystrophin in Duchenne muscular dystrophy mice. Nature Communications, 15(1), 6141. [CrossRef]
Figure 1.
Rational design of novel Midi-Dystrophin. A) Schematic representation of Midi-Dystrophin 1 (Midi-Dys1) which has a molecular weight of 247,1 kDa. B) Schematic representation of Midi-Dystrophin 2 (Midi-Dys2) with its molecular weight of 265,6 kDa. C) Scheme representing the domains included in Midi-Dystrophin 3 (Midi-Dys 3) and its molecular weight of 256,1 kDa For A) B) C): In yellow are represented binding domain with cytoskeletal protein, in pink the binding with sarcolemma lipids, in green the binding of signaling proteins, and in blue the binding of transmembrane proteins (β-DG = β dystroglycan). D) 3D structure of the central rod domain of Midi-Dys1 including the R1-3, hinge 2, R8-13, R16-17, and R24 with the original dystrophin hinge and after the shortening. E) 3D structure of the central rod domain of Midi-Dys2 including the R1, hinge 2, R8-12, R16-17, hinge 3 and R20-24 with the original dystrophin hinge and after the shortening. F) 3D structure of the central rod domain of Midi-Dys3 including the R1-3, hinge 2, R8-19, R16-17, hinge 3 and R20-24 with the original dystrophin hinge and after the shortening. For D), E), F): the green heatmaps represent the expected position error (if 0 is green and if 30 is white) for each aligned residue. The corresponding color of each portion of the protein indicates the level of confidence in the prediction: dark blue corresponds to high confidence in the prediction, light blue corresponds to medium confidence, yellow corresponds to low confidence, and orange corresponds to very low confidence in the prediction. G) Predicted template modeling (pTM) score for structural predictions of Midi-Dys1, 2 and 3 central rod domains for shorter and original H) Insertion sites of GP41-1 split-intein highlighting the ligation junction and the extein flanking amino acids. Created with BioRender.com.
Figure 1.
Rational design of novel Midi-Dystrophin. A) Schematic representation of Midi-Dystrophin 1 (Midi-Dys1) which has a molecular weight of 247,1 kDa. B) Schematic representation of Midi-Dystrophin 2 (Midi-Dys2) with its molecular weight of 265,6 kDa. C) Scheme representing the domains included in Midi-Dystrophin 3 (Midi-Dys 3) and its molecular weight of 256,1 kDa For A) B) C): In yellow are represented binding domain with cytoskeletal protein, in pink the binding with sarcolemma lipids, in green the binding of signaling proteins, and in blue the binding of transmembrane proteins (β-DG = β dystroglycan). D) 3D structure of the central rod domain of Midi-Dys1 including the R1-3, hinge 2, R8-13, R16-17, and R24 with the original dystrophin hinge and after the shortening. E) 3D structure of the central rod domain of Midi-Dys2 including the R1, hinge 2, R8-12, R16-17, hinge 3 and R20-24 with the original dystrophin hinge and after the shortening. F) 3D structure of the central rod domain of Midi-Dys3 including the R1-3, hinge 2, R8-19, R16-17, hinge 3 and R20-24 with the original dystrophin hinge and after the shortening. For D), E), F): the green heatmaps represent the expected position error (if 0 is green and if 30 is white) for each aligned residue. The corresponding color of each portion of the protein indicates the level of confidence in the prediction: dark blue corresponds to high confidence in the prediction, light blue corresponds to medium confidence, yellow corresponds to low confidence, and orange corresponds to very low confidence in the prediction. G) Predicted template modeling (pTM) score for structural predictions of Midi-Dys1, 2 and 3 central rod domains for shorter and original H) Insertion sites of GP41-1 split-intein highlighting the ligation junction and the extein flanking amino acids. Created with BioRender.com.
