1. Introduction
As was established by the World Health Organization (WHO) and the Food and Agriculture Organization of the United Nations (FAO) probiotics are live microorganisms that, when administered in adequate amounts, provide health benefits to the host (Hill et al. 2014). Health-promoting effects were predominantly demonstrated for specific probiotic strains of the bacteria genera Lactobacillus and Bifidobacterium (Fijan 2014), while the only yeast with a probiotic status evidenced clinically is Saccharomyces cerevisiae var. boulardii (Sb) (Łukaszewicz 2012). The probiotic yeast has unique physiological properties resulting in increased tolerance to stress conditions in the GI tract (Fietto et al. 2004). The mode of probiotic yeast action is not completely understood, whereby the beneficial effects have been confirmed in clinical studies (Czerucka et al. 2007). The clinical studies did not show any major side effects related to Sb. However, some cases of Sb fungemia are documented (Cesaro et al. 2000; Ellouze et al. 2016; Kara 2018; Santino et al. 2014; Thygesen et al. 2012). The most quoted cases concerned severely ill or immunocompromised patients.
Initially, Sb was categorized as a novel species of the genus Saccharomyces. The taxonomic position of Sb has been controversial over the years (Van Der Aa Kühle and Jespersen 2003; Cardinali and Martini 1994; Fietto et al. 2004; Kurtzman and Robnett 1998; McCullough et al. 1998; Molnar et al. 1995). McFarland indicated some crucial divergences at the physiological (i.e., lack of ability to use galactose as carbon source and lack of ability to produce ascospores) and molecular levels (i.e., individual chromosome and gene copy numbers) between Sb and Saccharomyces cerevisiae (Sc) (McFarland 1996). Using comparative genomic hybridization for whole-genome analysis, Sb's physiological and genotypic distinctive features were confirmed (Edwards-Ingram et al. 2007). The observations concerned the specific properties of Ty elements (yeast retrotransposons) and the copy number of genes in its subtelomeric regions. These were confirmed in the subsequent genomic comparative study, which additionally showed that Sb strains are closely related to Sc wine strains (Khatri et al. 2017). Pais and colleagues hypothesized that different phenotypes exhibited by Sb and Sc might result from variations in gene expression control. They demonstrated that Sb did not share conserved promoter regions and transcription factor binding sites with Sc (Pais et al. 2021).
So far, the search for methods of intraspecies differentiation of Saccharomyces cerevisiae has mainly involved clinical studies. By combining randomly amplified polymorphic DNA-PCR, restriction fragment length polymorphism analysis of rDNA spacer regions and pulsed-field gel electrophoresis, all examined Sc isolates were discriminated in clinical research. Moreover, probiotic Sb strains were demonstrated to form a separate cluster within the species (Mitterdorfer et al. 2002). Additionally, a powerful microsatellite-based technique was developed for intraspecies differentiation of probiotic and clinical Sc strains (Hennequin et al. 2001). Furthermore, a high level of discrimination was achieved by hybridization with retrotransposon Ty917 (Posteraro et al. 2005). Finally, a rapid multiplex PCR method unequivocally identified probiotic-derived Sc isolates (Imre et al. 2019).
A modern technique that allows differentiation of the amplicons at the single base resolution is the High-Resolution Melting (HRM) analysis of qPCR amplified DNA fragments. It is an alternative single-tube approach with no time-consuming post-PCR processing and the need for sequencing to detect DNA polymorphisms. HRM analysis was primarily used in clinical research and diagnostics, but its robust potential and technological progress allow it to enter other areas of life sciences. In food sciences, HRM analysis has already been used for the differentiation of food-derived yeast species including Saccharomyces spp (Nadai et al. 2018), sourdough yeast (Bazalová et al. 2022; Ripari et al. 2016) and spoilage yeast (Erdem et al. 2016; Kesmen et al. 2018a, b). All procedures presented a high potential for HRM analysis in yeast species differentiation based on the amplification of regions within rRNA genes or ITS non-coding sequences characterized by low intraspecific variability and high interspecific polymorphism (Baleiras-Couto et al. 1996; Valente et al. 1996). Most of the studies cited dealt with culture-dependent identification. Culture-independent identification of sourdough yeast by HRM analysis did not always give conclusive results (Bazalová et al. 2022; Ripari et al. 2016).
Probiotics have become increasingly popular over the past two decades as a result of proven health-promoting properties. Specialists have recommended probiotic supplements for disorders that frustrate conventional medicine, such as irritable bowel syndrome, or to delay the development of allergies in children. As a result, a wide range of probiotic-fortified dietary supplements have been available in European markets. According to EFSA food supplements are concentrated sources of nutrients or other substances with a nutritional or physiological effect that are marketed in “ dose ” form. Combining typical medicine dosage forms with misleading advertising can create a false impression of a supplement's therapeutic properties. Dietary supplements are currently regulated as foods and their legalization is facilitated by the law. The current state of the regulations and the expanded supplement market create an environment conducive to food adulterations, necessitating rapid testing to verify product status. The problem may soon affect a much wider range of foods. Concerning probiotic yeast, intense research activity arose in developing functional foods supplemented with Sb not only as a probiotic agent, but also as a key element for the generation of bioactives increasing the antioxidant capacity (Değirmencioğlu et al. 2016; Rekha and Vijayalakshmi 2010), improving nutritional value (Zamora-Vega et al. 2012), or stabilization of LAB strains throughout fermentation and storage (Chan et al. 2023; Karaolis et al. 2013).
It is crucial to verify the strain identity of Sb yeasts since their beneficial properties are considered to be strain-specific. Therefore, this study aimed to create a rapid genetic test to authenticate probiotic yeast strains. The Sb identification efficiency of interspecies and intragenus primer pairs was verified using qPCR-HRM analysis, conducted with reference Sc and Sb strains, along with Kluyveromyces marxianus and Pichia fermentans as negative controls. The utility of selected primer pairs previously optimised in culture-dependent studies was verified in dietary supplements and yeast cell mixtures corresponding in composition to kefir, which is a natural source of probiotic yeast origin (Goktas et al. 2021; Gut et al. 2019).
2. Materials and Methods
2.1. Biological Material
2.1.1. Strains
The following reference strains were used in the optimisation of qPCR-HRM analysis focused on identifying probiotic strains of Saccharomyces cerevisiae: (i) Saccharomyces boulardii CNCM I-745 (Enterol, Biocodex, Gentilly, France) (Sb745) and Saccharomyces boulardii CNCM-I-3799 (Oslonik max extra, TZF Polfa, Warsaw, Poland) (Sb3799) isolated from probiotic preparations under this project; (ii) and collection strains Saccharomyces cerevisiae ATCC 9763 (ScATCC9763) and Saccharomyces cerevisiae Ethanol Red (Lesaffre; France) (ScEtRed). Additionally, (i) Kluyveromyces marxianus DSM 5422 (German Collection of Microorganisms and Cell Cultures GmbH; Germany) (Km), (ii) food-derived isolates Saccharomyces cerevisiae (ScD) and Pichia fermentans (PfD) deposited in the microbial collection of Department Biotechnology and Food Microbiology (DBFM), Poznan University of Life Sciences (PULS) (iii) Lactobacillus delbrueckii subsp. lactis DSM 20072 (German Collection of Microorganisms and Cell Cultures GmbH; Germany) (Ldsubl), were used in the subsequent stages of testing the qPCR-HRM. All strains were maintained as glycerol stocks at -80°C until used. Yeast strains were recovered on YPD agar plates [(g L-1): yeast extract, 10 (Biomaxima, Lublin, Poland), bactopeptone, 20 (BTL, Łódź, Poland), glucose, 20 (POCh, Gliwice, Poland) and agar, 15 (BTL)] and lactic acid bacteria (LAB) were retrieved with de man, Rogosa and Sharpe (M.R.S) agar (BTL).
2.1.2. Probiotic Supplements
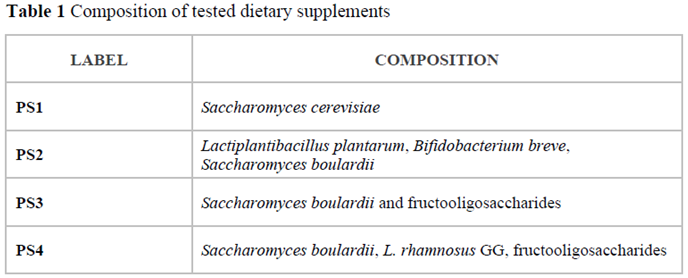
Four probiotic preparations classified as dietary supplements and available in local pharmacies were analysed with the optimised qPCR-HRM method. The composition of chosen products is shown in Table 1.
2.2. Microbiological Methods
2.2.1. S. boulardii Reference Strains Isolation
The reference strains of Sb used in this study were isolated from two different commercial products, one with proven therapeutic effects and the other classified as a dietary supplement. To revive the yeast, the contents of each capsule (250 mg) were suspended in 20 mL of YPD in a sterile 50 mL Falcon tube. The mixture was then shaken at 150 rpm at 30°C overnight. Afterwards, the log serial dilutions were made from the overnight culture. 100 µL of each dilution was transferred in duplicate onto Yeast extract Glucose Chloramphenicol agar (YGC) (BTL, Łódź, Poland). Incubation was carried out overnight at 30°C. Single colonies were then reinoculated onto fresh YGC agar. The yeast isolates were first verified based on morphological characteristics with microscopy and then subjected to MALDI TOF mass spectrometry identification.
2.2.2. Sporulation Test
2.2.2.1. Procedure of Sporulation Induction
5 mL of YPD medium in a 50 mL Falcon tube was inoculated with a fresh yeast colony for determination of the ascospore formation. Each culture was incubated at 30oC in a shaking incubator at 150 rpm for 18-20 hours. 200 µL of the overnight culture were transferred into 5mL of liquid YPD and incubated at 30oC in a shaking incubator until the cell suspension reached OD equal to 1. Then the culture was centrifuged at 1811 x g for 5 min and the supernatant was discarded. The pellet was resuspended in 5mL of pre-sporulation medium (g L-1: yeast extract, 10; pepton, 20; potassium acetate, 10 (POCh, Gliwice, Poland)) and grew for 18-24 hours at 30oC in shacking incubator. The culture was centrifuged at 1811 x g for 5 min and the supernatant was discarded. Finally, the pellet was resuspended in 5mL of sporulation medium (g L-1: potassium acetate, 10) and allowed to sporulate for 48 hours at 30oC in a shaking incubator. The tested strains were subjected to the sporulation procedure in two independent replicates.
