1. Introduction
Throughout the COVID-19 pandemic, regular surveillance of newly introduced SARS-CoV-2 variants allowed for better understanding of disease transmission patterns and ultimately better preparedness for possible outbreaks and surges [
1]. The emergence of new variants stems from SARS-CoV-2 replication within its hosts over successive cycles, leading to the accumulation of numerous mutations, to include, but not limited to the receptor-binding domain [
2,
3]. Collectively, these mutations in the genetic makeup of SARS-CoV-2 provide fitness advantages such as improved transmission, infectivity, different tropism, modulated virulence, and evasion of the immune response triggered by vaccination or prior infection [
4]. When evidence emerged in November 2021 of B.1.1.529 SARS-CoV-2 Omicron variant transmission, Omicron was highlighted as a variant of concern by the World Health Organization (WHO) [
5]. When initial reports suggested that the Omicron variant was associated with increased transmissibility and potential immune evasion, an expansive international response of increased prevention, control and surveillance measures was triggered [
6], including focused attempts to confirm the presence of the variant in wastewater.
At the peak of the pandemic, wastewater surveillance proved to be a highly effective tool for early warning of increased community transmission of SARS-CoV-2, allowing for timely public health responses in areas with increasing concentrations of the virus in wastewater [
7,
8,
9]. The relevance of this surveillance method for detecting genetic variants of SARS-CoV-2 was also highlighted during the emergence of different variants including the Delta (B.1.617.2) variant in mid-2021 as well as subsequent variants in later periods [
10,
11,
12]. In general, variant-specific wastewater surveillance provided data for prevention and control approaches, and important knowledge for understanding the transmission dynamics of each variant. For the Omicron variant, its specific genetic characteristic comprises the ‘S-gene target failure’ (SGTF) – a mutation in the S-gene which resulted in a deletion of amino acids at position 69 and 70 [
13,
14]. The SGTF is characteristic for both the Omicron and the originally circulating Alpha (B.1.1.7) variants; however, it was not found in the previously abundant Delta variant. Therefore, detection of the SGTF in samples at this time was highly suggestive of the presence of Omicron.
Next-generation sequencing (NGS) has traditionally been used for genetic sequencing to confirm genotypes and to assess genetic mutations of bacterial or viral genomes found in human clinical samples and has also proven to be reliable for similar analysis of wastewater samples [
15]. However, this analytical method is time consuming, often expensive, and requires expert knowledge in computational biology. Furthermore, this method may not result in detection of variants found in low-abundance and can yield inconclusive, low-quality results on samples extracted from wastewater [
16]. As an alternative to genetic sequencing, some laboratories have relied on Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR) assays which generate results in real-time and can indicate the presence of variant-specific mutations in gene targets of samples isolated from either human samples or wastewater [
17,
18,
19].
Both NGS and RT-qPCR were successfully used to identify the SARS-CoV-2 Omicron variant in wastewater collected from several cities across the US, Europe, Asia and Africa [
20,
21,
22,
23,
24].
In this paper we report the initial detection of Omicron in wastewater from several locations throughout Oklahoma and demonstrate how targeted RT-qPCR can be used routinely for assessing real-time spread and dynamics of the variant in different communities.
2. Materials and Methods
2.1. Wastewater Sampling and Sample Preparation
During November 1, 2021, to January 31, 2022, we collected wastewater samples twice per week from wastewater treatment plants (WWTP) in the City of Anadarko (one), the City of Tulsa (three), Oklahoma City (four), as well as 13 sewersheds in the city of Oklahoma City. The WWTP samples were collected as either time-weighted composite or flow-weighted composite by employees at each respective WWTP and transported appropriately to our lab for processing. All sewershed samples were collected in a similar manner as previously described by Kuhn et al. [
7]. Modifications to sample collection were limited to collection type (either time weighted composite sample or flow-weighted composite sample).
