Submitted:
05 September 2024
Posted:
06 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Mathematical Modelling
3. Mathematical analysis of the COVID-19 model
3.1. Positivity and Boundedness of the Solution of the Model
1.2. Local Asymptotic Stability of the DFE
3.3. Endemic Equilibrium Point (EEP)
3.4. Backward Bifurcation Analysis of the COVID-19 Model
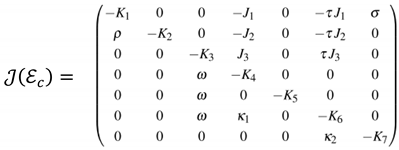
3.4.1. Local asymptotic stability of Endemic Equilibrium Point (EEP) for the COVID-19 model
3.5. Global Stability of DFE When There Is No Re-Infection
3.6. Sensitivity Analysis
4. Numerical Simulations
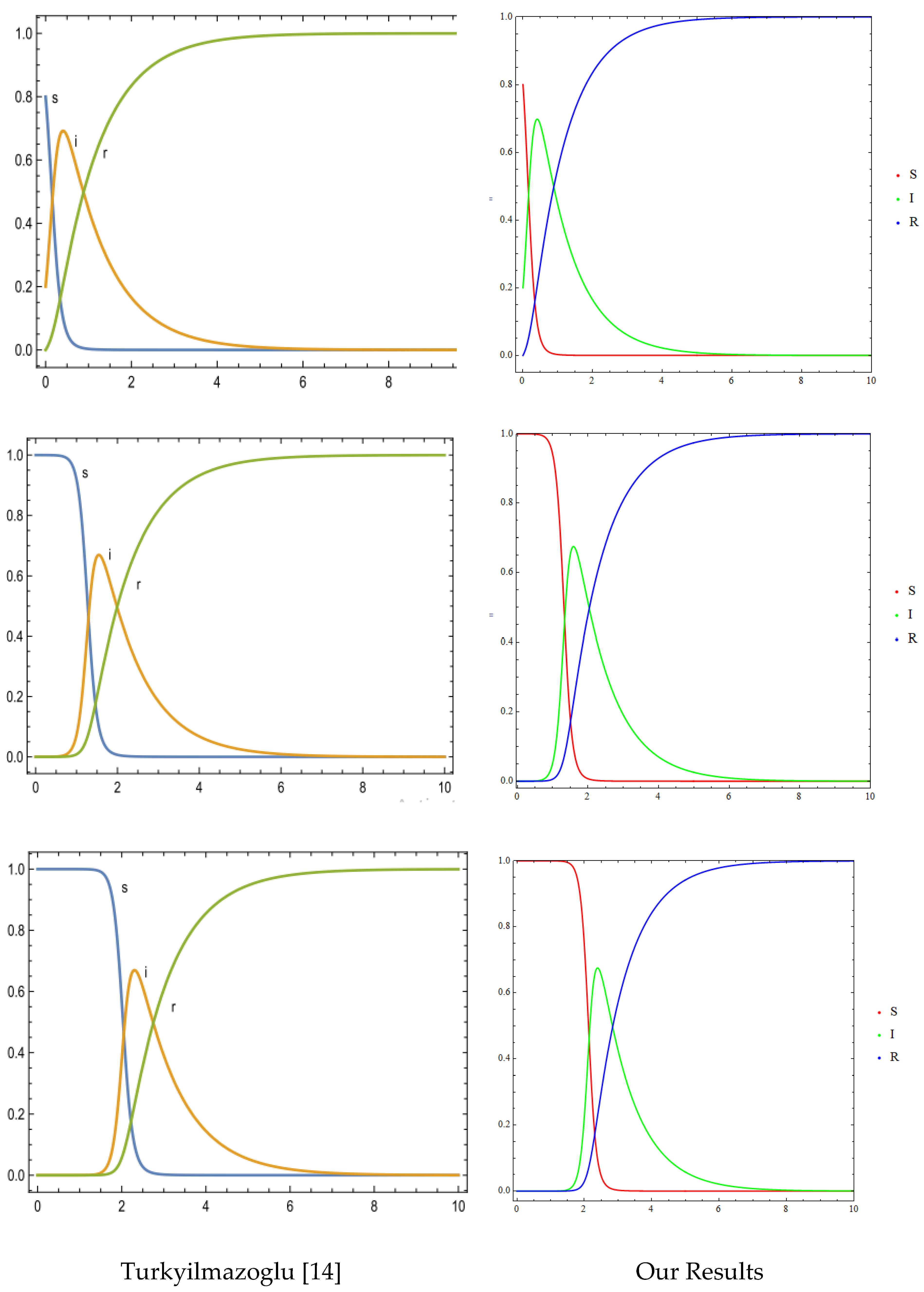
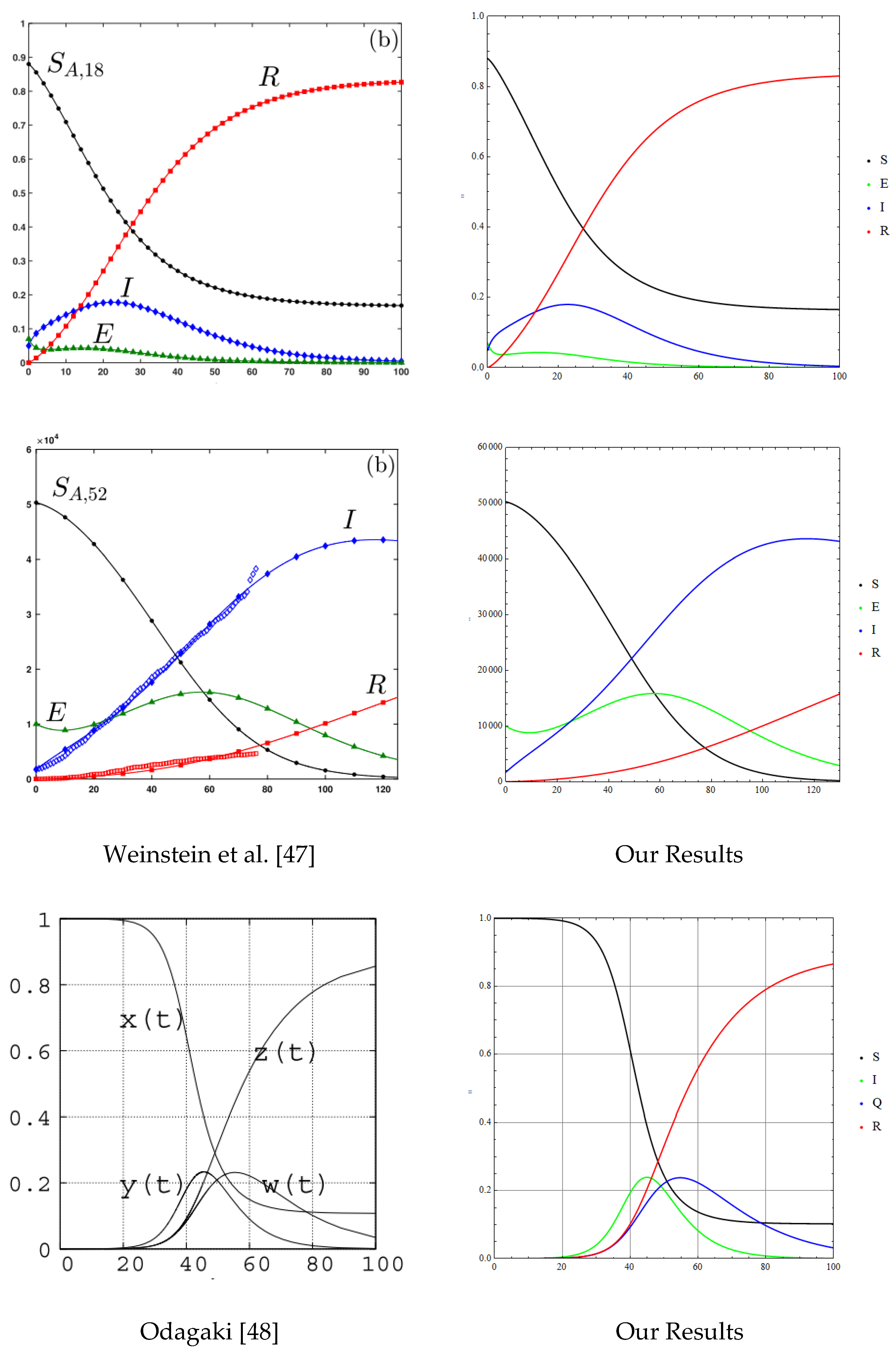
4.1. Numerical Method and Validations
4.2. Effect of Vaccines
4.2.1. Effect of COVID-19 Vaccine Coverage
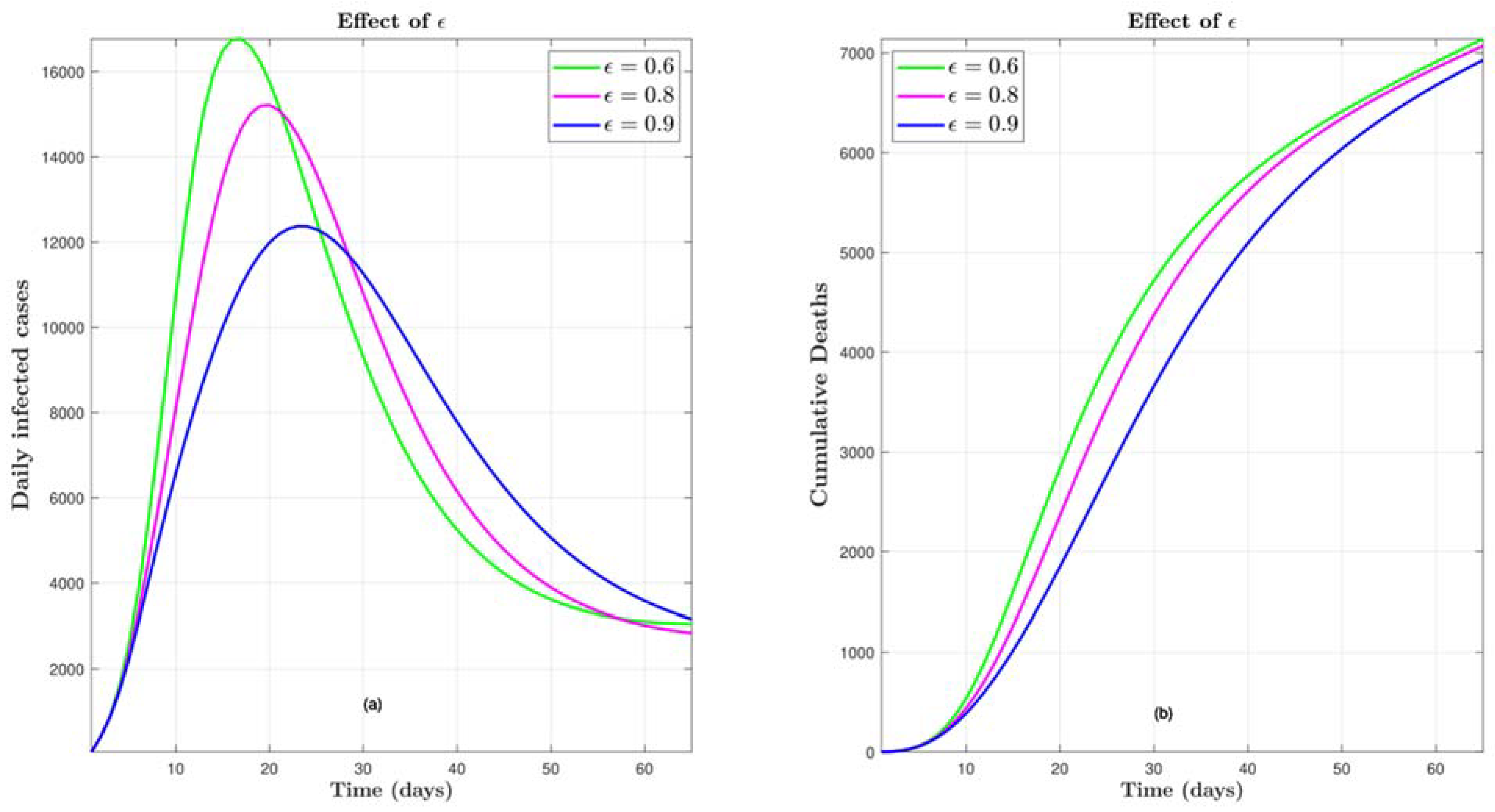
4.2.2. Effect of COVID-19 Vaccine Efficacy
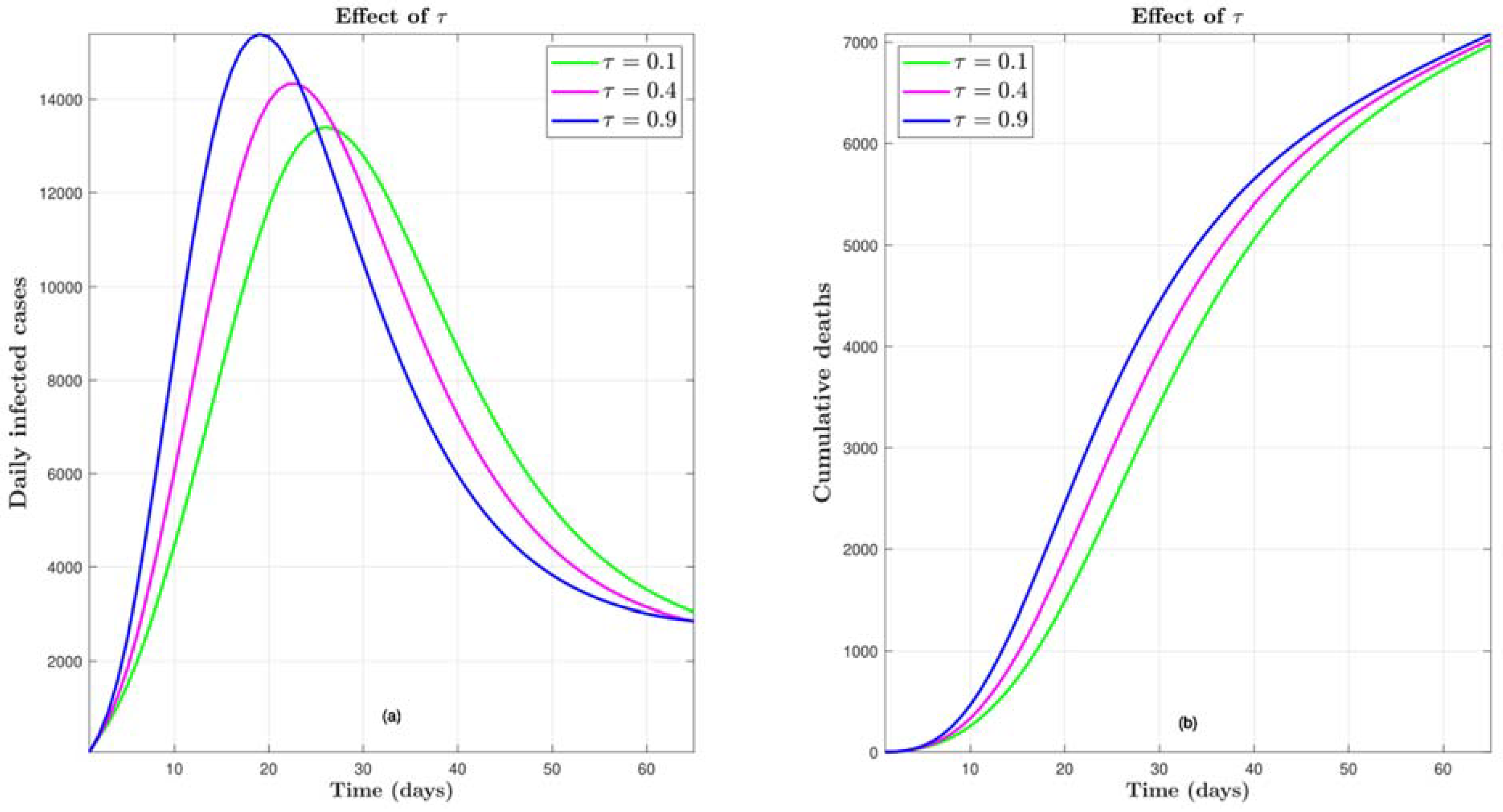
4.3. Effect of τ
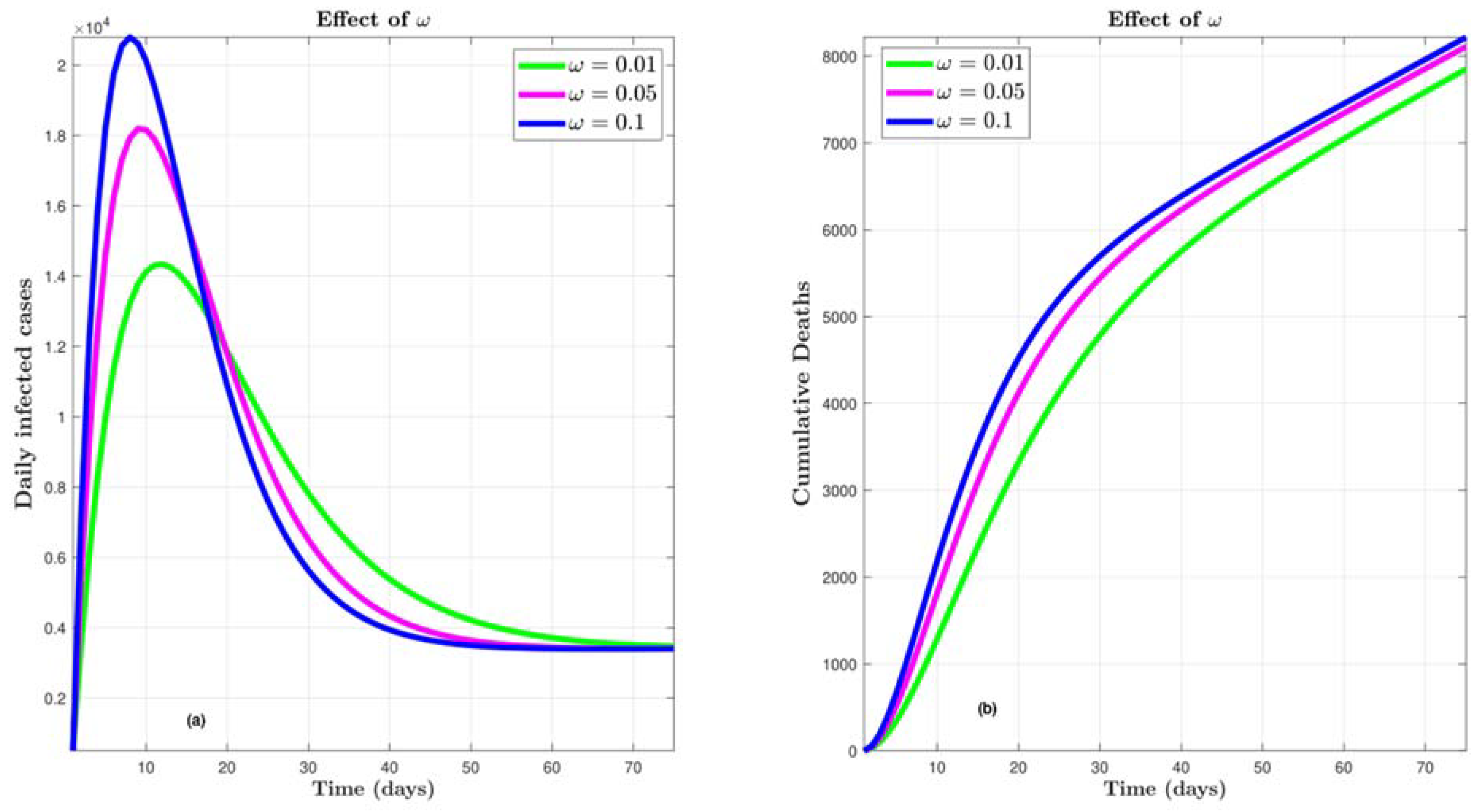
4.4. Effect of ω
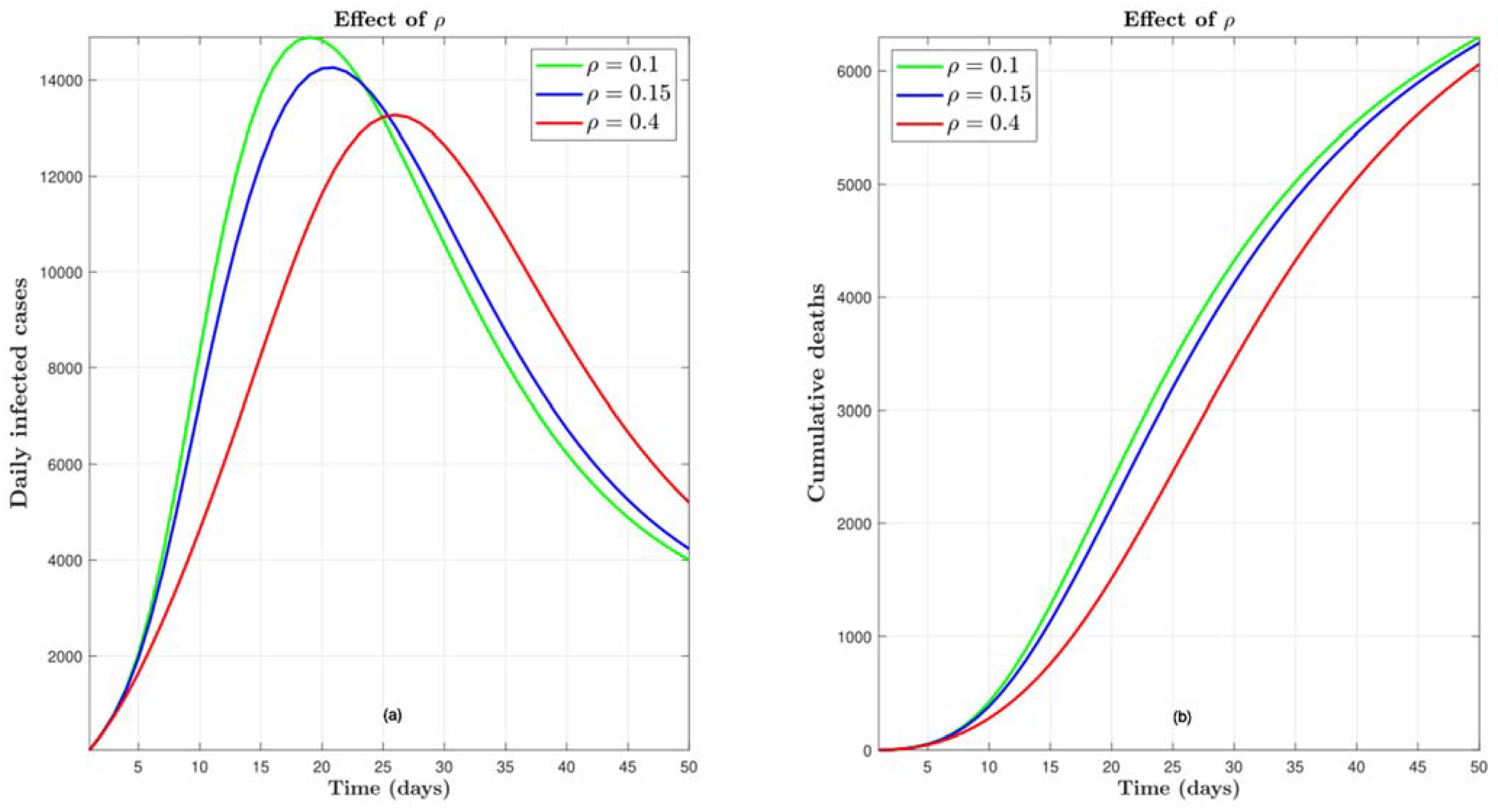
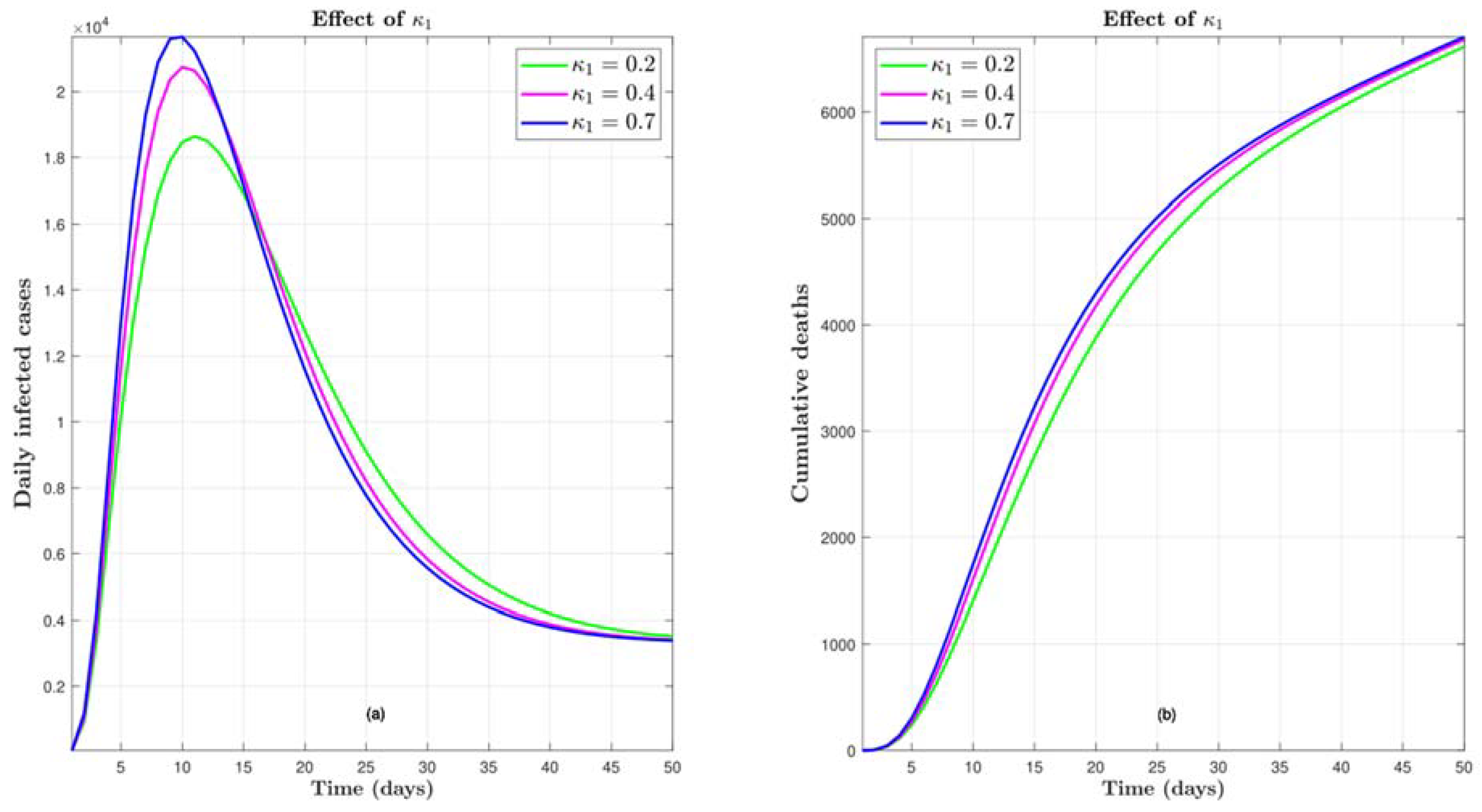
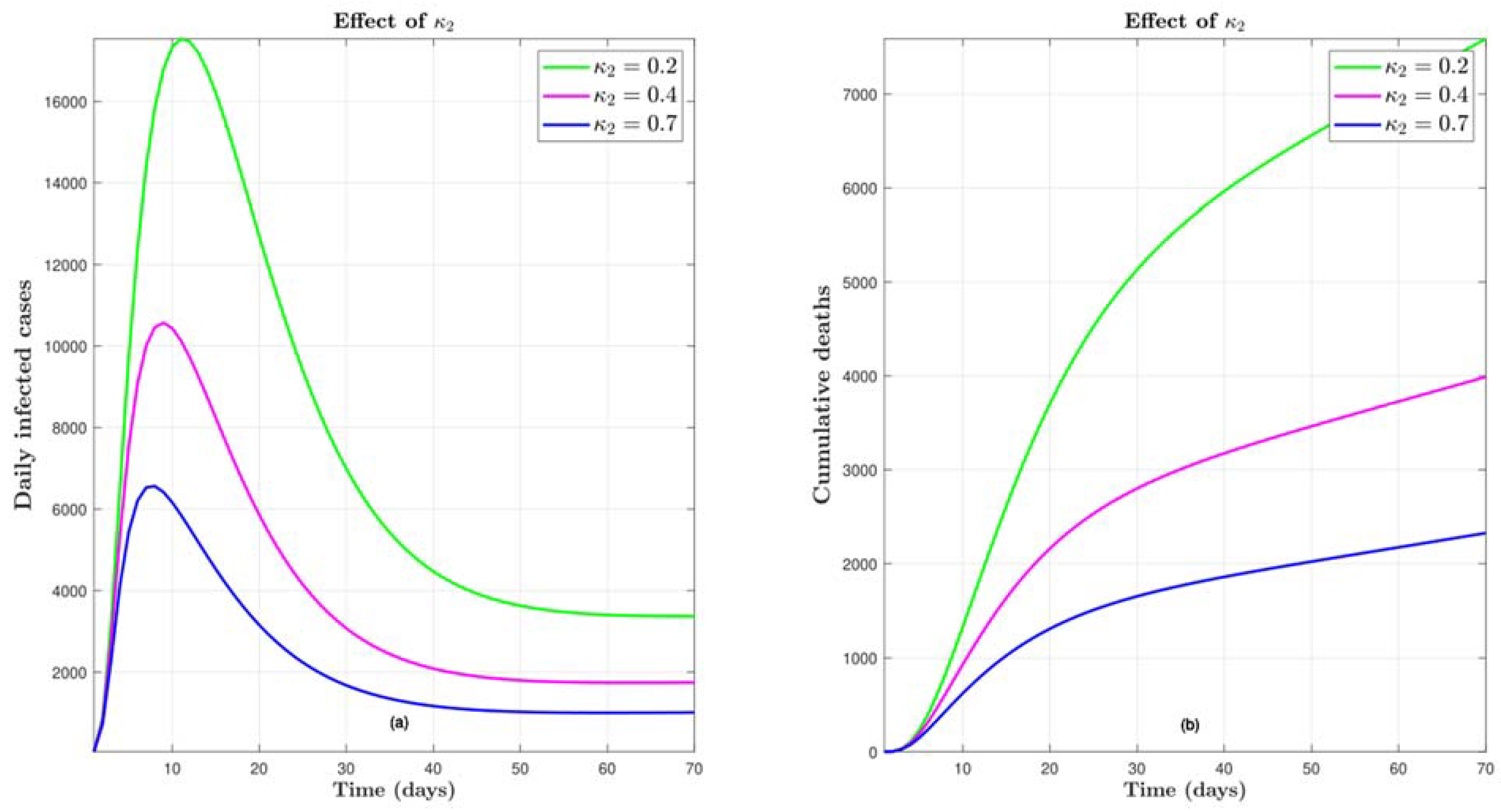
4.5. Effect of
4.6. Effect of
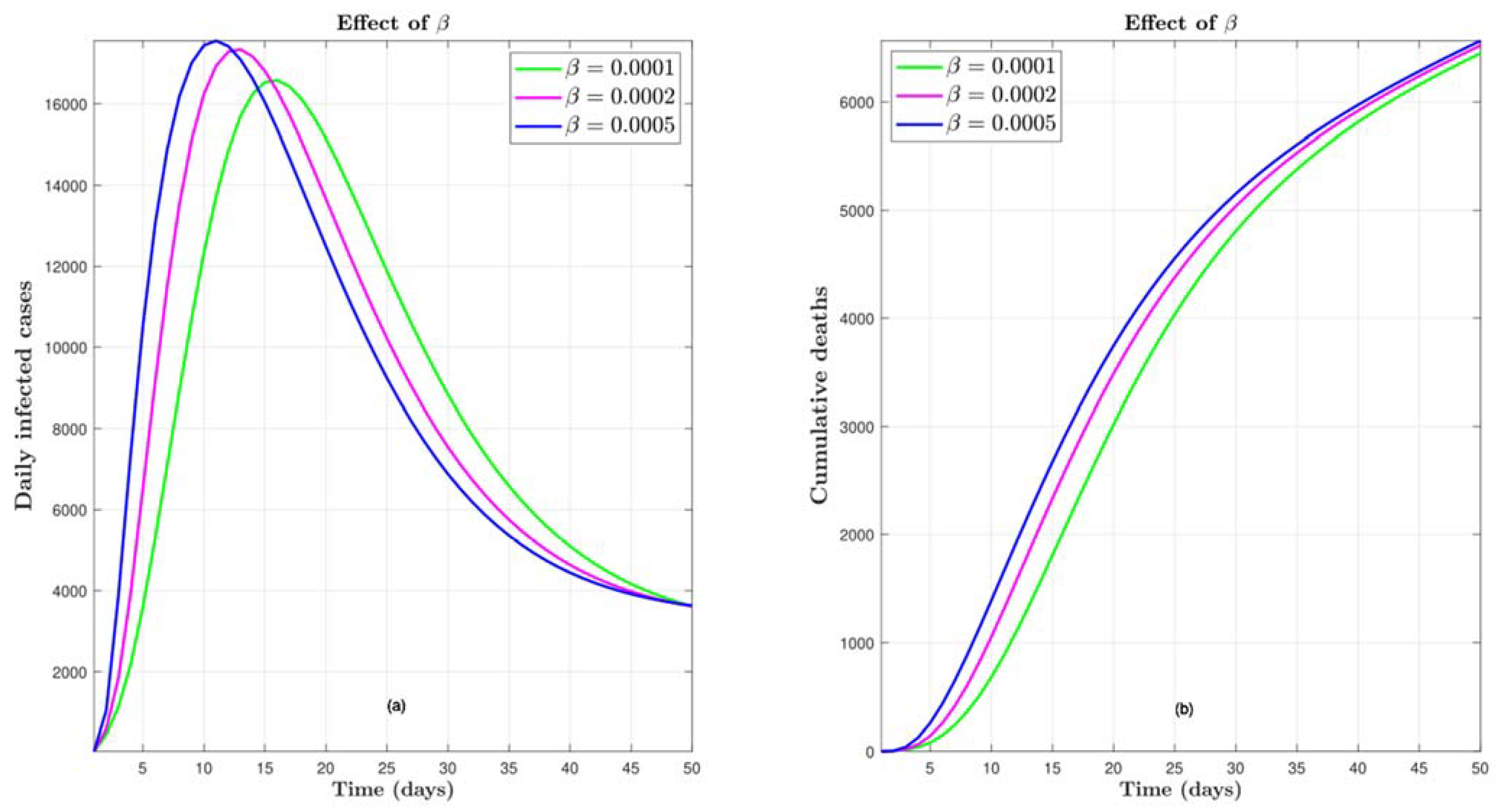
4.7. Effect of Transmission Rate
5. Conclusion
References
- Barlow, N.S.; Weinstein, S.J. Accurate closed-form solution of the SIR epidemic model. Physica. D 2020, 408, 132540–132540. [Google Scholar] [CrossRef] [PubMed]