2.2.2.2. Cells’ Ziehl-Neelsen Staining
The basic dye, concentrated carbol fuchsin (g L-1: fuchsin, 33,3 (Chempur, Piekary Śląskie, Poland), phenol, 66,7 (Chempur) and 167 mL of ethanol (POCh, Gliwice, Poland)) was applied to the fixed smear of yeast on a degreased basic slide for 15 min. During staining the slide was heated with a burner (to the so-called "three pairs"). Then the preparation was discoloured in a 3% solution of hydrochloric acid (Honeywell, Charlotte, USA) in ethanol (POCh, Gliwice, Poland) (acid alcohol). Afterwards, the contrast dye 0.1% (w/v) methylene blue (Chempur) was applied for 10 min. Finally, the preparation was observed in the light microscope Primo star (Zeiss, Oberkochen, Germany) under 1000x magnification using immersion.
2.2.3. The Mixtures of Lactic Acid Bacteria and Yeast Cells
5 mL of fresh M.R.S Broth or liquid YPD in a 50 mL Falcon tube was inoculated with a bacterial (
Ldsub
l) or yeast colony (
ScD,
Sb745,
Km, PfD), respectively. The cultures were incubated at 30°C with 250 rpm shaking for 20 h. The average cells’ concentration of each overnight culture was determined using CellDrop FL (DeNovix INC, Wilmington, USA). Subsequently, mixtures of microorganisms were prepared in saline water (0.85% w/v NaCl). Suspensions included
ScD and
Sb745 cells, where
Sb accounted for 10% (Mx_0.1), 50% (Mx_0.5) and 90% (Mx_0.9) of the total amount of yeast cells. Another group of suspensions consisted of those that contained a fixed number of
Ldsub
l cells and yeast cells with a 60% share of
ScD (Mx_
Sc) or
Sb (Mx_
Sb), 38% of
Km and 2% of
PfD. In this series of mixtures, the ratio of
Sc cells to
Sb cells was the same as in suspensions composed only of
Sc cells (
Figure S1).
2.3. MALDI-TOF Mass Spectrometry
Mass spectrometry analysis was conducted in the Microbiological Laboratory of the Jagiellonian Center of Innovation (Cracow, Poland). MALDI-TOF mass spectrometer Microflex LT (Bruker Daltonics, Bremen, Germany) was used for matrix-assisted laser desorption/ionization with time-of-flight analysis. Identification of the microorganisms was based on the unique ribosomal proteins’ profile compared to a representative database of bacteria, yeast-like fungi, filamentous fungi and dermatophytes, using the MALDI Biotyper system (Bruker Daltonics). Identification confidence was expressed by identification indicator (Ii). An identification indicator higher or equal to 2,00 means high-confidence identification. Ii within the range 1,70 - 1,99 means low-confidence identification, and below 1,69, no species identification is possible.
2.4. Total Genomic DNA Extraction Procedures
2.4.1. Total Genomic DNA Isolation from Yeast Culture
Prior to genomic DNA extraction, all the yeast strains were freshly cultured on YPD agar at 30°C for 20 h. The biomass was collected with a 10 µl inoculation loop and resuspended in 0,5 ml saline solution, and the cells were pelleted by centrifugation. DNA extraction from cells’ pellets was performed using a Genomic mini AX yeast spin kit (A&A Biotechnology, Gdynia, Poland) according to the manufacturer’s protocol, including an optional step of yeast lysis with lyticase at 30°C, followed by vigorous vortexing. The concentration and purity of the isolated DNA were determined using a UV spectrophotometer NanoDrop ND-1000 (Thermo Fisher Scientific, Wilmington, USA). The OD260/OD280 ratio of the DNA samples, reflecting their average purity, ranged between 1,8 and 2,0. The quality and integrity of the DNA samples were verified through agarose gel electrophoresis according to a standard method (Sambrook and Russell, 2001). Only DNA samples exhibiting sharp and intensive bands were forwarded to qPCR. The extracted DNA was stored at –20°C.
2.4.2. Total Genomic DNA Isolation from Dietary Supplements and Microbial Mixtures
The capsule contents of each dietary supplement were crushed in liquid nitrogen in a mortar. The fresh yeast and bacterial cultures were used for preparing mixtures (
Section 2.2.3). 2 mL of each mixture was pelleted by centrifugation. Total genomic DNA was extracted from 100 mg of each food supplement and each mixture pellet using Genomic mini AX Food kit (A&A Biotechnology, Gdynia, Poland). The procedure was conducted according to the manufacturer's protocol. The first step was lysis with proteinase K at 30°C, followed by vigorous vortexing. DNA was isolated and purified from the lysate with column work through gravity and then eluted, precipitated and dissolved in sterile water. The concentration and purity of the isolated DNA were determined using a UV spectrophotometer NanoDrop ND-1000 (Thermo Fisher Scientific, Wilmington, USA). The quality and integrity of the DNA samples were verified through agarose gel electrophoresis according to a standard method (Sambrook and Russell, 2001). The extracted DNA was stored at –20°C.
2.5. Quantitative Real-Time PCR – High-Resolution Melting Analysis (qPCR-HRM)
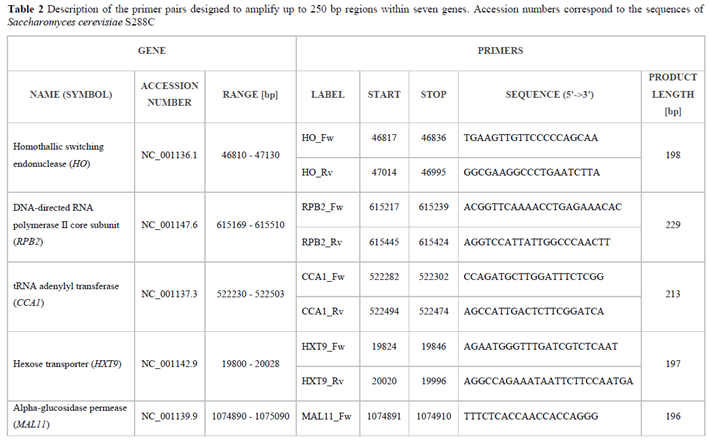
2.5.1. Primer Pairs
rDNA cluster sequences (
18S rRNA,
26S rRNA and ITS region) of the yeast strains under study were amplified with primer pairs designed previously (Borkowska and Celińska 2023). As part of this work, additional primer pairs were designed.
S. cerevisiae S288C genes’ sequences were retrieved from the NCBI database. Their accession numbers are listed in Table 2. The targeted sequences were aligned and compared to
Saccharomyces spp using multiple sequence alignment tool from NCBI. The multiple sequence alignment was searched for conservative sequence segments allowing the attachment of intragroup primers, flanking DNA regions showing intragroup heterogeneity. Amplicon length was kept within the range of 100 to 250 bp. The primers were designed with the Primer3-BLAST tool, and synthesized by Merck KGaA (Darmstadt, Germany). Initially, annealing temperature optimisation of all primer pairs was performed using Perpetual OptiTaq PCR MasterMix according to the manufacturer’s instructions (EURx, Gdansk, Poland) in a temperature gradient of 58 - 63°C. The amplification efficiency was verified through agarose gel electrophoresis according to a standard method (Sambrook and Russell 2001).
TDA8 primer pair was eliminated from the further study due to a lack of amplification on
Sc samples in the optimisation step. Furthermore, amplification products of all studied primer pairs were positively verified for expected lengths (
Figure S2).
2.5.2. qPCR Protocol
PCR mixtures contained 5 µL of commercial qPCR master mix with Eve Green dye (Bio-Rad Laboratories, Inc., CA, USA), 0,5 µL of 10 µM forward and reverse primer and 1- 4 µL of DNA sample (5 ng of single-yeast DNA or 20 ng of microbial-mix DNA per reaction) in a total volume of 10 µL. The amplification reactions were carried out in clear-walled PCR 96-well plates using the CFX96 cycler (Bio-Rad Laboratories, CA, USA). The cycling conditions were as follows: 95°C for 3 min was followed by 30 - 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C (primer pairs for sequence segment within 18SrRNA, 26SrRNA, ITS, TEF1alpha, HO, RPB2, MAL11 and HXT9) or 58°C (primer pairs for CCA1) for 30 s and extension at 72°C for 30 s. Melting curves were obtained by cycling within a range of 65 to 95°C. Data acquisition was performed in 0,2°C increments, with a 10 s step. All DNA preparations were analysed in technical duplicates in at least two independent runs. To check the purity of reagents, No Template Control (NTC) for each primer pair was run in parallel.
2.5.3. HRM Analysis
The emerging qPCR amplicons were analyzed using Precision Melt Analysis Software (PMAS) (Bio-Rad Laboratories, Inc.). The pre-and post-melt regions, as well as the intersection point of the melt curves on temperature-shifted view, were carefully evaluated (and adjusted where necessary) for each amplicon. Clustering settings were adjusted to increase the sensitivity of clustering based on the shape of the melt curves. A Tm difference threshold of 0,2°C was set for all the melt curve analyses. Clustering confidence scores were assigned automatically by PMAS.
2.6. Statistical Analysis
The statistical significance of the observed differences based on clustering in HRM analysis of yeast strains for the four examined regions (18SrRNA, 26SrRNA, ITS, TEF1alpha) was evaluated using the software DarWin version 6.0.21. Afterwards, the strains were grouped using the unweighted pair-group method with the arithmetic averages (UPGMA) clustering algorithm.