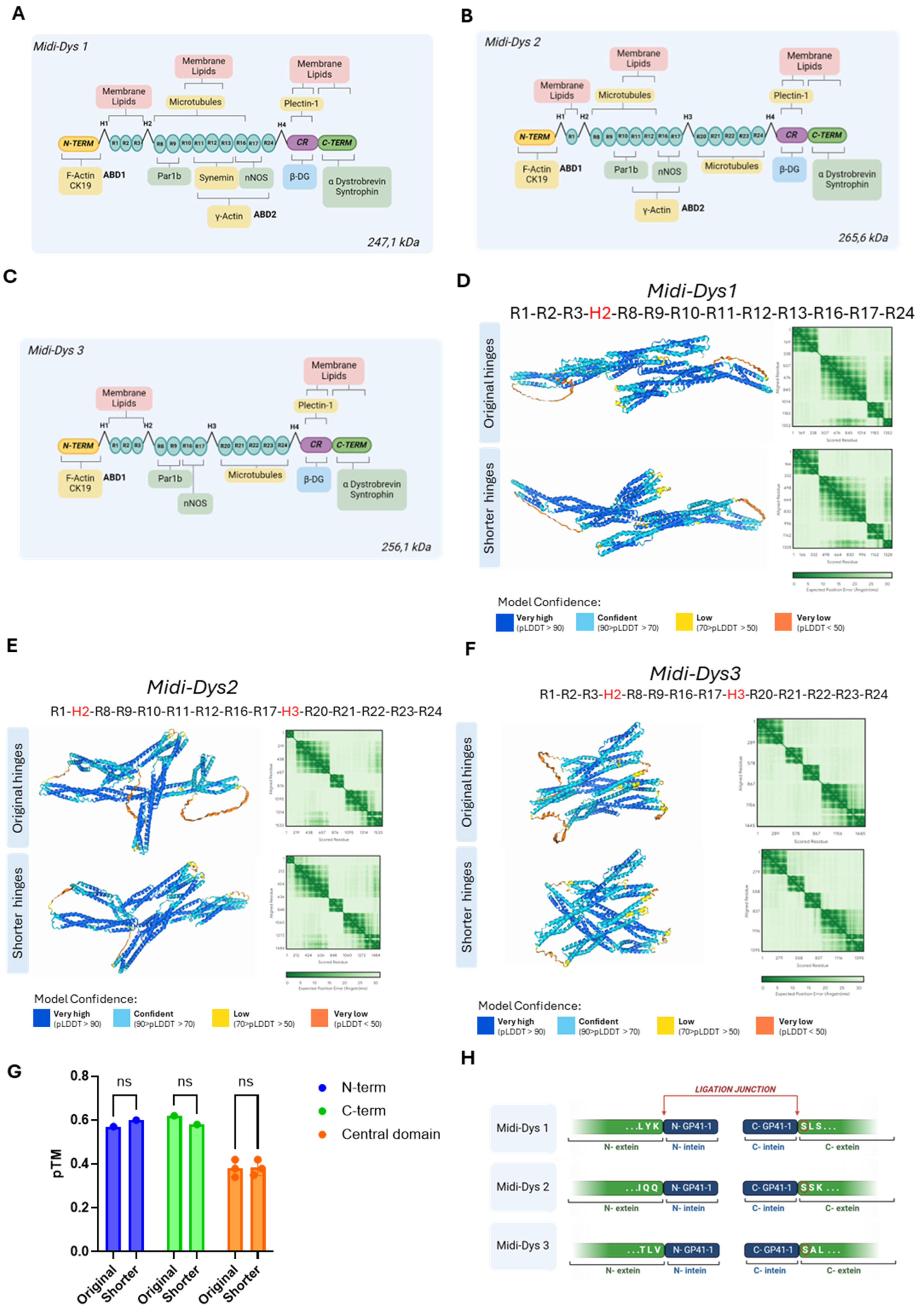
Figure 2.
Deep learning-based predictions of intein residue interactions and in vitro validation of GFP intein-split PTS. A) Schematic representation of the dual plasmid GFP GP41-1 split-intein, followed by a HA tag. B) Predicted structure of gp41—1 intein and GFP colored by predicted local distance difference test (pLDDT) (red, 0–50; yellow, 50–60; green, 60–80; cyan, 80-90 and blue, more than 90). C) The same prediction colored by structured protein chain. GP41-1 intein is highlighted by a square box. Light pink -200aa region, pink N-term gp41-1 intein, blue, C-term intein, light blue +200aa region. D) PAE matrix highlighting the predicted error of interaction between the intermolecular domains. E) Fluorescent images of HEK293T lipofected with single GFP plasmid (containing the GFP in one plasmid sequence), or with N-terminal GFP plasmid and C-terminal GFP plasmid. Single vector controls are included. Condition 1: N-term GFP + C-term GFP. Condition 2: N-term GFP alone. Condition 3: C-term GFP alone. Condition 4: Single plasmid GFP alone F) FACS analysis of HEK293T lipofected with GFP GP41-1 split intein dual plasmid. GFP mean fluorescent intensity (MFI) over GFP-positive cells is indicated in each experimental condition. Condition 1: N-term GFP + C-term GFP. Condition 2: N-term GFP alone. Condition 3: C-term GFP alone. Condition 4: Single plasmid GFP alone. Created with BioRender.com.
Figure 2.
Deep learning-based predictions of intein residue interactions and in vitro validation of GFP intein-split PTS. A) Schematic representation of the dual plasmid GFP GP41-1 split-intein, followed by a HA tag. B) Predicted structure of gp41—1 intein and GFP colored by predicted local distance difference test (pLDDT) (red, 0–50; yellow, 50–60; green, 60–80; cyan, 80-90 and blue, more than 90). C) The same prediction colored by structured protein chain. GP41-1 intein is highlighted by a square box. Light pink -200aa region, pink N-term gp41-1 intein, blue, C-term intein, light blue +200aa region. D) PAE matrix highlighting the predicted error of interaction between the intermolecular domains. E) Fluorescent images of HEK293T lipofected with single GFP plasmid (containing the GFP in one plasmid sequence), or with N-terminal GFP plasmid and C-terminal GFP plasmid. Single vector controls are included. Condition 1: N-term GFP + C-term GFP. Condition 2: N-term GFP alone. Condition 3: C-term GFP alone. Condition 4: Single plasmid GFP alone F) FACS analysis of HEK293T lipofected with GFP GP41-1 split intein dual plasmid. GFP mean fluorescent intensity (MFI) over GFP-positive cells is indicated in each experimental condition. Condition 1: N-term GFP + C-term GFP. Condition 2: N-term GFP alone. Condition 3: C-term GFP alone. Condition 4: Single plasmid GFP alone. Created with BioRender.com.