- Tomchin, D.A.; Fradkov, A.L. Prediction of the COVID-19 spread in Russia based on SIR and SEIR models of epidemics. IFAC PapersOnLine 2020, 53, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Marinov, T.T.; Marinova, R.S. Dynamics of COVID-19 using inverse problem for coefficient identification in SIR epidemic models. Chaos, Solitons & Fractals: X 2020, 5, 100041–100041. [Google Scholar] [CrossRef]
- Neves, A.G.M.; Guerrero, G. Predicting the evolution of the COVID-19 epidemic with the A-SIR model: Lombardy, Italy and São Paulo state, Brazil. Physica. D 2020, 413, 132693–132693. [Google Scholar] [CrossRef]
- Alenezi, M.N.; Al-Anzi, F.S.; Alabdulrazzaq, H. Building a sensible SIR estimation model for COVID-19 outspread in Kuwait. Alexandria engineering journal 2021, 60, 3161–3175. [Google Scholar] [CrossRef]
- Din, R.u.; Algehyne, E.A. Mathematical analysis of COVID-19 by using SIR model with convex incidence rate. Results in physics 2021, 23, 103970–103970. [Google Scholar] [CrossRef]
- Malavika, B.; Marimuthu, S.; Joy, M.; Nadaraj, A.; Asirvatham, E.S.; Jeyaseelan, L. Forecasting COVID-19 epidemic in India and high incidence states using SIR and logistic growth models. Clinical epidemiology and global health 2021, 9, 26–33. [Google Scholar] [CrossRef]
- Kozyreff, G. Hospitalization dynamics during the first COVID-19 pandemic wave: SIR modelling compared to Belgium, France, Italy, Switzerland and New York City data. Infectious disease modelling 2021, 6, 398–404. [Google Scholar] [CrossRef]