3. Results
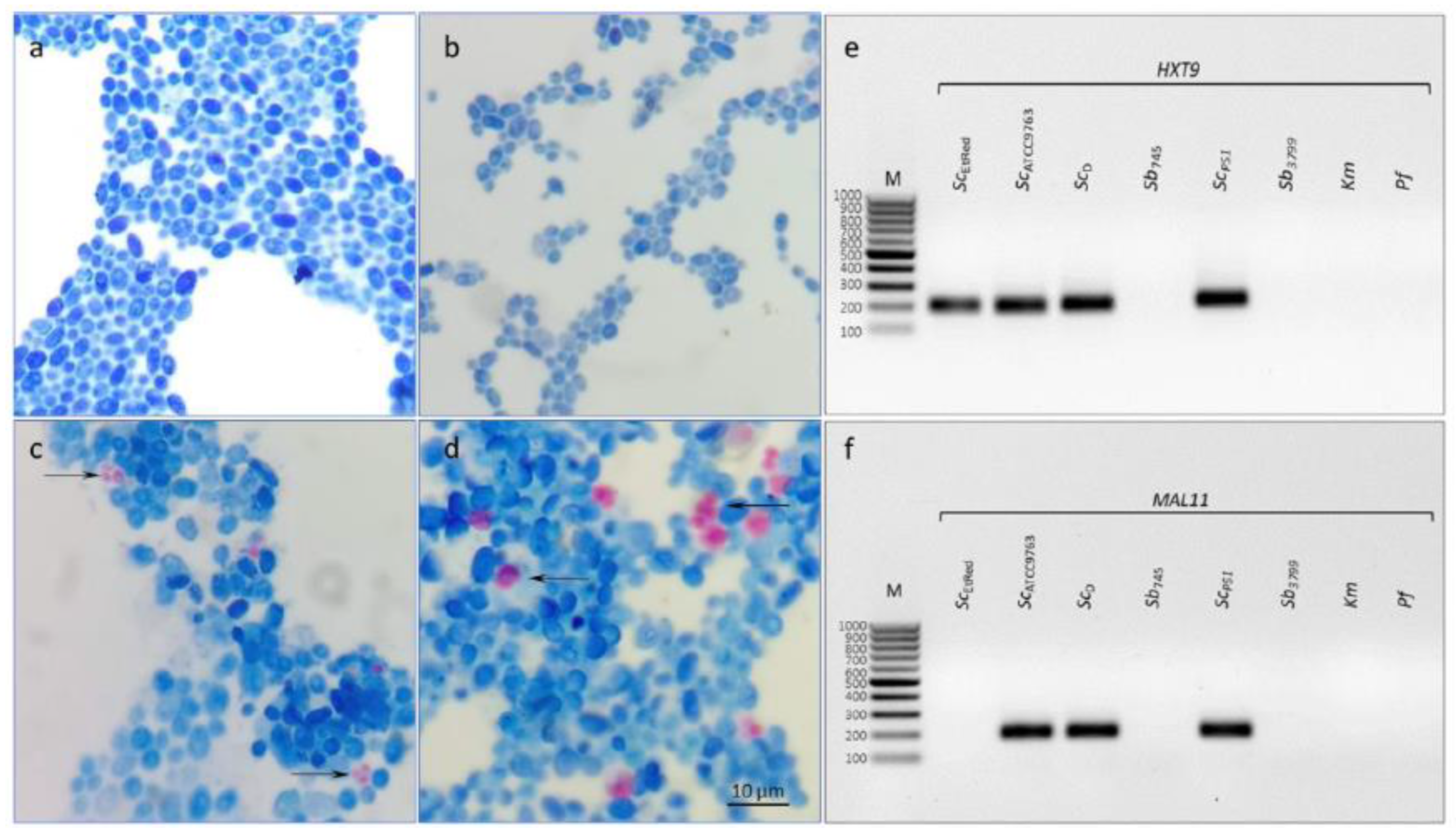
3.1. Isolation and Identification of Reference Strains Saccharomyces Cerevisiae var. boulardii
Two reference strains were identified with high confidence by MALDI-TOF mass spectrometry as
Saccharomyces cerevisiae. Ii was 2,16 and 2,09 for
Sb745 and
Sb3799, respectively. The reference strains did not show the ability to sporulate (
Figure 1a, b), as well as no amplification of regions
MAL11 and
HXT9 was detected for both (
Figure 1e, f).
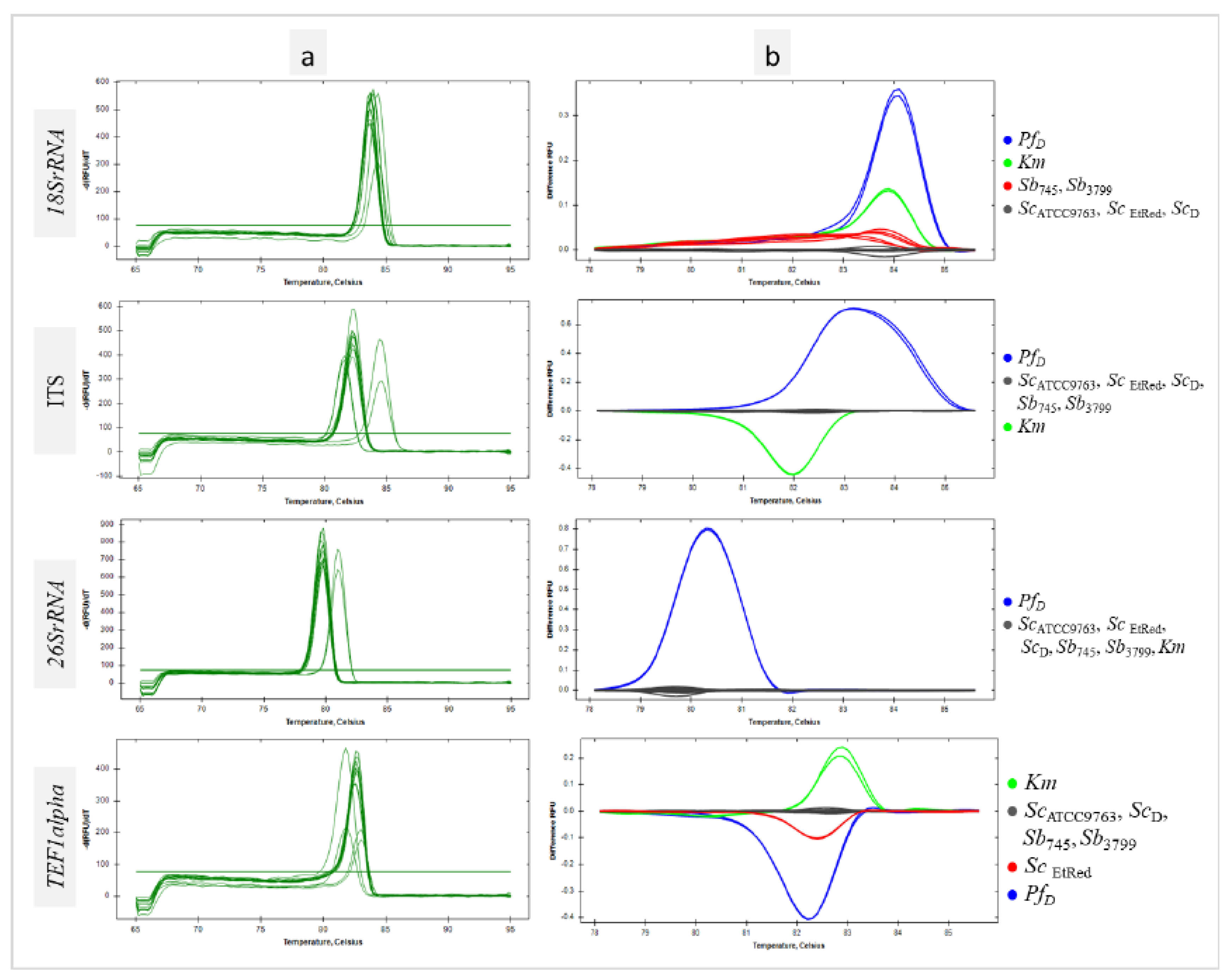
3.2. Differentiation of Saccharomyces Cerevisiae Strains Using Interspecies Primer Pairs in qPCR-HRM Analysis
For all sequences tested, a homogeneous product was obtained for each sample as evidenced by single peaks (
Figure 2a). HRM analysis of the
18SrRNA region amplicon showed individual clustering of
Sc and probiotic
Sb strains at the differentiation thresholds used. The other species used in the study, the
Km collection strain and the
PfD isolate were grouped separately (
Figure 2b
, 18SrRNA). The maximum RFU difference between the
Sb and
Sc clusters was 0,03, a slightly larger difference of 0,1 was between the
Sb and
Sc clusters combined and the
PfD cluster, and the largest difference of 0,3 was between the
Sb and
Sc clusters combined and the
Km cluster. For the non-coding ITS sequence, all strains belonging to
Sc species, including the probiotic ones, were in one cluster.
Km and
PfD control strains formed separate clusters (
Figure 2b, ITS). The maximum RFU difference was 0,6 between the
Sb/
Sc cluster and
Km. Slightly less, 0,4, was shown by comparing the
Sb/
Sc and
PfD clusters. (
Figure 2b, ITS). According to HRM analysis, the qPCR reaction with the primer pair for the
26SrRNA region yielded a uniform product for all
Sc strains as well as the
Km strain. In this case, only
PfD was grouped separately (
Figure 2b,
26SrRNA). Amplification of a region selected within
TEF1alpha resulted in a product whose denaturation profile was the same for all
Sc and
Sb tested except
ScEtRed. The
TEF1alpha region products obtained for the negative control strains were clustered individually (
Figure 2b,
TEF1alpha). The melting temperatures of the amplicons of each of the genomic DNA regions tested did not differ within the
Sc species significantly (
Table S1). HRM-based clustering of the selected sequences analyzed with DarWin software showed that the reference probiotic strains tested are genetically very similar and phylogenetically a variant of the species
Sc (
Figure S3).