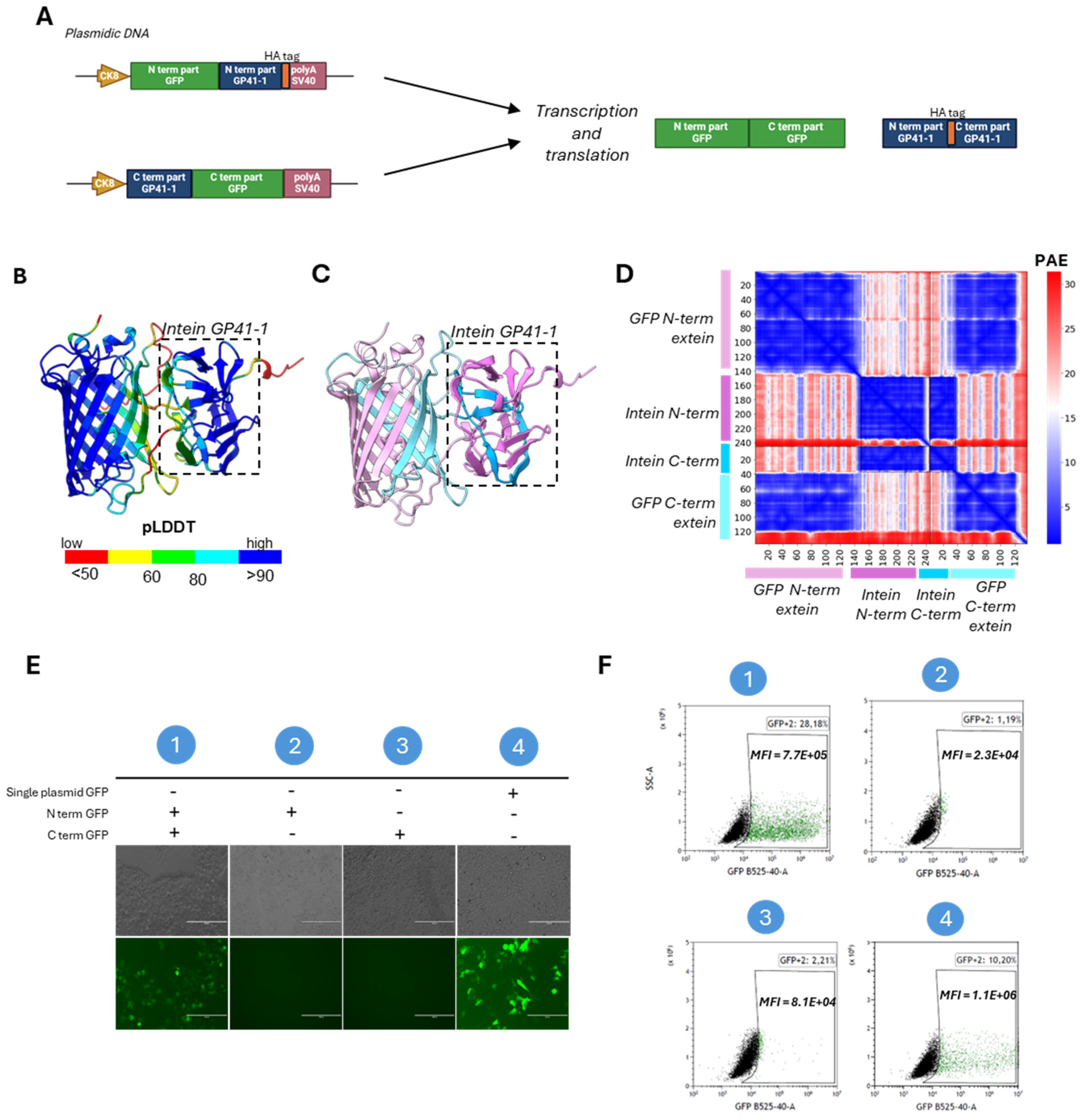
Figure 3.