- Campos, E.L.; Cysne, R.P.; Madureira, A.L.; Mendes, G.L.Q. Multi-generational SIR modeling: Determination of parameters, epidemiological forecasting and age-dependent vaccination policies. Infectious disease modelling 2021, 6, 751–765. [Google Scholar] [CrossRef]
- Kudryashov, N.A.; Chmykhov, M.A.; Vigdorowitsch, M. Analytical features of the SIR model and their applications to COVID-19. Applied Mathematical Modelling 2021, 90, 466–473. [Google Scholar] [CrossRef]
- Cooper, I.; Mondal, A.; Antonopoulos, C.G. A SIR model assumption for the spread of COVID-19 in different communities. Chaos, solitons and fractals 2020, 139, 110057–110057. [Google Scholar] [CrossRef] [PubMed]
- Nesteruk, I. Statistics-Based Procedure of Parameter Identification for the Classical SIR Model; Springer: Singapore, 2021; pp. 33–36. [Google Scholar] [CrossRef]
- Nesteruk, I. Applications of the General SIR Model for Calculations of the COVID-19 Epidemic Waves in Ukraine; Springer: Singapore, 2021; pp. 141–146. [Google Scholar] [CrossRef]
- Turkyilmazoglu, M. Explicit formulae for the peak time of an epidemic from the SIR model. Physica. D 2021, 422, 132902–132902. [Google Scholar] [CrossRef] [PubMed]