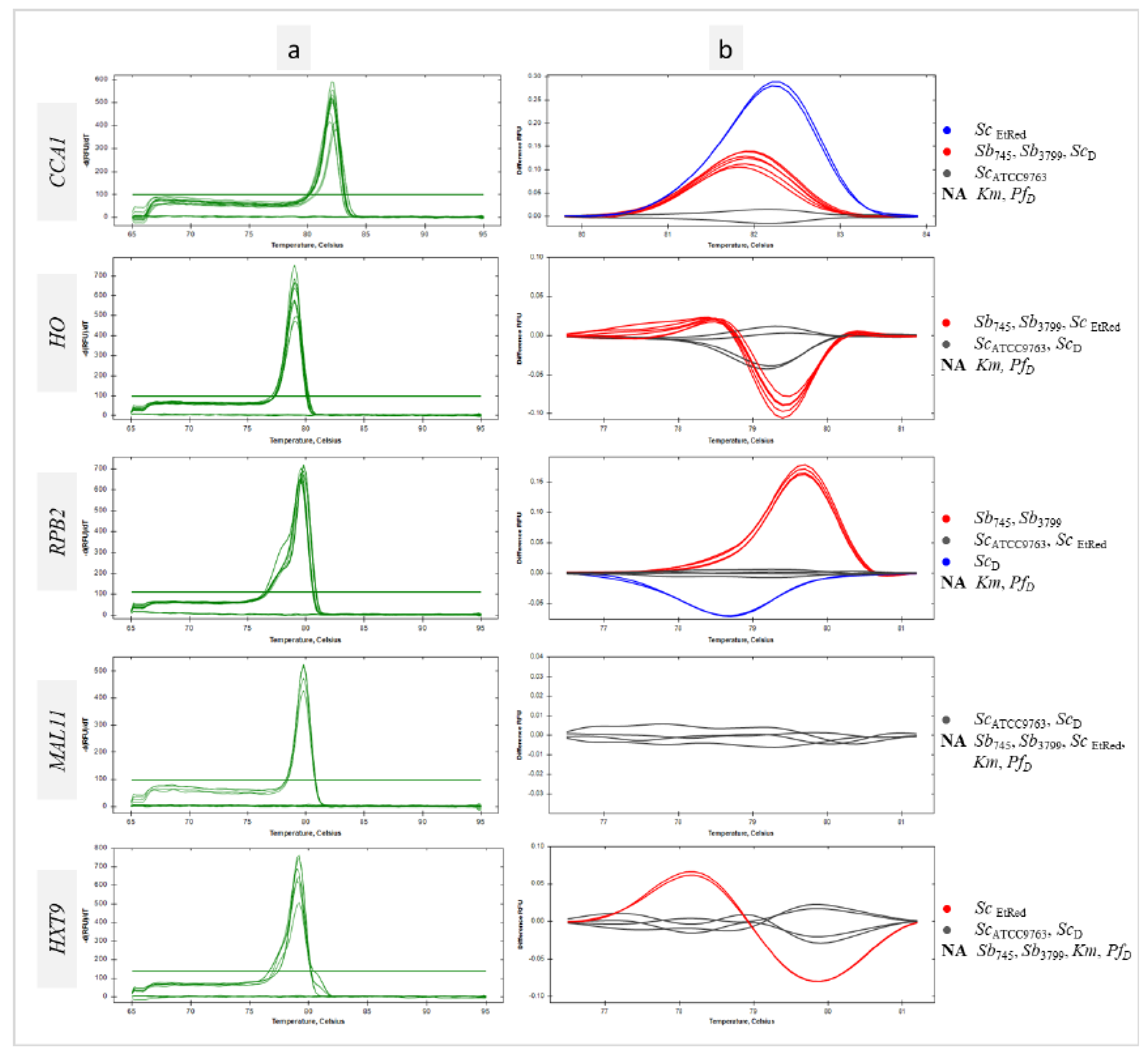
3.3. Differentiation of Saccharomyces Cerevisiae Strains Using Intragenus Primer Pairs in qPCR-HRM Analysis
No products were demonstrated for
Km and
PfD control templates using primer pairs designed for the genus
Saccharomyces (Figures 3 and S2). Furthermore, no amplification of the
MAL11 and
HXT9 regions was observed for the reference strains
Sb (
Figure 3,
MAL11,
HXT9,
Figure 1e, f). In addition, no sequence amplification in the
MAL11 region was detected for the
ScEtRed template (
Figure 3 MAL11,
Figure S2). For all sequences tested, a homogeneous product was obtained for the remaining samples as evidenced by single peaks (
Figure 3a). The peak melting curves of
RPB2 amplicons for
ScD had shoulders, indicative of polymorphisms in amplified sequence. HRM software grouped
Sb probiotic strains and
ScD isolate in one cluster at
CCA1 region analysis (
Figure 3 CCA1). A similar result was the joint clustering of probiotic strains and
ScEtRed regarding a melting temperature and profile of
HO sequence (
Figure 3 HO). The maximum RFU difference in the
HO melting profile was 0,1 between the
Sb745 cluster and the
ScATCC9763 cluster (
Figure 3b
HO). The amplicon obtained with the primer pair designed for
RPB2 was essential in the differentiation of the probiotic strains against the other
Sc tested in this study. The RFU difference between the clusters was 0,2 (
Figure 3b
RPB2). Significant differences were found in the melting temperatures of
CCA1 PCR products for all
Sc strains tested, including the two probiotic strains. Significantly the melting temperature of
RPB2 amplicon was the same for both
Sb strains and differed from the others by 0,2°C (
Table S1).
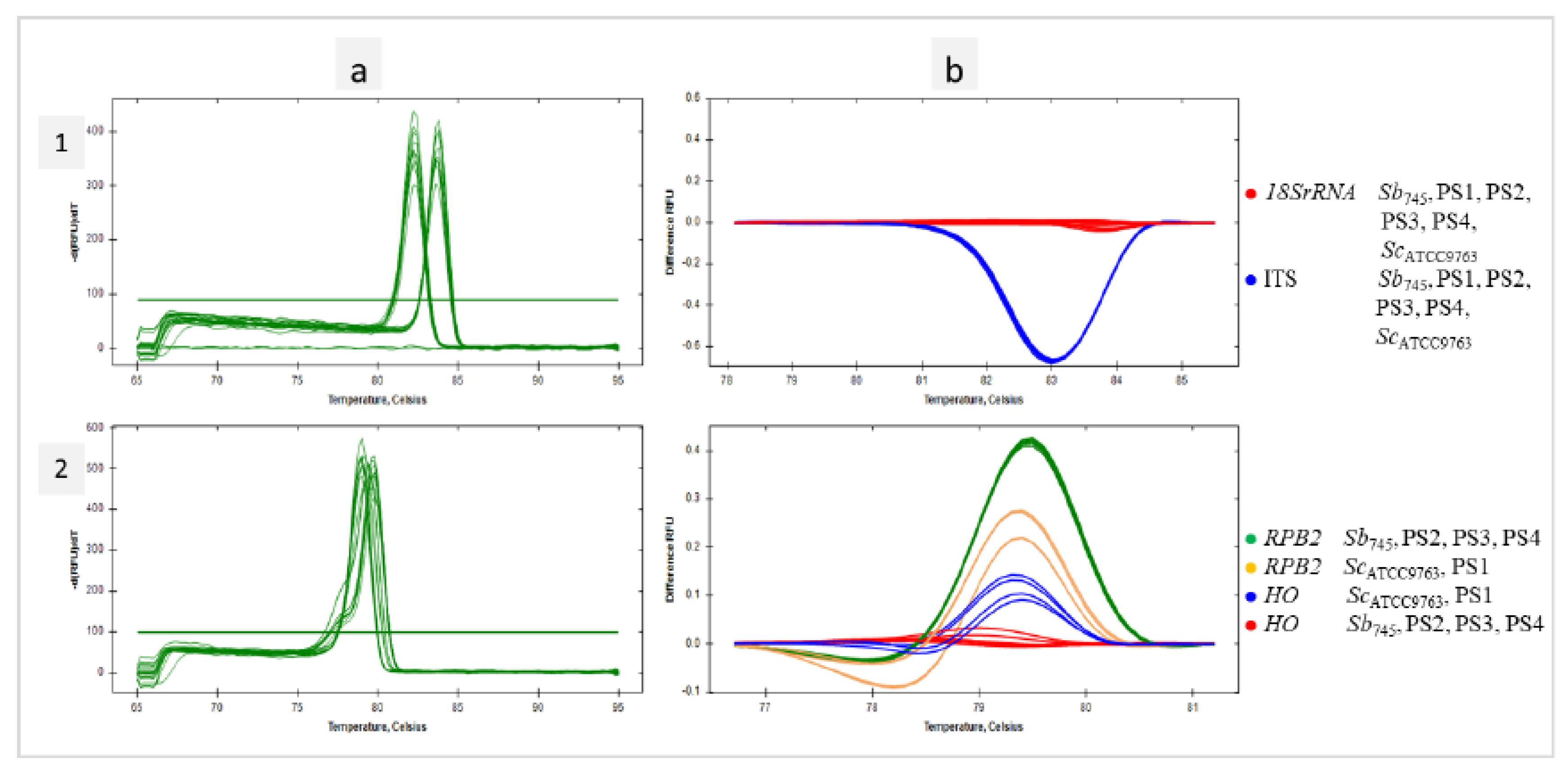
3.4. Identification of Probiotic Yeast in Dietary Supplements with qPCR-HRM Based on a Verified Set of Primer Pairs
In the present study, a genetic analysis of four probiotic supplements available in pharmacies was performed. Their composition is presented in Table 1. DNA extracts obtained from the preparations along with reference templates of
Sb745 and
ScATCC9763 were subjected to the qPCR reactions with the selected primer pairs. According to HRM analysis, the
18SrRNA sequences for each of the test samples were grouped with the references. The same effect was observed for the amplicons of the ITS region (
Figure 4.1a, b). If
Saccharomyces-specific primer pairs were used, only PR2, PR3 and PR4 samples were found to cluster with the
Sb745 reference for
HO sequences, as well as
RPB2. The PR1 sample remained in separate clusters for both studied sequences with
ScATCC9763 amplicons (
Figure 4.2a, b). PS1 strain was identified by MALDI-TOF mass spectrometry as
Saccharomyces cerevisiae with high confidence (
Ii was 2,15). The amplification of
MAL11 and
HXT9 regions on the DNA template of the PR1 supplement was found (
Figure 1e, f). In addition, the isolated strain of the PR1 supplement was able to sporulate (
Figure 1c).