Three technical replicates, each with a volume of 32 mL, were prepared for nucleic acid extraction from each wastewater sample. Each sample was passed through a 70 µm nylon mesh cell strainer and amended with Bovine Coronavirus (BCoV) as an internal control by adding 100 uL of a 1:100 dilution of the reconstituted Bovine Rota-Coronavirus Vaccine (Calf Guard
®, Zoetis). Virus particles were concentrated via a PEG precipitation modified after Wu et al. [
25], in which 8 mL of a solution containing 125 uM PEG8000 and 2M NaCl2 was added to each sample and vortexed to mix. After overnight incubation at 4
oC (12-16 h), virus particles were collected via centrifugation (14,500 x g, 4
oC, 45 min). Supernatant was decanted and pelleted solids were used for total nucleic acid extractions following a protocol modified from the Bio On Magnetic Beads (BOMB) platform [
26], described in Kuhn et al. [
7].
2.2. Molecular Analysis
Nucleic acids from extractions were quantified using the Reverse Transcriptase Quantitative Polymerase Chain Reaction (RT-qPCR) with primers and probes (nCOV_N1; Integrated DNA Technologies, Inc. Coralville, Iowa, USA) used in the Centers for Disease Control and Prevention (CDC) 2019-nCoV Real-Time RT-PCR Diagnostic Panel instructions for use under CDC’s Emergency Use Authorization [
27] as previously described by Kuhn et al. [
7].
Known variants of SARS-CoV-2 were identified with the TaqMan SARS-CoV-2 Mutation Research Panel (ThermoFisher) [
28] which targeted two different alleles: an S-gene target containing a 69-70del S-gene mutation (i.e., SGTF) and a ‘Wild Type’ (WT) allele that does not contain said deletion. Detection of targets containing the 69-70del S-gene mutation were identified as the Omicron variant and WT alleles were presumed to be the Delta variant. The mutation research panel was designed according to the protocol (Revision C.0) developed by the manufacturer [
28] but modified by replacing the TaqPath mastermix CG with the TaqPath™ 1-Step Multiplex Master Mix No ROX (Applied Biosystems, Waltham, MA, USA). The reaction mix of 2.5µL TaqPath™ 1-Step Multiplex Master Mix No ROX, 0.25 µL TaqMan™ SARS-CoV-2 Mutation Panel Assay (40X) (S.delH69V70), and 4.75 µL Nuclease-free treated water was added to each well of a 0.1-ml, 96-well plate. Each reaction contained; 2.5 μL wastewater nucleic acid extract diluted 1:4, synthetic RNA (positive control), or nuclease-free treated water (negative control) was added for a total volume of 10 µL. The following thermocycling parameters were used for PCR amplification: 60°C for 30 s, 50°C for 10 min, and 95°C for 2 min; then 45 cycles of 95°C for 30 s and 60
oC for 30 s; and a final elongation step of 60
oC for 30 s. The quantity of template molecules (SARS-CoV-2 viral genome equivalents) in each reaction was estimated using a standard curve of triplicate reactions containing 10
1, 10
2, 10
3, 10
4, and 10
5 copies of the Twist Alpha control 14 and the Twist Delta control 23 [
29].
2.3. SARS-CoV-2 Genome Alignment and Variant Detection
To confirm the presence of the Omicron variant in the wastewater samples analyzed in this study, we performed high-throughput sequencing across the entire genome of SARS-CoV-2. We concentrated specifically on the period between December 2021 and January 2022, as it marked the transition from the Delta variant to Omicron. For this purpose, we extracted total nucleic acids from eight wastewater samples collected in Oklahoma City and submitted them for paired-end (150 bp) high-throughput sequencing. cDNA was synthesized with LunaScript
® RT SuperMix Kit (NEB) per manufacturer’s protocol. Samples were then prepared for sequencing using xGen™ SARS-CoV-2 Amplicon Panel with xGen Amplicon UDI Primer Plate 1 (IDT) per manufacturer’s protocol. Samples were checked for quality and sequenced at the Oklahoma Medical Research Foundation using Illumina NextSeq 500 Mid-output 300 cycle runs with PE150 reads. Paired-end 150 bp FASTQ files were processed using the C-WAP bioinformatics pipeline (
https://www.github.com/CFSAN-Biostatistics/C-WAP) [
30], referencing the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate Wuhan-Hu-1 genome (Genbank ID: NC_045512.2). Variant prevalence was calculated using Freyja version 1.4.2 (github.com/andersen-lab/Freyja) [
31].