- Anand, N.; Sabarinath, A.; Geetha, S.; Somanath, S. Predicting the Spread of COVID-19 Using SIR Model Augmented to Incorporate Quarantine and Testing. Transactions of the Indian National Academy of Engineering (Online) 2020, 5, 141–148. [Google Scholar] [CrossRef]
- Lounis, M.; Bagal, D.K. Estimation of SIR model’s parameters of COVID-19 in Algeria. Bulletin of the National Research Centre 2020, 44, 180–180. [Google Scholar] [CrossRef]
- Ramos, A.M.; Ferrández, M.R.; Vela-Pérez, M.; Kubik, A.B.; Ivorra, B. A simple but complex enough θ-SIR type model to be used with COVID-19 real data. Application to the case of Italy. Physica. D 2021, 421, 132839–132839. [Google Scholar] [CrossRef] [PubMed]
- Shahrear, P.; Rahman, S.M.S.; Nahid, M.M.H. Prediction and mathematical analysis of the outbreak of coronavirus (COVID-19) in Bangladesh. Results in Applied Mathematics 2021, 10, 100145–100145. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.B.; Hasan, M.; Harun-Al-Rashid, A.; Ahsan, M.I.; Imran, M.A.S.; Ahmed, S.S.U. Ancestral origin, antigenic resemblance and epidemiological insights of novel coronavirus (SARS-CoV-2): Global burden and Bangladesh perspective. Infection, genetics and evolution 2020, 84, 104440–104440. [Google Scholar] [CrossRef]
- Ahmed, J.; Jaman, M.H.; Saha, G.; Ghosh, P. Effect of environmental and socio-economic factors on the spreading of COVID-19 at 70 cities/provinces. Heliyon 2021, 7, e06979–e06979. [Google Scholar] [CrossRef]