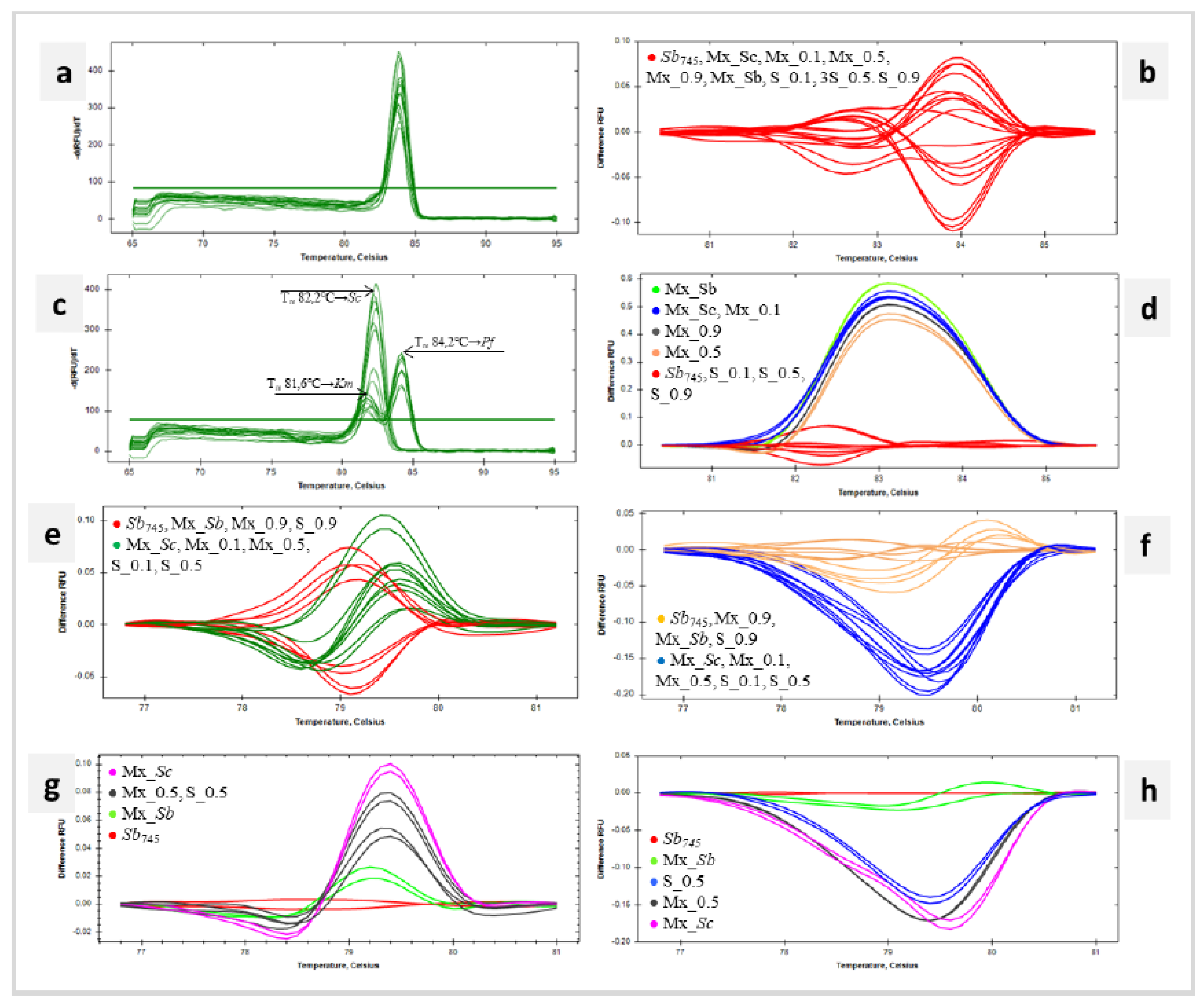
3.5. Identification of Probiotic Yeasts in Microbial Mixtures with qPCR-Based HRM Analysis
The mixed suspensions (Mx) for DNA isolation containing a fixed amount of
Ldsub
l (log 8 CFU mL
-1),
Km (3,8 log 6 CFU mL
-1) and
PfD (2 log 5 CFU mL
-1) cells were prepared. Successively, the suspensions Mx_Sc, Mx_0.1, Mx_0.5, Mx_0.9 and Mx_Sb contained increasing numbers of
Sb745 cells. In addition, suspensions S_0.1, S_0.5, and S_0.9 were prepared, containing only
ScD and
Sb745 cells, which were combined in analogous ratios to suspensions Mx (
Figure S1). DNA was extracted from the obtained cells’ pellets with a kit designed for foods. The resulting templates and
Sb745 positive control DNA were amplified using the primer pairs for
18SrRNA, ITS,
HO and
RPB2 regions. Considering the
18SrRNA sequence, the melting temperatures (T
m) of the amplicons obtained for the following samples were in the range of 83,80 – 84,0°C (
Figure 5a). In the same range remained the T
m of the
18SrRNA amplification products of the yeast species studied, namely
ScD,
Km and
PfD (
Table S1). The software grouped all tested samples into one cluster (
Figure 5b). In HRM analysis of ITS sequences, more clusters were obtained (
Figure 5d). The first grouped the reference strain
Sb745 and mixtures with
Sb745 and
ScD (S_0.1, S_0.5 i S_0.9). In this cluster were amplicons with a T
m of 82,4°C (
Figure 5c), which corresponded to the melting point of the ITS product for
ScD (
Table S1). The remaining clusters included samples containing
Km,
PfD,
ScD and
Sb745 in different proportions (Figure 5d). These amplification products showed double peaks with T
m of 81,6°C and 84,2°C. (
Figure 5c). The T
m indicated in the figure corresponded to the melting temperatures of ITS sequences of
Km and
PfD species, respectively (
Table S1). In each case, the product with a melting point corresponding to
PfD was quantitatively predominant. Exploring the
HO sequence, the Mx_Sb, Mx_0.9 and S_0.9 samples were clustered with the positive control in the HRM analysis. The remaining Mx_Sc, Mx_0.1, Mx_0.5, S_0.1, and S_0.5 were grouped in a separate cluster (
Figure 5e). The same result was obtained for HRM analysis of
RPB2 amplicons (
Figure 5f). For both marker sequences, a gradational distribution of differential melting curves was observed depending on the initial
Sb745 cell content of the sample. The smaller the proportion of
Sb745 cells in the suspension mix, the further the differential melting curve was from the reference curve. Regarding the adopted clustering parameters, the denaturation curve profiles of Mx_0.5 and S_0.5 products were significantly different from the
Sb745 reference, as shown in the detailed graphs (
Figure 5g, h). The melting point of the
HO product of all the mixtures tested was 79,0°C, which is consistent with
Table S1 data on
HO T
m for
ScD and
Sb745 samples. T
m of
RBP2 products amplified with
Sb745 and Mx_Sb DNA templates was 79,8°C and corresponded to
Sb745 amplicons.
RPB2 amplification products of the other mixed samples achieved T
m of 79,4 – 79,6°C, in which range the melting point of the
ScD sequence falls (
Table S1).
4. Discussion
In the early 2000s molecular data obtained by DNA analyses, including rDNA sequences, strongly indicated a close relatedness of Sb to Sc and thereby supported the recognition of Sb as a subtype of Sc, not as a separate species regardless of some genotypic differences (Van Der Aa Kühle and Jespersen 2003; Edwards-Ingram et al. 2007; Fietto et al. 2004; Posteraro et al. 2005). The comparative genomic research demonstrated that Sb and Sc share more than 99% genomic relatedness as determined by Average Nucleotide identity (ANI). The results revealed that the Sb probiotic strains are closer to wine strains of Sc than industrial or baking strains (Khatri et al. 2017). In the present work, a dendrogram was generated based on the clustering obtained in the HRM analysis. The clustering was conducted based on the melt curves progression obtained for the four analyzed regions: 18SrRNA, 26SrRNA, ITS and TEF1alpha. All Sc strains were grouped and reference probiotic Sb strains created a separate sub-cluster within as indicated.
Since the health-promoting effects of Sb are strain-dependent, the preparations referred to as probiotics must include strains with appropriate phenotypic and genetic characteristics. The purpose of this study was to develop a qPCR-HRM-based analysis to detect and identify Sb in probiotic-fortified foods. Initially, two probiotic strains described in detail by the manufacturers were isolated to serve as references. The specific features such as the non-sporulation phenotype and absence of selected genes, including MAL11 and HXT9, confirmed the strains to be probiotic subtypes. After prolonged incubation of Sb on a sporulation medium some changes in cell morphology were observed. Cells appeared enlarged and highly granular. An analogous effect was observed earlier, presumed to be the result of the initiation of the sporulation process combined with the inability to complete meiosis (Edwards-Ingram et al. 2007).
Reviewing the melting temperatures of each rDNA amplicon, there were no significant differences between the Sc and Sb strains tested. A pair of primers designed for the 18SrRNA region tested on pure cultures enabled the clustering of subtype Sb outside the Sc cluster. HRM analysis results concerning 26SrRNA and ITS sequences resulted in joint clustering of Sb and Sc strains. In comparative genomic studies, the sequence ITS1-5.8S rDNA-ITS2 of Sb displayed some subtle differences and 99% resemblance. As regards the sequence of the D1/D2 domain of the 26SrRNA had a similarity value of 100% compared to the Sc genomes. The probability of constructing an effective differentiation sequence within rDNA is very low, with the scarcity of polymorphisms and the strict requirements for HRM analysis (Van Der Aa Kühle and Jespersen 2003; Fietto et al. 2004; Posteraro et al. 2005). The problematic rDNA regions were used in attempts to differentiate species of the genus Saccharomyces using HRM analysis. Four primer combinations were designed spanning the 26SrRNA polymorphic region of 10 Saccharomyces species aligned. The highest discrimination level was achieved with five clusters for 10 type strains examined (Nadai et al. 2018). The combination of 26SrRNA and ITS regions in another HRM analysis of Saccharomyces species enabled discrimination of the most studied yeasts at the species level. However, differentiation of Sc, Sb and Saccharomyces uvarum in the pure cultures or the mixed samples failed (Bazalová et al. 2022).