2.4. Epidemiological Data and Analyses
We calculated 10-day moving averages for SARS-CoV-2, Omicron and Delta concentrations at sewersheds and WWTPs. Concentrations of Omicron and Delta variants at sewersheds and WWTPs were used to calculate the relative proportion of Omicron over time. For each sewershed, we obtained estimates on population sizes and demographic variables (e.g., ethnic composition, proportion of population aged 65 years or older and median income) using census-traction portion estimates clipped to sewershed/facility polygon boundaries [
7]. The average number of days for introduction of and dominance of Omicron was calculated and compared between explanatory variables (e.g., location type, demographic characteristics) with descriptive statistics and t-tests. We analyzed days to introduction and dominance of Omicron and concentrations of Omicron in relation to demographic information using generalized linear univariate and multivariate modeling. All statistical analyses were performed in STATA 17 [
32].
3. Results
Between November 1, 2021, and January 31, 2022, we collected and analyzed 466 wastewater samples from 13 neighborhood sewersheds and 9 wastewater WWTPs for concentrations of total SARS-CoV-2, sequences that are wild type at amino acids H69 and V70 in the virus S gene (presumed to be the Delta variant based on circulating variants at that time), and sequences conferring a deletion of amino acids H69 and V70 in the virus S gene (SGTF variant; Omicron).
3.1. SARS-CoV-2 Variant Detection
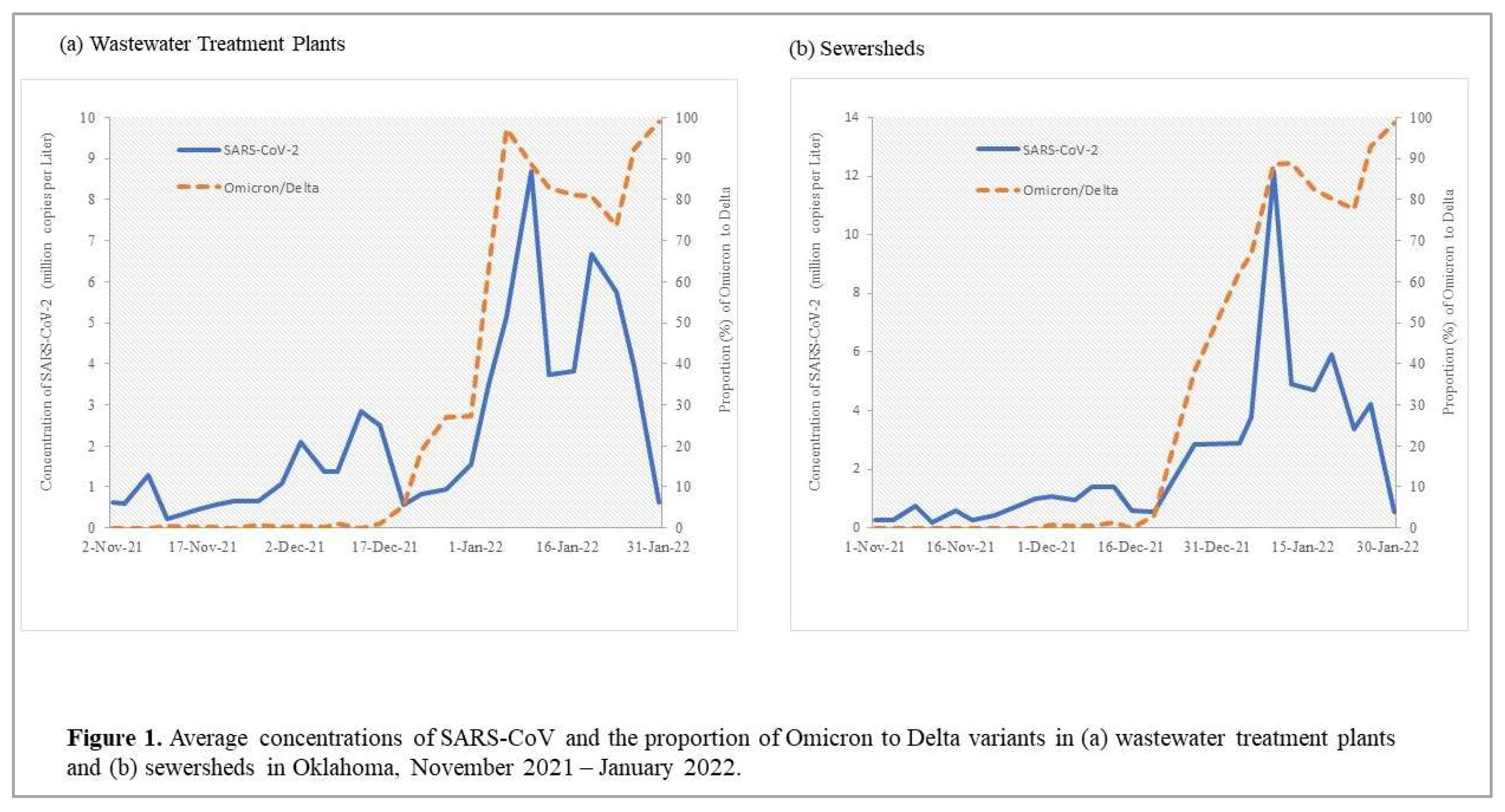
Allele-specific RT-qPCR analysis resulted in the first detection of SGTF in early December 2021. Subsequently, there was a rapid increase in SGTF detection through January 2022 in both WWTPs and sewersheds while Delta detection decreased beginning January 2022 (
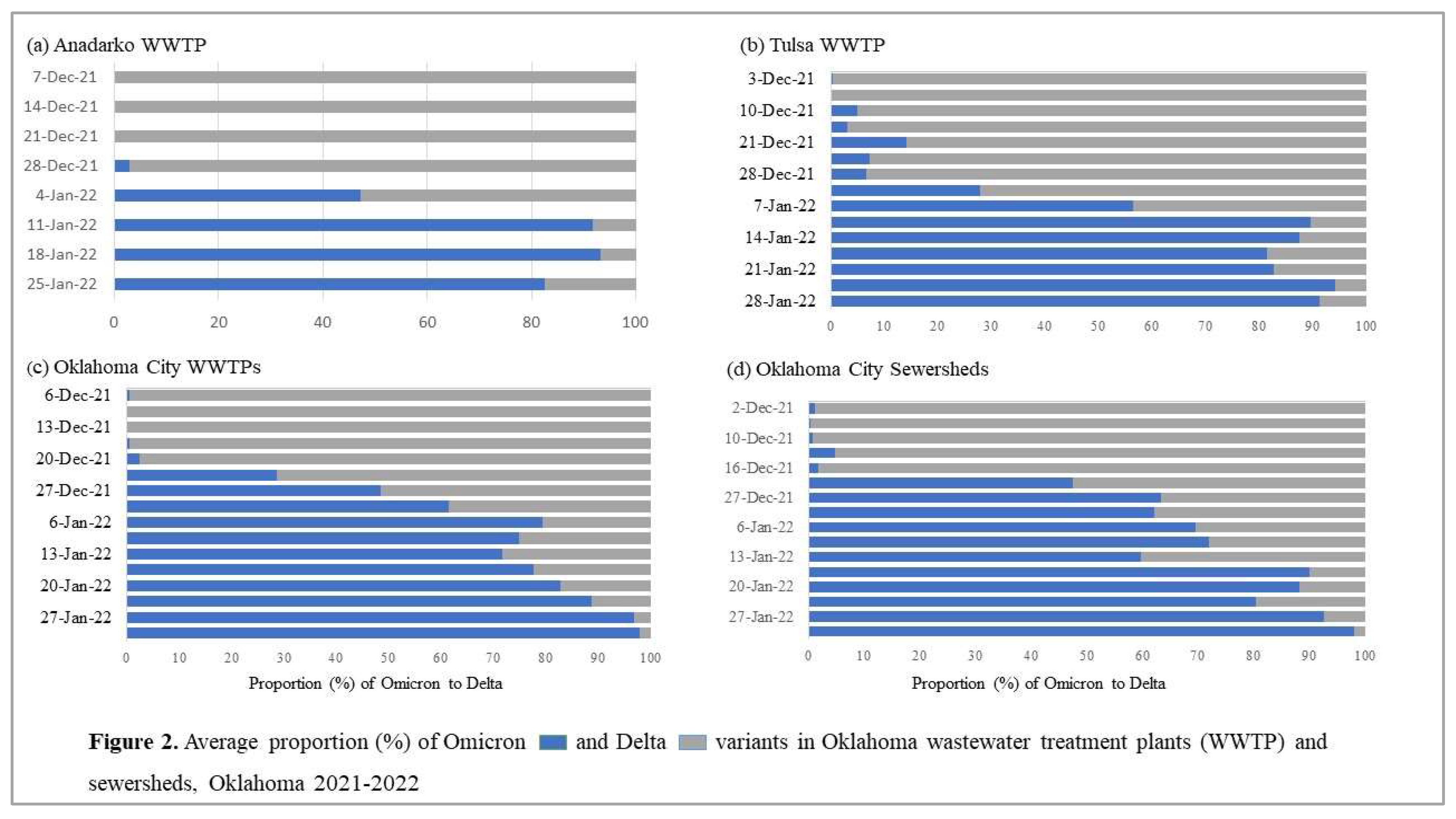
Figure 1). We identified a faint but positive SGTF detection in the Oklahoma City WWTP and associated sewershed in early December 2022 (
Figure 2) which was confirmed to be the Omicron variant through NGS. The presence of SGTF increased, becoming the overwhelmingly abundant genotype found in both Oklahoma City and Tulsa WWTPs and sewersheds between December 2021 and January 2022 (
Figure 2).