- Alrabaiah, H.; Arfan, M.; Shah, K.; Mahariq, I.; Ullah, A. A comparative study of spreading of novel corona virus disease by ussing fractional order modified SEIR model. Alexandria Engineering Journal 2021, 60, 573–585. [Google Scholar] [CrossRef]
- López, L.; Rodó, X. A modified SEIR model to predict the COVID-19 outbreak in Spain and Italy: Simulating control scenarios and multi-scale epidemics. Results in physics 2021, 21, 103746–103746. [Google Scholar] [CrossRef]
- Hussain, T.; Ozair, M.; Ali, F.; Rehman, S.u.; Assiri, T.A.; Mahmoud, E.E. Sensitivity analysis and optimal control of COVID-19 dynamics based on SEIQR model. Results in physics 2021, 22, 103956–103956. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhong, J.; Ji, Y.-M.; Yang, F. A new SEIAR model on small-world networks to assess the intervention measures in the COVID-19 pandemics. Results in physics 2021, 25, 104283–104283. [Google Scholar] [CrossRef]
- Youssef, H.; Alghamdi, N.; Ezzat, M.A.; El-Bary, A.A.; Shawky, A.M. Study on the SEIQR model and applying the epidemiological rates of COVID-19 epidemic spread in Saudi Arabia. Infectious disease modelling 2021, 6, 678–692. [Google Scholar] [CrossRef]
- Yu, Z.; Arif, R.; Fahmy, M.A.; Sohail, A. Self organizing maps for the parametric analysis of COVID-19 SEIRS delayed model. Chaos, solitons and fractals 2021, 150, 111202–111202. [Google Scholar] [CrossRef]