In silico predictions of intein residue interactions and validation of Midi-Dys intein PTS. A) Predicted structure of GP41—1 intein, and Midi-Dys1 200aa around the split sites colored by pLDDT (red, 0–50; yellow, 50–60; green, 60–80; cyan, 80-90 and blue, more than 90). B) The same prediction colored by structured protein chain. GP41-1 intein is highlighted by a square box. Light pink -200aa region, pink N-term gp41-1 intein, blue, C-term intein, light blue +200aa region. C) PAE matrix highlighting the predicted error of interaction between the intermolecular domains. D) Predicted structure of gp41—1 intein and Midi-Dys2 200aa around the split sites colored by pLDDT (red, 0–50; yellow, 50–60; green, 60–80; cyan, 80-90 and blue, more than 90). E) The same prediction colored by chain. GP41-1 intein is indicated by a square box. Light pink -200aa region, pink N-term gp41-1 intein, blue, C-term intein, light blue +200aa region. F) PAE matrix highlighting the predicted error of interaction between the N-terminal chain and the C-terminal chain. G) Predicted structure of gp41—1 intein and Midi-Dys3 200aa around the split sites colored by pLDDT (red, 0–50; yellow, 50–60; green, 60–80; cyan, 80-90 and blue, more than 90. H) The same prediction colored by chain. GP41-1 intein is inside a square box. Light pink -200aa region, pink N-term gp41-1 intein, blue, C-term intein, light blue +200aa region. I) PAE matrix highlighting the predicted error of interaction between the two split chains. J) Schematic representations of Midi-Dystrophins lipofection in HEK293T and capillary western blot. K) Capillary western blot of HEK293T lipofected with Midi-Dys 1-2-3 with N-term and C-term specific antibody. Reconstituted Midi-Dys 1-2 are visible around 220-230 kDa, while not-reconstituted Midi-Dys 1-2-3 are visible around 120-130 kDa. Created with BioRender.com.
Figure 3.
In silico predictions of intein residue interactions and validation of Midi-Dys intein PTS. A) Predicted structure of GP41—1 intein, and Midi-Dys1 200aa around the split sites colored by pLDDT (red, 0–50; yellow, 50–60; green, 60–80; cyan, 80-90 and blue, more than 90). B) The same prediction colored by structured protein chain. GP41-1 intein is highlighted by a square box. Light pink -200aa region, pink N-term gp41-1 intein, blue, C-term intein, light blue +200aa region. C) PAE matrix highlighting the predicted error of interaction between the intermolecular domains. D) Predicted structure of gp41—1 intein and Midi-Dys2 200aa around the split sites colored by pLDDT (red, 0–50; yellow, 50–60; green, 60–80; cyan, 80-90 and blue, more than 90). E) The same prediction colored by chain. GP41-1 intein is indicated by a square box. Light pink -200aa region, pink N-term gp41-1 intein, blue, C-term intein, light blue +200aa region. F) PAE matrix