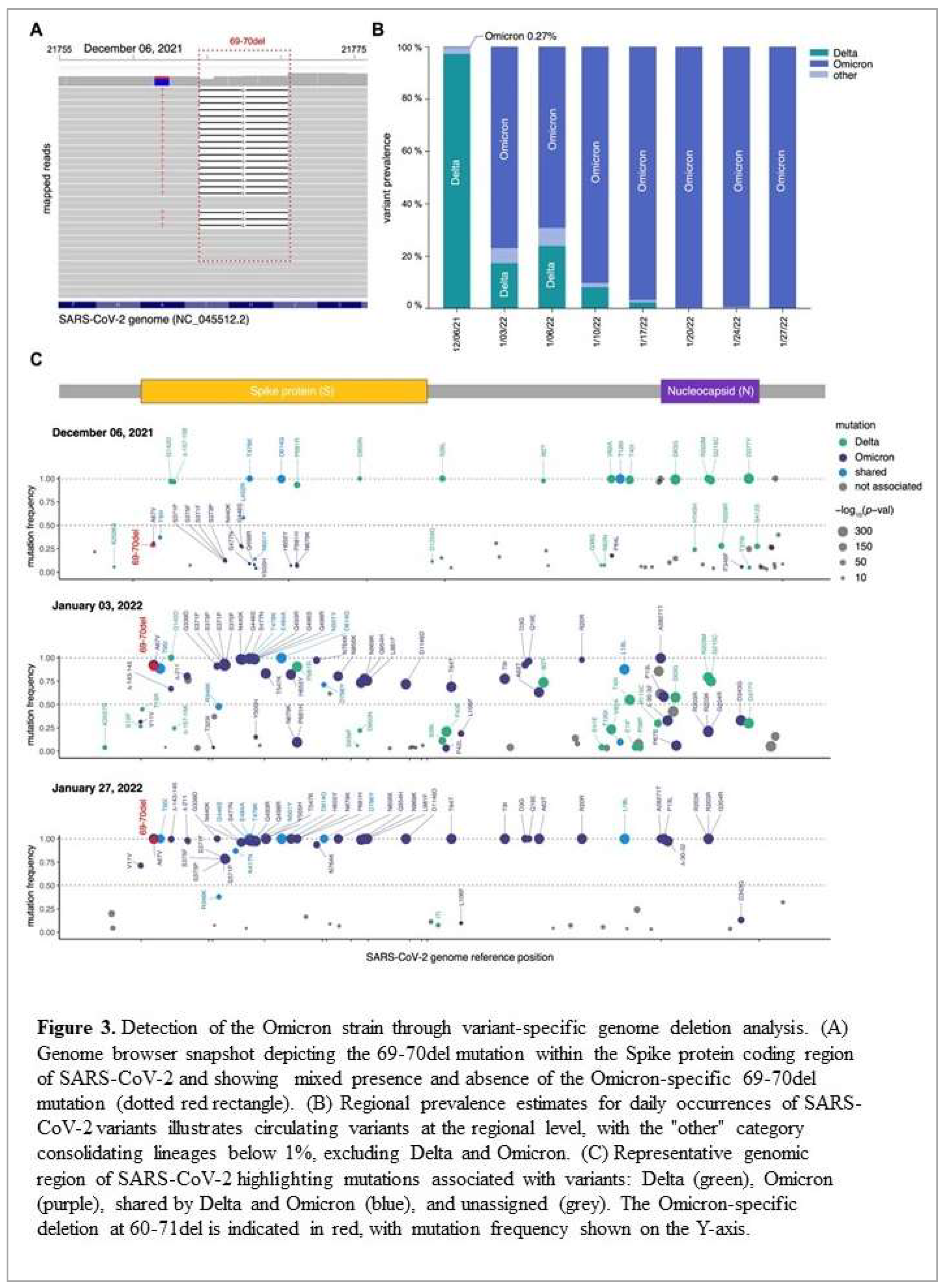
For the sequencing, all sequenced samples had an average of 1.52 x10
5 mapped paired-end reads and a genome coverage exceeding 80% with at least 10 reads per nucleotide (10x). Visual inspection of the mapped reads from the December 6th, 2021, sample confirmed the presence of the six-nucleotide deletion corresponding to histidine-69 (S:H69) and valine-70 (S:V70) (
Figure 3A, red dotted line). Analysis of variant prevalence across the eight samples revealed the Omicron variant at a frequency of 0.27% on December 6th, 2021, which had completely taken over by January 20th, 2022 (> 99%) (
Figure 3B). Further examination of sequences from the December 6th, 2021, sample revealed additional Omicron-specific mutations within the Spike coding region, including A69V, S375F, Q498R, E484A, Y505H, and H655Y (
Figure 3C, purple points). Similarly, samples from January 2022 showed a marked increase in mutations specific to the Omicron variant, particularly within the spike (S) and nucleocapsid (N) genes (
Figure 3C, purple points in middle and bottom panel). Additional affirmation that our PCR-based assay detected the Omicron variant was provided by the absence of Alpha-specific mutations, such as A570D, T716I, and S982A (
Figure 3C, purple points).
Collectively, these findings strongly support the suitability of our PCR-based assay to monitor the emergence of Omicron in our particular wastewater samples by targeting the two successive amino acid deletions (S:H69 and S:V70).