- Adhikary, A.; Pal, A. A six compartments with time-delay model SHIQRD for the COVID-19 pandemic in India: During lockdown and beyond. Alexandria engineering journal 2022, 61, 1403–1412. [Google Scholar] [CrossRef]
- Akindeinde, S.O.; Okyere, E.; Adewumi, A.O.; Lebelo, R.S.; Fabelurin, O.O.; Moore, S.E. Caputo fractional-order SEIRP model for COVID-19 Pandemic. Alexandria engineering journal 2022, 61, 829–845. [Google Scholar] [CrossRef]
- Basnarkov, L. SEAIR Epidemic spreading model of COVID-19. Chaos, solitons and fractals 2021, 142, 110394–110394. [Google Scholar] [CrossRef] [PubMed]
- Khedher, N.b.; Kolsi, L.; Alsaif, H. A multi-stage SEIR model to predict the potential of a new COVID-19 wave in KSA after lifting all travel restrictions. Alexandria engineering journal 2021, 60, 3965–3974. [Google Scholar] [CrossRef]
- Pekpe, K.M.; Zitouni, D.; Gasso, G.; Dhifli, W.; Guinhouya, B.C. From SIR to SEAIRD: A novel data-driven modeling approach based on the Grey-box System Theory to predict the dynamics of COVID-19. Applied intelligence (Dordrecht, Netherlands) 2022, 52, 71–80. [Google Scholar] [CrossRef]
- Zhu, W.-j.; Shen, S.-f. An improved SIR model describing the epidemic dynamics of the COVID-19 in China. Results in physics 2021, 25, 104289–104289. [Google Scholar] [CrossRef]