highlighting the predicted error of interaction between the N-terminal chain and the C-terminal chain. G) Predicted structure of gp41—1 intein and Midi-Dys3 200aa around the split sites colored by pLDDT (red, 0–50; yellow, 50–60; green, 60–80; cyan, 80-90 and blue, more than 90. H) The same prediction colored by chain. GP41-1 intein is inside a square box. Light pink -200aa region, pink N-term gp41-1 intein, blue, C-term intein, light blue +200aa region. I) PAE matrix highlighting the predicted error of interaction between the two split chains. J) Schematic representations of Midi-Dystrophins lipofection in HEK293T and capillary western blot. K) Capillary western blot of HEK293T lipofected with Midi-Dys 1-2-3 with N-term and C-term specific antibody. Reconstituted Midi-Dys 1-2 are visible around 220-230 kDa, while not-reconstituted Midi-Dys 1-2-3 are visible around 120-130 kDa. Created with BioRender.com.
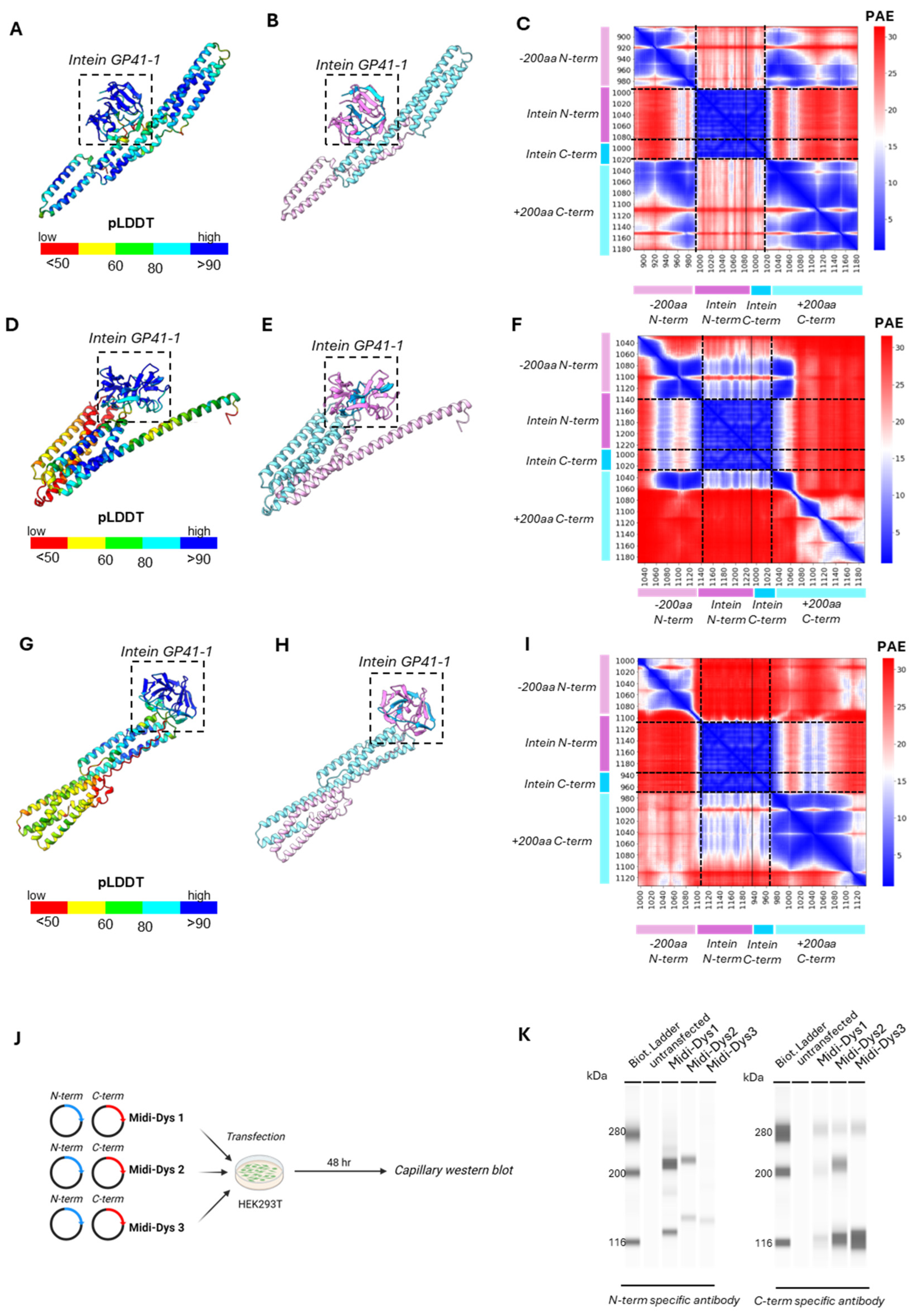
Figure 4.
Efficacy of reconstitution of Midi-Dystrophins in Dba2-mdx mouse model. A) Schematic representation of the in vivo protocol. One month old Dba2/mdx mice were injected with Midi-Dys1, Midi-Dys2 and Midi-Dys3 at the dose of 2.0E+13 vg/kg each vector (total dose 4.0E+13 vg/kg). Tissues were collected after 7 weeks from the intravenous injection. B) Capillary western blot of protein lysates of diaphragm at 7 weeks post-injection in Dba2-mdx mice with N-term and C-term specific antibodies. Reconstituted Midi-Dys are visible around 200 kDa, while non-reconstituted Midi-Dys are visible around 116 kDa. C) Quantification of Midi-Dys1-2-3 and Micro-Dys protein expression after capillary western blot using N-term specific antibody in gastrocnemius, tibialis anterior and diaphragm at 7 weeks post-injection in Dba2-mdx mice. Values are represented as normalized area under the curve (AUC). D) Quantification of Midi-Dys1-2-3 (reconstituted and non-reconstituted) protein expression after capillary western blot using C-term Midi-Dys specific antibody in gastrocnemius, tibialis anterior and diaphragm at 7 weeks post-injection in Dba2-mdx mice. Values are represented as normalized area under the curve (AUC). Blue values represent the percentage of reconstitution, calculated as protein expression of reconstituted Midi-Dys over the total. E) Representative Dystrophin IHF pictures of the diaphragm at 7 weeks post-injection in Dba2-mdx mice using N-term specific antibody. F) Quantification of Dystrophin positive myofibers in gastrocnemius, tibialis anterior and diaphragm at 7 weeks post-injection in Dba2-mdx mice. The percentage of dystrophin + fibers is represented of number of dystrophin positive fibers over fibers positive for laminin. For all the panels: N=4-5. One-way ANOVA statistic test. *p<0.05, **p<0.005, ***p<0.001 ns= not significant.
Figure 4.