3.2. Timing of Omicron Introduction
Our first detection of the SARS-CoV-2 Omicron variant was an average of 48 days after the start of the study period (ranging from 11 to 73 days,
Table 1). The variant was first detected in an Oklahoma City wastewater treatment facility on November 11, 2021, and in an Oklahoma City sewershed on December 2, 2021 (
Table 1). For the Tulsa and Anadarko WWTPs, we first detected the Omicron variant on December 10 and December 28, 2021, respectively (
Table 1).
Generally, concentrations of SARS-CoV-2 and the proportion of Omicron detected in wastewater samples increased during the study period, with SARS-CoV-2 peaking and then decreasing from January 10,2022, while the proportion of Omicron continued to increase (
Figure 2,
Table 1).
3.3. Timing of Omicron Dominance
Throughout the study period, the Omicron vs. Delta proportion increased markedly in all WWTPs and sewersheds (
Figure 1), reaching 100% in several locations at the end of January 2022. For all locations, it lasted an average of 63 days from November 1, 2021, until the proportion of Omicron exceeded 50% (
Table 1). This progression was significantly longer in sewersheds compared to treatment plants (t=4.4, df=423, p<0.001). We also observed a reverse relationship between Omicron introduction and the time until the proportion of Omicron exceeded 50% where locations with a longer introduction time experienced more rapid dominance of Omicron and vice versa (z=-6.1, 95%CI: -0.3- -0.1, p<0.05).