Genes encoding translation elongation factor (TEF1alpha), actin (ACT1), RNA polymerase subunit (RPB1) and COX2, were stated to be more adequate in correct differentiation of the problematic/closely related species (Cappello et al. 2010; Eizaguirre et al. 2018; Hulin et al. 2014). Moreover, TEF1alpha sequencing and MALDI-TOF mass-spectrometry were found more relevant for differentiation within the Pichia cactophila clade, than sequencing of standard barcoding regions ITS and D1/D2 (Guitard et al. 2015). Nowadays, Sc promoter regions were found not to be fully conserved, in terms of nucleotide sequence nor predicted transcription factor (TF) binding sites, in homolog Sb genes. Some of the differentially expressed genes in Sb strains were found to have gained or lost TF binding sites in their promoter regions (Pais et al. 2021). Therefore, a pair of primers for the yeast polymorphic region within TEF1alpha was included in this work. It was effective in interspecies discrimination while showing a very low degree of intraspecies differentiation. Only the ScEtRed strain remained in a distinct cluster from the other Sc and Sb strains tested.
As intended, the primer pairs designed for this work for regions within CCA1, HO, RPB2, HXT9 and MAL11 were not amplified in species other than Sc. Amplification of HXT9 and MAL11 sequences was found only in Sc strains. The exception was ScEtRed, which showed no MAL11 amplification. ScEtRed closely relates to wine strains therefore it might show high similarities to Sb (Gronchi et al. 2022). MLST (Multilocus Sequence Typing) involved the sequencing of four nuclear genes CCA1, CYT1 (ubiquinol-cytochrome-c reductase catalytic subunit gene), HMX1 (heme oxygenase gene), NUP116 (FG-nucleoporin gene) and ITS region resulted in the uniform clade of clinical isolates and commercial probiotic yeasts (Imre et al. 2019). In this study, a primers pair designed for the CCA1 sequence was not sufficient to provide differentiation of probiotic strains. In contrast, the expected effect was found for the RPB2 sequence. Moreover, HRM analysis with RPB2 amplicon effectively clustered separately newly isolated Saccharomyces paradoxus strain (data not shown). HO sequence clustered Sb strains with ScEtRed indicating the close affinity of Sb and Sc wine strains.
In further research, 18S rRNA and ITS sequences were selected for qPCR-HRM analysis of collected dietary supplements. Comparative analysis with rDNA regions confirmed the presence of Sc strains in the dietary supplements tested. Moreover, the selected intraspecies sequences, HO and RPB2, confirmed the presence of Sb in PS2, PS3, PS4, and Sc in PS1, according to their composition as declared by the manufacturers. The preparations did not include detailed specifications of the yeast strains declared. As established, MAL11, MAL13 (transcription factor gene), and ARN2 (siderophore transporter gene) were present in more than 70% of the strains of different subgroups of Sc strains but were absent in all the probiotic strains. The large hexose transporter family comprises HXT11 and HXT9 which were absent from all strains of Sb (Khatri et al. 2017). Therefore, HXT9 and MAL11 sequences were amplified on PS2 - PS4 templates. As a result, no amplification products of the MAL11 region were confirmed, while only for PS2 the HXT9 amplicon was not detected. Thus, the strains included in the examined supplements except PS2, do not completely correspond to those with health-promoting properties.
HRM analysis of DNA extracted from seed mixtures showed reduced sensitivity detection of the selected template compared to the analysis of mixed DNA samples (Emenyeonu et al. 2018). The yield of bacterial DNA from sourdough fermented with different strains of the same species may differ up to 100,000-fold even if the organisms are present at the same cell counts (Zheng et al. 2015). The observations support the assumption that the structure of the food matrix could lower the recovery of the nuclear or organellar DNA. It was declared that qPCR-HRM analysis detects but does not identify organisms if they account for 0.1–1% of the bacterial and yeast population, respectively (Lin and Gänzle 2014). Therefore, the effectiveness of selected sequences in identifying probiotic strains was tested in mixtures containing three yeast species. Mixtures were prepared that microbiologically corresponded in composition to kefirs (Goktas et al. 2021; Kalamaki and Angelidis 2017; Nejati et al. 2020; Özer and Kirmaci 2014; Wang et al. 2008). The application of commercial kefirs in the study failed. The reason was the very low yeast content, no more than log 2 CFU mL-1 of cells was determined in commercial products (data not shown). Due to very small differences in melting temperature (Tm) and melting profile of 18SrRNA amplicons for Sb745, ScD, Km and PfD all mix-yeast samples were clustered together. If the ITS sequence was considered, common clustering of samples containing only strains of Sc species was observed (Sb745 and ScD). In contrast, the other samples with additional Km and PfD presence formed several separate clusters. ITS region was the most variable in DNA primary structure amongst analyzed rDNA sequences. Consequently, it has the highest strength of interspecies differentiation in the HRM analysis. Amplification with a pair of ITS primers on DNA extracted from mixtures yielded heterogeneous products as a result of the increased affinity of the primers for the Pf sequence, as a consequence of nucleotide changes in the annealing regions of the Sc and Km sequences (Borkowska and Celińska 2023).
The designed intragenus primer pairs for
HO or
RPB2 regions were the best in the identification of
Sb in the mixtures. The
RPB2 sequence had the highest differentiation power. The reference probiotic strain was detectable and identifiable by qPCR-HRM when the mixture contained at least log 7 CFU mL
-1 of
Sb cells, and
Sb745 significantly exceeded the
ScD strain quantitatively.
Figure 5h shows that the presence of other yeast species does not significantly change the product melting profile for M_Sb. In contrast, a reduction in the number of
Sb745 cells to
ScD resulted in a significant increase in difference RFU of 0,2 to the reference. The limits for accurate quantification of
Sc in wine artificially contaminated, with real-time PCR using specific primers, were established for log 5 CFU mL
-1 in sweet wine and log 6 CFU mL
-1 in red wine (Martorell et al. 2005). Such high detection thresholds by qPCR indicate that the stated threshold for identification of
Sb cells using qPCR-HRM analysis is plausible. A required minimum dose of
health-boosting microorganisms is log 6 CFU mL
-1 or CFU g
-1 for the food product to be labelled as a probiotic (White and Hekmat 2018). Since the viability of microorganisms is the key to achieving health benefits (log 6 – log 7 CFU per g during the expected shelf-life of the probiotic food or beverage, according to WHO/FAO, 2006), some researchers even suggest increasing the dose up to log 7 CFU mL
-1 or CFU g
-1 (Fiocco et al. 2020). This is sufficient enough to detect and identify the probiotic strain of
Sb yeast in probiotic-enriched foods with qPCR-HRM analysis using the
RPB2 marker.
5. Conclusions
qPCR-HRM analysis using interspecies 18SrRNA and ITS sequences optimized in culture-dependent analysis identified Sb at the species level, while intraspecies HO and RPB2 sequences at the variety level in single-yeast dietary supplements. Enhancement of S. cerevisiae var. boulardii differentiation was achieved by amplification of HXT9 and MAL11 regions with presence-absence variation. The low variability in sequences amplified by interspecies primer pairs in qPCR-HRM analysis prevented the differentiation of Sb in mixtures of three yeast species. In contrast, the identification of Sb using designed intragenus primer pairs was successful in Sc mixtures and mixtures with kefir microbial composition. The RPB2 sequence showed the highest intraspecies differentiation power. However, qPCR-HRM analysis identified Sb only with the variety predominance in the microbial mix. Therefore, the presented qPCR-HRM analysis can be considered an appropriate tool for identifying Sb in probiotic-enriched food matrices.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Monika Borkowska, Michał Kułakowski and Kamila Myszka. The first draft of the manuscript was written by Monika Borkowska and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a subvention from the PULS fund (number 506.771.03.00) received from the Ministry of Science and Higher Education.
Data Availability statement
The datasets generated during the current study are available from the corresponding author upon request.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
References
- Baleiras-Couto MM, Eijsma B, Hofstra H, Huis In’T Veld JH, Van Der Vossen JM (1996) Evaluation of Molecular Typing Techniques to Assign Genetic Diversity among Saccharomyces Cerevisiae Strains. Applied and Environmental Microbiology 62(1), 41 – 46. DOI: 10.1128/aem.62.1.41-46.1996.
- Bazalová O, Cihlář JZ, Dlouhá Z, Bár L, Dráb V, Kavková M (2022) Rapid Sourdough Yeast Identification Using Panfungal PCR Combined with High Resolution Melting Analysis. Journal of Microbiological Methods 199. DOI: 10.1016/j.mimet.2022.106522.
- Borkowska M, Celińska E (2023) Multiple Region High Resolution Melting-Based Method for Accurate Differentiation of Food-Derived Yeasts at Species Level Resolution. Food Microbiology 109. DOI: 10.1016/j.fm.2022.104120.
- Cappello MS, Poltronieri P, Blaiotta G, Zacheo G (2010) Molecular and Physiological Characteristics of a Grape Yeast Strain Containing Atypical Genetic Material. International Journal of Food Microbiology 144(1), 72 – 80. DOI: 10.1016/j.ijfoodmicro.2010.08.013.
- Cardinali G, Martini A (1994) Electrophoretic Karyotypes of Authentic Strains of the Sensu Strict0 Group of the Genus Saccharomyces? International Journal of Systematic Bacteriology 44(4), 791 – 797. DOI: 10.1099/00207713-44-4-791.
- Cesaro S, Chinello P, Rossi L, Zanesco L (2000) Saccharomyces Cerevisiae Fungemia in a Neutropenic Patient Treated with Saccharomyces Boulardii. Supportive Care in Cancer 8(6), 504 – 5. DOI: 10.1007/s005200000123.
- Chan MZA, Tan LT, Heng SWQ, Liu SQ (2023) Effect of Co-Fermentation of Saccharomyces Boulardii CNCM-I745 with Four Different Probiotic Lactobacilli in Coffee Brews on Cell Viabilities and Metabolic Activities. Fermentation 9(3). DOI: 10.3390/fermentation9030219.
- Czerucka D, Piche T, Rampal P (2007) Review article: Yeast as Probiotics - Saccharomyces Boulardii. Alimentary Pharmacology and Therapeutics 26(6), 767 – 78. DOI: 10.1111/j.1365-2036.2007.03442.x.
- Değirmencioğlu N, Gurbuz O, Şahan Y (2016) The Monitoring, Via an In Vitro Digestion System, of the Bioactive Content of Vegetable Juice Fermented with Saccharomyces Cerevisiae and Saccharomyces Boulardii. Journal of Food Processing and Preservation 40(4), 798 – 811. DOI: 10.1111/jfpp.12704.