Efficacy of reconstitution of Midi-Dystrophins in Dba2-mdx mouse model. A) Schematic representation of the in vivo protocol. One month old Dba2/mdx mice were injected with Midi-Dys1, Midi-Dys2 and Midi-Dys3 at the dose of 2.0E+13 vg/kg each vector (total dose 4.0E+13 vg/kg). Tissues were collected after 7 weeks from the intravenous injection. B) Capillary western blot of protein lysates of diaphragm at 7 weeks post-injection in Dba2-mdx mice with N-term and C-term specific antibodies. Reconstituted Midi-Dys are visible around 200 kDa, while non-reconstituted Midi-Dys are visible around 116 kDa. C) Quantification of Midi-Dys1-2-3 and Micro-Dys protein expression after capillary western blot using N-term specific antibody in gastrocnemius, tibialis anterior and diaphragm at 7 weeks post-injection in Dba2-mdx mice. Values are represented as normalized area under the curve (AUC). D) Quantification of Midi-Dys1-2-3 (reconstituted and non-reconstituted) protein expression after capillary western blot using C-term Midi-Dys specific antibody in gastrocnemius, tibialis anterior and diaphragm at 7 weeks post-injection in Dba2-mdx mice. Values are represented as normalized area under the curve (AUC). Blue values represent the percentage of reconstitution, calculated as protein expression of reconstituted Midi-Dys over the total. E) Representative Dystrophin IHF pictures of the diaphragm at 7 weeks post-injection in Dba2-mdx mice using N-term specific antibody. F) Quantification of Dystrophin positive myofibers in gastrocnemius, tibialis anterior and diaphragm at 7 weeks post-injection in Dba2-mdx mice. The percentage of dystrophin + fibers is represented of number of dystrophin positive fibers over fibers positive for laminin. For all the panels: N=4-5. One-way ANOVA statistic test. *p<0.05, **p<0.005, ***p<0.001 ns= not significant.
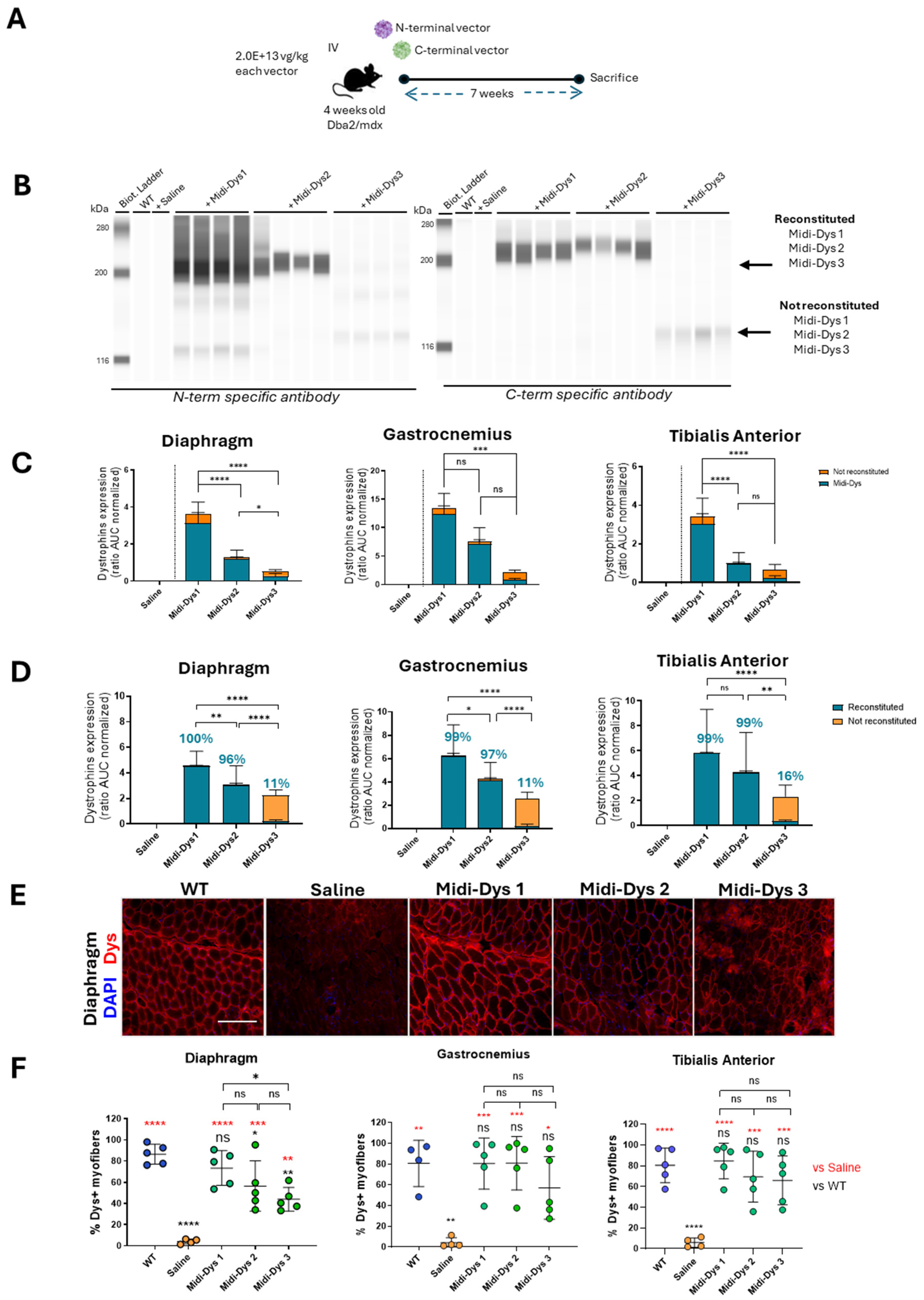
Figure 5.