- Chen, M.; Li, M.; Hao, Y.; Liu, Z.; Hu, L.; Wang, L. The introduction of population migration to SEIAR for COVID-19 epidemic modeling with an efficient intervention strategy. Information fusion 2020, 64, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Contreras, S.; Villavicencio, H.A.; Medina-Ortiz, D.; Biron-Lattes, J.P.; Olivera-Nappa, Á. A multi-group SEIRA model for the spread of COVID-19 among heterogeneous populations. Chaos, solitons and fractals 2020, 136, 109925–109925. [Google Scholar] [CrossRef]
- Avila-Ponce de León, U.; Pérez, Á.G.C.; Avila-Vales, E. An SEIARD epidemic model for COVID-19 in Mexico: Mathematical analysis and state-level forecast. Chaos, solitons and fractals 2020, 140, 110165–110165. [Google Scholar] [CrossRef]
- Diekmann, O.; Heesterbeek, H.; Metz, J.A.J. On the definition and the computation of the basic reproduction ratio R0 in models for infectious diseases in heterogeneous populations. Journal of mathematical biology 1990, 28, 365–382. [Google Scholar] [CrossRef] [PubMed]
- van den Driessche, P.; Watmough, J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Mathematical biosciences 2002, 180, 29–48. [Google Scholar] [CrossRef]
- Chavez, C.C.; Feng, Z.; Huang, W. On the computation of R0 and its role on global stability. Mathematical approaches for emerging and re-emerging infection diseases: an introduction 2002, 125, 31–65. [Google Scholar]
- Hethcote, H.W. The Mathematics of Infectious Diseases. SIAM review 2000, 42, 599–653. [Google Scholar] [CrossRef]