- Edwards-Ingram L, Gitsham P, Burton N, Warhurst G, Clarke I, Hoyle D, Oliver SG, Stateva L (2007) Genotypic and Physiological Characterization of Saccharomyces Boulardii, the Probiotic Strain of Saccharomyces Cerevisiae. Applied and Environmental Microbiology 73(8), 2458 – 67. DOI: 10.1128/AEM.02201-06.
- Eizaguirre JI, Peris D, Rodríguez ME, Lopes CA, De Los Ríos P, Hittinger CT, Libkind D (2018) Phylogeography of the Wild Lager-Brewing Ancestor (Saccharomyces Eubayanus) in Patagonia. Environmental Microbiology 20(10), 3732 – 43. DOI: 10.1111/1462-2920.14375.
- Ellouze O, Berthoud V, Mervant M, Parthiot JP, Girard C (2016) Septic Shock Due to Saccharomyces Boulardii. Medecine et Maladies Infectieuses 46(2), 104 – 5. DOI: 10.1016/j.medmal.2015.12.003.
- Emenyeonu LC, Croxford AE, Wilkinson MJ (2018) The Potential of Aerosol EDNA Sampling for the Characterisation of Commercial Seed Lots. PLoS ONE 13(8). DOI: 10.1371/journal.pone.0201617.
- Erdem M, Kesmen Z, Özbekar E, Çetin B, Yetim H (2016) Application of High-Resolution Melting Analysis for Differentiation of Spoilage Yeasts. Journal of Microbiology 54(9), 618 – 25. DOI: 10.1007/s12275-016-6017-8.
- Fietto JLR, Araújo RS, Valadão FN, Fietto LG, Brandão RL, Neves MJ, Gomes FCO, Nicoli JR, Castro IM, (2004) Molecular and Physiological Comparisons between Saccharomyces Cerevisiae and Boulardii. Canadian Journal of Microbiology 50(8), 615 – 21. DOI: 10.1139/w04-050.
- Fijan S (2014) Microorganisms with Claimed Probiotic Properties: An Overview of Recent Literature. International Journal of Environmental Research and Public Health 11(5), 4745 – 67. DOI: 10.3390/ijerph110504745.
- Fiocco D, Longo A, Arena MP, Russo P, Spano G, Capozzi V (2020) How Probiotics Face Food Stress: They Get by with a Little Help. Critical Reviews in Food Science and Nutrition 60(9), 1552 – 80. DOI: 10.1080/10408398.2019.1580673.
- Goktas H, Dikmen H, Demirbas F, Sagdic O, Dertli E (2021) Characterisation of Probiotic Properties of Yeast Strains Isolated from Kefir Samples. International Journal of Dairy Technology 74(4), 715 – 22. DOI: 10.1111/1471-0307.12802.
- Gronchi N, De Bernardini N, Cripwell RA, Treu L, Campanaro S, Basaglia M, Foulquié-Moreno MR, Thevelein JM, Van Zyl WH, Favaro L, Casella S (2022) Natural Saccharomyces Cerevisiae Strain Reveals Peculiar Genomic Traits for Starch-to-Bioethanol Production: The Design of an Amylolytic Consolidated Bioprocessing Yeast. Frontiers in Microbiology 12. DOI: 10.3389/fmicb.2021.768562.
- Guitard J, Atanasova R, Brossas JY, Meyer I, Gits M, Marinach C, Vellaissamy S, Angoulvant A, Mazier D, Hennequin C (2015) Candida Inconspicua and Candida Norvegensis: New Insights into Identification in Relation to Sexual Reproduction and Genome Organization. Journal of Clinical Microbiology 53(5), 1655 – 61. DOI: 10.1128/JCM.02913-14.
- Gut AM, Vasiljevic T, Yeager T, Donkor ON (2019) Characterization of Yeasts Isolated from Traditional Kefir Grains for Potential Probiotic Properties. Journal of Functional Foods 58, 56 – 66. DOI: 10.1016/j.jff.2019.04.046.
- Hennequin C, Thierry A, Richard GF, Lecointre G, Nguyen HV, Gaillardin C, Dujon B (2001) Microsatellite Typing as a New Tool for Identification of Saccharomyces Cerevisiae Strains. Journal of Clinical Microbiology 39(2), 551 – 59. DOI: 10.1128/JCM.39.2.551-559.2001.
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME (2014) Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nature Reviews Gastroenterology and Hepatology 11(8), 506 – 14. DOI: 10.1038/nrgastro.2014.66.
- Hulin M, Harrison E, Stratford M, Wheals AE (2014) Rapid Identification of the Genus Dekkera/Brettanomyces, the Dekkera Subgroup and All Individual Species. International Journal of Food Microbiology 187, 7 – 14. DOI: 10.1016/j.ijfoodmicro.2014.06.028.
- Imre A, Rácz HV, Antunovics Z, Rádai Z, Kovács R, Lopandic K, Pócsi I, Pfliegler WP (2019) A New, Rapid Multiplex PCR Method Identifies Frequent Probiotic Origin among Clinical Saccharomyces Isolates. Microbiological Research 227. DOI: 10.1016/j.micres.2019.126298.
- Kalamaki MS, Angelidis AS (2017) Isolation and Molecular Identification of Yeasts in Greek Kefir. International Journal of Dairy Technology 70(2), 261 – 68. DOI: 10.1111/1471-0307.12329.
- Kara I, Yıldırım F, Özgen Ö, Erganiş S, Aydoğdu M, Dizbay M, Gürsel G, Kalkanci A (2018) Saccharomyces Cerevisiae Fungemia after Probiotic Treatment in an Intensive Care Unit Patient. J Mycol Med 28(1), 218 – 21. DOI: 10.1016/j.mycmed.2017.09.003.
- Karaolis C, Botsaris G, Pantelides I, Tsaltas D (2013) Potential Application of Saccharomyces Boulardii as a Probiotic in Goat’s Yoghurt: Survival and Organoleptic Effects. International Journal of Food Science and Technology 48(7), 1445 – 52. DOI: 10.1111/ijfs.12111.
- Kesmen Z, Özbekar E, Büyükkiraz ME (2018a) Multifragment Melting Analysis of Yeast Species Isolated from Spoiled Fruits. Journal of Applied Microbiology 124(2), 522–34. DOI: 10.1111/jam.13645.
- Kesmen Z, Büyükkiraz ME, Özbekar E, Çelik M, Özkök FÖ, Kılıç Ö, Çetin B, Yetim H (2018b) Assessment of Multi Fragment Melting Analysis System (MFMAS) for the Identification of Food-Borne Yeasts. Current Microbiology 75(6), 716 – 25. DOI: 10.1007/s00284-018-1437-9.
- Khatri I, Tomar R, Ganesan K, Prasad GS, Subramanian S (2017) Complete Genome Sequence and Comparative Genomics of the Probiotic Yeast Saccharomyces Boulardii. Scientific Reports 7(1). DOI: 10.1038/s41598-017-00414-2.
- Kurtzman CP, Robnett CJ (1998) Identification and Phylogeny of Ascomycetous Yeasts from Analysis of Nuclear Large Subunit (26S) Ribosomal DNA Partial Sequences. Antonie Van Leeuwenhoek 73(4), 331 – 71. DOI: 10.1023/a:1001761008817.
- Lin XB, Gänzle MG (2014) Quantitative High-Resolution Melting PCR Analysis for Monitoring of Fermentation Microbiota in Sourdough. International Journal of Food Microbiology 186, 42 – 48. DOI: 10.1016/j.ijfoodmicro.2014.06.010.
- Łukaszewicz M (2012) Saccharomyces Cerevisiae Var. Boulardii – Probiotic Yeast. Probiotics. InTech Press, US. DOI: 10.5772/50105.
- Martorell P, Querol A, Fernández-Espinar MT (2005) Rapid Identification and Enumeration of Saccharomyces Cerevisiae Cells in Wine by Real-Time PCR. Applied and Environmental Microbiology 71(11), 6823 – 30. DOI: 10.1128/AEM.71.11.6823-6830.2005.
- McCullough MJ, Clemons KV, McCusker JH, Stevens DA (1998) Species Identification and Virulence Attributes of Saccharomyces Boulardii (Nom. Inval.). Journal of Clinical Microbiology 36(9), 2613 – 2617. DOI: 10.1128/JCM.36.9.2613-2617.1998.
- McFarland LV (1996) Saccharomyces boulardii is not Saccharomyces cerevisiae. Clin. Infect. Dis. 22, 200 – 201. DOI: 10.1093/clinids/22.1.200.
- Mitterdorfer G, Mayer HK, Kneifel W, Viernstein H (2002) Clustering of Saccharomyces Boulardii Strains within the Species S. Cerevisiae Using Molecular Typing Techniques. J Appl Microbiol. 93(4), 521 – 30. DOI: 10.1046/j.1365-2672.2002.01710.x.
- Molnar O, Messner R, Prillinger H, Stahl U, Slavikova E (1995) Genotypic Identification of Saccharomyces Species Using Random Amplified Polymorphic DNA Analysis. Systematic and Applied Microbiology 18(1), 136 – 45. DOI: 10.1016/S0723-2020(11)80461-3.
- Nadai C, Bovo B, Giacomini A, Corich V (2018) New Rapid PCR Protocol Based on High-Resolution Melting Analysis to Identify Saccharomyces Cerevisiae and Other Species within Its Genus. Journal of Applied Microbiology 124(5), 1232 – 42. DOI: 10.1111/jam.13709.
- Nejati F, Junne S, Kurreck J, Neubauer P (2020) Quantification of Major Bacteria and Yeast Species in Kefir Consortia by Multiplex TaqMan QPCR. Frontiers in Microbiology 11. DOI: 10.3389/fmicb.2020.01291.
- Özer B, Kirmaci HA (2014) Fermented Milks: Products of Eastern Europe and Asia. Encyclopedia of Food Microbiology: Second Edition, 900 – 907. Elsevier Inc. DOI: 10.1016/B978-0-12-384730-0.00123-3.
- Pais P, Oliveira J, Almeida V, Yilmaz M, Monteiro PT, Teixeira MC (2021) Transcriptome-Wide Differences between Saccharomyces Cerevisiae and Saccharomyces Cerevisiae Var. Boulardii: Clues on Host Survival and Probiotic Activity Based on Promoter Sequence Variability. Genomics 113(2), 530 – 39. DOI: 10.1016/j.ygeno.2020.11.034.