Functional and histopathological therapeutic efficacy in Dba2-mdx mice. A) CPK (Creatine phospho kinase) concentration in the sera of Dba2-mdx mice injected with Midi-Dys1-2-3. Results are shown as concentration defined units/liter (U/L). B) Myom3 (Myomesin 3) concentration in the sera of Dba2-mdx mice injected with Midi-Dys 1-2-3. Results are represented as Myom3 concentrations defined as pg/µl. C) Representative images of Sirius red coloration in the diaphragm of Dba2-mdx mice 7 weeks post injection. Red-colored regions are highly fibrotic and contain collagen deposits. D) Quantification of collagen deposits area in in gastrocnemius, tibialis anterior and diaphragm at 7 weeks post-injection in Dba2-mdx mice. Results are shown as percentage of collagen deposits area over the total area of the muscle cut. E) Representative images of Red alizarin coloration in the gastrocnemius of Dba2-mdx mice 7 weeks post injection. Red areas are indicated a region containing calcium deposits. F) Quantification of calcium deposits area in gastrocnemius of Dba-mdx injected mice. Results are shown as percentage of calcium positive regions over total area of the muscle cut. G) Gene expression analysis of Tmem8c, TGF-β, and Pdgfr-α in tibialis anterior and gastrocnemius of WT mice and DBA2/mdx mice injected with saline, Midi-Dys1, Midi-Dys2 or Midi-Dys3. Results are shown as heatmap of relative abundance of gene expression over P0 normalizer gene. For all the panels: N=4-5. For panel A-F: One-way ANOVA statistic test. *p<0.05, **p<0.005, ***p<0.001 ns= not significant. For panel G: Two-way ANOVA statistic test and multiple comparisons with main row effect. *p<0.05, **p<0.005, ***p<0.001, ****<0.0001, ns= not significant.
Figure 5.
Functional and histopathological therapeutic efficacy in Dba2-mdx mice. A) CPK (Creatine phospho kinase) concentration in the sera of Dba2-mdx mice injected with Midi-Dys1-2-3. Results are shown as concentration defined units/liter (U/L). B) Myom3 (Myomesin 3) concentration in the sera of Dba2-mdx mice injected with Midi-Dys 1-2-3. Results are represented as Myom3 concentrations defined as pg/µl. C) Representative images of Sirius red coloration in the diaphragm of Dba2-mdx mice 7 weeks post injection. Red-colored regions are highly fibrotic and contain collagen deposits. D) Quantification of collagen deposits area in in gastrocnemius, tibialis anterior and diaphragm at 7 weeks post-injection in Dba2-mdx mice. Results are shown as percentage of collagen deposits area over the total area of the muscle cut. E) Representative images of Red alizarin coloration in the gastrocnemius of Dba2-mdx mice 7 weeks post injection. Red areas are indicated a region containing calcium deposits. F) Quantification of calcium deposits area in gastrocnemius of Dba-mdx injected mice. Results are shown as percentage of calcium positive regions over total area of the muscle cut. G) Gene expression analysis of Tmem8c, TGF-β, and Pdgfr-α in tibialis anterior and gastrocnemius of WT mice and DBA2/mdx mice injected with saline, Midi-Dys1, Midi-Dys2 or Midi-Dys3. Results are shown as heatmap of relative abundance of gene expression over P0 normalizer gene. For all the panels: N=4-5. For panel A-F: One-way ANOVA statistic test. *p<0.05, **p<0.005, ***p<0.001 ns= not significant. For panel G: Two-way ANOVA statistic test and multiple comparisons with main row effect. *p<0.05, **p<0.005, ***p<0.001, ****<0.0001, ns= not significant.