- Carr, J. Applications of Centre Manifold Theory; Applied Mathematical Sciences; Springer, 2012; Volume 35. [Google Scholar]
- Castillo-Chavez, C.; Song, B. Dynamical models of tuberculosis and their applications. Math. Biosci. Eng 2004, 1, 361–404. [Google Scholar] [CrossRef] [PubMed]
- La Salle, J.P., The stability of dynamical systems. 1976: SIAM.
- Ngonghala, C.N.; Iboi, E.; Eikenberry, S.; Scotch, M.; MacIntyre, C.R.; Bonds, M.H.; Gumel, A.B. Mathematical assessment of the impact of non-pharmaceutical interventions on curtailing the 2019 novel Coronavirus. Mathematical biosciences 2020, 325, 108364–108364. [Google Scholar] [CrossRef]
- Saha, A.K.; Podder, C.N.; Niger, A.M. Dynamics of novel COVID-19 in the presence of Co-morbidity. Infectious disease modelling 2022, 7, 138–160. [Google Scholar] [CrossRef]
- Mancuso, M.; Eikenberry, S.E.; Gumel, A.B. Will vaccine-derived protective immunity curtail COVID-19 variants in the US? Infectious disease modelling 2021, 6, 1110–1134. [Google Scholar] [CrossRef] [PubMed]
- Burden and Faires, Numerical Analysis. 2015: Cengage Learning.
- Weinstein, S.J.; Holland, M.S.; Rogers, K.E.; Barlow, N.S. Analytic solution of the SEIR epidemic model via asymptotic approximant. Physica. D 2020, 411, 132633–132633. [Google Scholar] [CrossRef] [PubMed]
- Odagaki, T. Exact properties of SIQR model for COVID-19. Physica A 2021, 564, 125564–125564. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