- Posteraro B, Sanguinetti M, Romano L, Torelli R, Novarese L, Fadda G (2005) Molecular Tools for Differentiating Probiotic and Clinical Strains of Saccharomyces Cerevisiae. International Journal of Food Microbiology 103(3), 295 – 304. DOI: 10.1016/j.ijfoodmicro.2004.12.031.
- Rekha CR, Vijayalakshmi G (2010) Bioconversion of Isoflavone Glycosides to Aglycones, Mineral Bioavailability and Vitamin B Complex in Fermented Soymilk by Probiotic Bacteria and Yeast. Journal of Applied Microbiology 109(4), 1198 – 1208. DOI: 10.1111/j.1365-2672.2010.04745.x.
- Ripari V, Gänzle MG, Berardi E (2016) Evolution of Sourdough Microbiota in Spontaneous Sourdoughs Started with Different Plant Materials. International Journal of Food Microbiology 232, 35 – 42. DOI: 10.1016/j.ijfoodmicro.2016.05.025.
- Sambrook J, Russell D (2001) Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press, New York.
- Santino L, Alari’ A, Bono S, Tetp E, Bernardini A, Magrini L, Di Somma S, Teggi A (2014) Saccharomyces cerevisiae fungemia, a possible consequence of the treatment of Clostridium difficile colitis with a probioticum. International Journal of Immunopathology and Pharmacology 27 (1), 143 – 146. DOI: 10.1177/039463201402700120.
- Thygesen JB, Glerup H, Tarp B (2012) Saccharomyces Boulardii Fungemia Caused by Treatment with a Probioticum. BMJ Case Reports. DOI: 10.1136/bcr.06.2011.4412.
- Valente P, Gouveia C, De Lemos GA, Pimentel ’ D, Van Elsas JD, Mendonqa-Hagler LC, Hagler AN (1996) PCR Amplification of the RDNA Internal Transcribed Spacer Region for Differentiation of Saccharomyces Cultures. FEMS Microbiol Lett. 137(2-3), 253 – 6. DOI: 10.1111/j.1574-6968.1996.tb08114.x.
- Van Der Aa Kühle A, Jespersen L (2003) The Taxonomic Position of Saccharomyces Boulardii as Evaluated by Sequence Analysis of the D1/D2 Domain of 26S RDNA, the ITS1-5.8S RDNA-ITS2 Region and the Mitochondrial Cytochrome-c Oxidase II Gene. Systematic and Applied Microbiology 26(4), 564 – 71. DOI: 10.1078/072320203770865873.
- Wang SY, Chen HC, Liu JR, Lin YC, Chen MJ (2008) Identification of Yeasts and Evaluation of Their Distribution in Taiwanese Kefir and Viili Starters. Journal of Dairy Science 91(10), 3798 – 3805. DOI: 10.3168/jds.2007-0468.
- White J, Hekmat S (2018) Development of Probiotic Fruit Juices Using Lactobacillus Rhamnosus GR-1 Fortified with Short Chain and Long Chain Inulin Fiber. Fermentation 4(2). DOI: 10.3390/fermentation4020027.
- Zamora-Vega R, Montañez-Soto JL, Martínez-Flores HE, Flores-Magallón R, Muñoz-Ruiz CV, Venegas-González J, De Jesús Ariza Ortega T (2012) Effect of Incorporating Prebiotics in Coating Materials for the Microencapsulation of Sacharomyces Boulardii. International Journal of Food Sciences and Nutrition 63(8), 930 – 35. DOI: 10.3109/09637486.2012.687364.
- Zheng J, Ruan L, Sun M, Gänzle M (2015) A Genomic View of Lactobacilli and Pediococci Demonstrates That Phylogeny Matches Ecology and Physiology. Applied and Environmental Microbiology 81(20), 7233 – 43. DOI: 10.1128/AEM.02116-15.
Figure 1.
Verification of sporulation and HXT9 or MAL11 presence in Sc strains. Microscopic images of yeast cells of Sb745 (a), Sb3799 (b), ScPS1 (c), ScD (d) strains. The cells’ suspensions were induced to sporulate under starvation conditions and then stained with the Ziehl-Neelsen method. Electrophoretic separation of HXT9 (e) and MALL1 (f) amplicons obtained in PCR for S. cerevisiae var. boulardii reference strains (Sb745, Sb3799), S. cerevisiae strains (ScATCC9763, ScEtRed, ScD and ScPS1), K. marxianus (Km) and P. fermentans (Pf). M – DNA Marker 100bp LOAD (Syngen Biotech, Wroclaw, Poland).
Figure 1.
Verification of sporulation and HXT9 or MAL11 presence in Sc strains. Microscopic images of yeast cells of Sb745 (a), Sb3799 (b), ScPS1 (c), ScD (d) strains. The cells’ suspensions were induced to sporulate under starvation conditions and then stained with the Ziehl-Neelsen method. Electrophoretic separation of HXT9 (e) and MALL1 (f) amplicons obtained in PCR for S. cerevisiae var. boulardii reference strains (Sb745, Sb3799), S. cerevisiae strains (ScATCC9763, ScEtRed, ScD and ScPS1), K. marxianus (Km) and P. fermentans (Pf). M – DNA Marker 100bp LOAD (Syngen Biotech, Wroclaw, Poland).
Figure 2.
Differentiation of yeast with interspecies primer pairs in qPCR-HRM analysis. Melt peaks (a) and difference curves grouped as colour-marked clusters (b) were obtained by qPCR-HRM analysis of 18SrRNA, ITS, 26SrRNA, and TEF1alpha regions. DNA templates of S. cerevisiae var. boulardii reference strains (Sb745, Sb3799), S. cerevisiae strains (ScATCC9763, ScEtRed, ScD), K. marxianus (Km) and P. fermentans (PfD) strains were amplified in technical duplicate.
Figure 2.
Differentiation of yeast with interspecies primer pairs in qPCR-HRM analysis. Melt peaks (a) and difference curves grouped as colour-marked clusters (b) were obtained by qPCR-HRM analysis of 18SrRNA, ITS, 26SrRNA, and TEF1alpha regions. DNA templates of S. cerevisiae var. boulardii reference strains (Sb745, Sb3799), S. cerevisiae strains (ScATCC9763, ScEtRed, ScD), K. marxianus (Km) and P. fermentans (PfD) strains were amplified in technical duplicate.
Figure 3.
Differentiation of Sc strains using intragenus primer pairs in qPCR-HRM analysis. Melt peaks (a) and difference curves grouped as colour-marked clusters (b) were obtained by qPCR-HRM analysis of CCA1, HO, RPB2, MAL11 and HXT9 regions. DNA templates of S. cerevisiae var. boulardii reference strains (Sb745, Sb3799), S. cerevisiae strains (ScATCC9763, ScEtRed, ScD), K. marxianus (Km) and P. fermentans (PfD) strains were amplified in technical duplicate. NA – not amplified.
Figure 3.
Differentiation of Sc strains using intragenus primer pairs in qPCR-HRM analysis. Melt peaks (a) and difference curves grouped as colour-marked clusters (b) were obtained by qPCR-HRM analysis of CCA1, HO, RPB2, MAL11 and HXT9 regions. DNA templates of S. cerevisiae var. boulardii reference strains (Sb745, Sb3799), S. cerevisiae strains (ScATCC9763, ScEtRed, ScD), K. marxianus (Km) and P. fermentans (PfD) strains were amplified in technical duplicate. NA – not amplified.
Figure 4.
Identification of Sb in dietary supplements with qPCR-HRM. Melt peaks (a) and difference curves grouped as colour-marked clusters (b) were obtained by qPCR-HRM analysis of 18SrRNA and ITS regions (1), HO and RPB2 regions (2). DNA template of S. cerevisiae var. boulardii reference strain (Sb745) as positive control and the dietary supplements’ DNA templates (PS1, PS2, PS3, PS4) were amplified in technical duplicate.
Figure 4.
Identification of Sb in dietary supplements with qPCR-HRM. Melt peaks (a) and difference curves grouped as colour-marked clusters (b) were obtained by qPCR-HRM analysis of 18SrRNA and ITS regions (1), HO and RPB2 regions (2). DNA template of S. cerevisiae var. boulardii reference strain (Sb745) as positive control and the dietary supplements’ DNA templates (PS1, PS2, PS3, PS4) were amplified in technical duplicate.
Figure 5.
Identification of Sb745 in microbial mixtures with qPCR-based HRM analysis. Melt peaks (a) and difference curves grouped as colour-marked clusters (b) were obtained by qPCR-HRM analysis of 18SrRNA. Melt peaks (c) – the arrows indicate species-specific peaks at Tm 81,6℃, 82,2℃ and 84,2℃ for Km, ScD and PfD respectively and difference curves (d) grouped as colour-marked clusters detected for ITS amplicon. Difference curves grouped as colour-marked clusters obtained by qPCR-HRM analysis of HO sequence presented for all samples (e) and selected samples (f), and RPB2 sequence presented for all samples (g) and selected samples (h). DNA templates of S. cerevisiae var. boulardii reference strain (Sb745) as positive control and the microbial mixtures’ DNA extracts (Mx_Sb, Mx_0.9, Mx_0.5, Mx_0.1, Mx_Sc, S_0.9, S_0.5, S_0.1) were amplified in technical duplicate.
Figure 5.
Identification of Sb745 in microbial mixtures with qPCR-based HRM analysis. Melt peaks (a) and difference curves grouped as colour-marked clusters (b) were obtained by qPCR-HRM analysis of 18SrRNA. Melt peaks (c) – the arrows indicate species-specific peaks at Tm 81,6℃, 82,2℃ and 84,2℃ for Km, ScD and PfD respectively and difference curves (d) grouped as colour-marked clusters detected for ITS amplicon. Difference curves grouped as colour-marked clusters obtained by qPCR-HRM analysis of HO sequence presented for all samples (e) and selected samples (f), and RPB2 sequence presented for all samples (g) and selected samples (h). DNA templates of S. cerevisiae var. boulardii reference strain (Sb745) as positive control and the microbial mixtures’ DNA extracts (Mx_Sb, Mx_0.9, Mx_0.5, Mx_0.1, Mx_Sc, S_0.9, S_0.5, S_0.1) were amplified in technical duplicate.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).