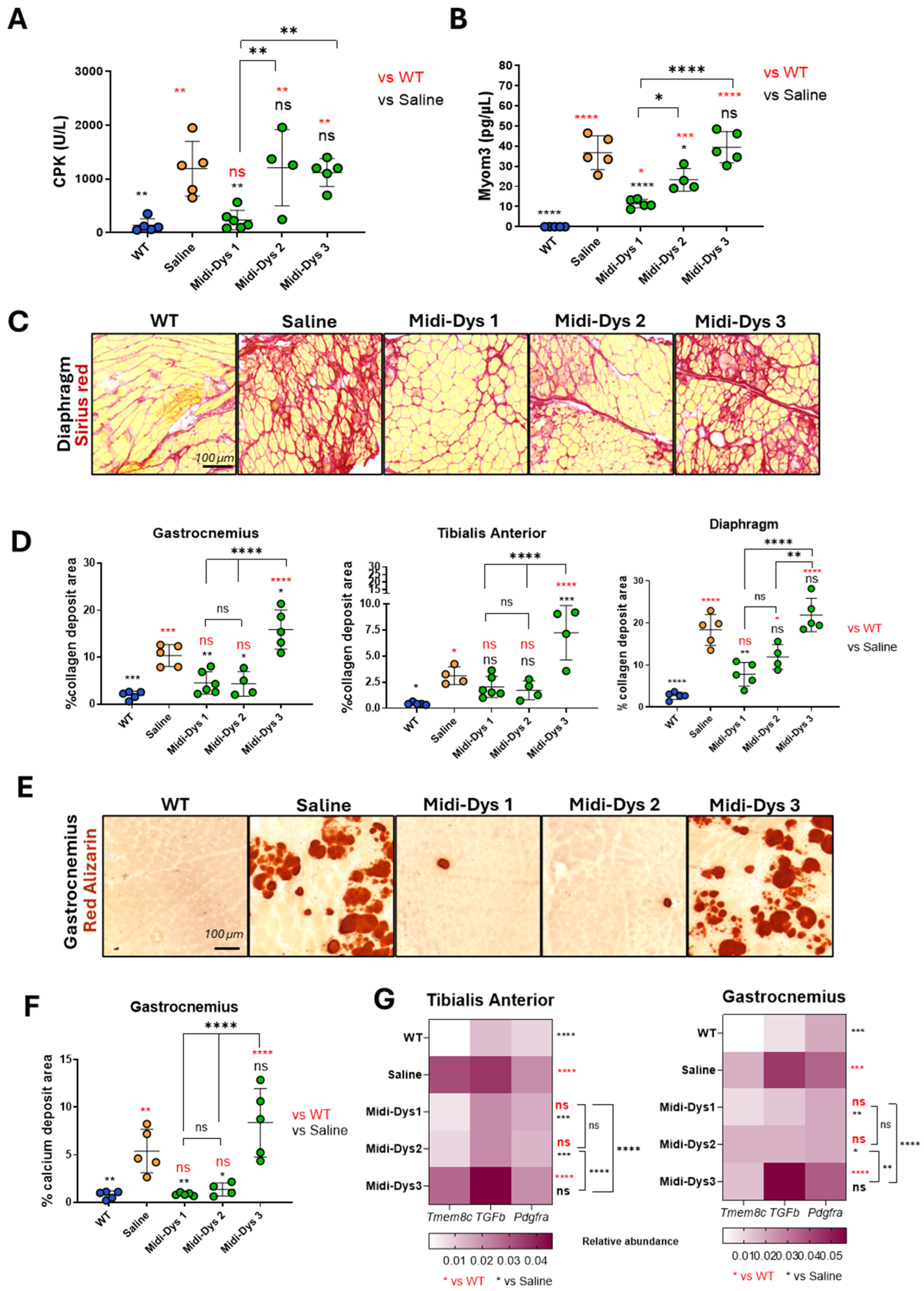
Table 1.
List of commercial antibodies used for Capillary western blot, immunofluorescence and Myom-3 ELISA.
Table 1.
List of commercial antibodies used for Capillary western blot, immunofluorescence and Myom-3 ELISA.
| Antigene |
Epitope |
Ref |
Concentration |
| Capillary western blot |
Immunofluore-scence |
ELISA |
| Dystrophin |
N-term specific antibody |
Leica
Cat#NCL-DYSB, RRID:AB_563691 |
1/20 |
1/10 |
/ |
| C-term specific antibody |
DSHB
Cat#MANCHO11(9F2), RRID:AB_2618131 |
1/100 |
/ |
/ |
| Laminin |
Polyclonal |
Thermo Fisher Scientific
Cat# PA5-115490,
RRID:AB_2900126 |
/ |
1/100 |
/ |
| Myomesin-3 |
Polyclonal |
Proteintech
Cat#17692-1-AP,
RRID:AB_2146624 |
/ |
/ |
1/100 |
Table 2.
List of primers used for viral genome copy number (VGCN) in ddPCR.
Table 2.
List of primers used for viral genome copy number (VGCN) in ddPCR.
| Amplified region |
Name |
Sequence (5’ ->3’) |
Probe |
Target vector |
| Spectrin-like region 1 |
R1_Forward |
CCAGGGAGAGATCAGCAATG |
/ |
Midi-Dys1
Midi-Dys2
Midi-Dys3 |
| R1_Reverse |
GGCTGTTAGGTCCATCATGTAG |
/ |
| R1_Probe |
GGCTGTTAGGTCCATCATGTAG |
FAM |
| Spectrin-like region 24 |
R24_Forward |
ACAGATGAGCTGGACCTGAA |
/ |
Midi-Dys1
Midi-Dys2
Midi-Dys3 |
| R24_Reverse |
TGGTCCTGTAGGCTGTCAAT |
/ |
| R24_Probe |
CTGAGGCAGGCTGAGGTGATCAAG |
VIC |
| mouse Titin exon 5 |
TitinMex5_Forward |
TTCAGTCATGCTGCTAGCGC |
/ |
genomic DNA |
| TitinMex5_Reverse |
AAAACGAGCAGTGACGTGAGC |
/ |
| TitinMex5_Probe |
TGCACGGAAGCGTCTCGTCTCAGTC |
5Cy5 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).