3.4. Demography and SARS-CoV-2 Variants
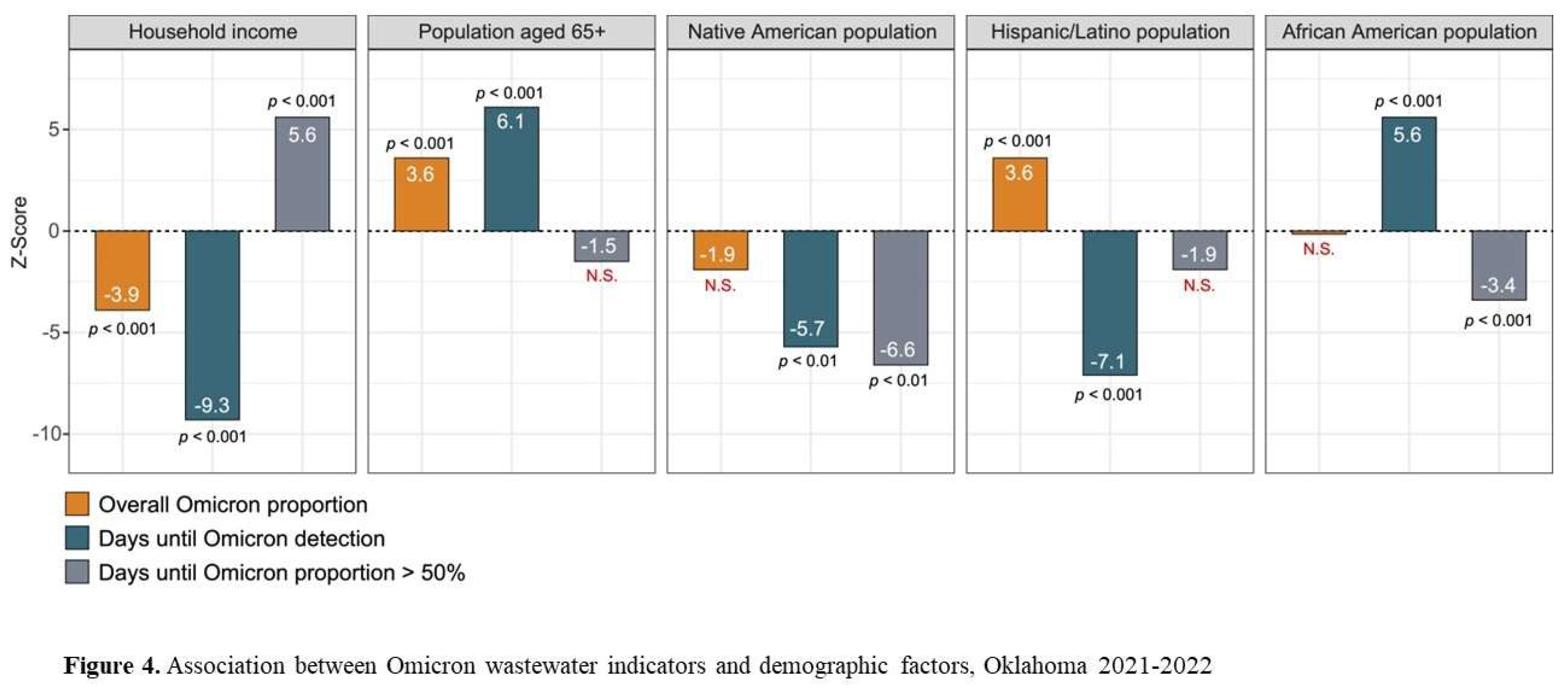
We explored univariate relationships between demographic characteristics and the indicators of Omicron and Delta in sewersheds (
Figure 4). The ethnic composition of monitoring locations significantly impacted when Omicron was first detected. Detection of Omicron occurred later in areas with larger African American and American Indian populations, smaller Hispanic/Latnio populations, lower household incomes and more persons aged 65+. Conversely these same areas also had a more rapid progression to Omicron dominance and some evidence of higher Omicron concentrations in general (
Figure 4).
Combining the wastewater indicators and demographic variables in multivariate models showed that later detection of Omicron was observed in areas with larger African American and American Indian populations and more persons aged 65+ (z=17.6, 95%CI:1.6-2.0, p<0.001). On the other hand, the number of days before the proportion of Omicron exceeded 50% was lower in those same areas and higher in areas with a high household income (z=64.8, 95%CI:4.3-4.5, <0.001).
4. Discussion
In this paper we use evidence from routine wastewater surveillance of SARS-CoV-2 in several locations across the State of Oklahoma to highlight the dynamic transition of a SARS-CoV-2 variant that was newly introduced in November 2021. The strength and sustainability of our results are founded in the use of sensitive PCR techniques and their reliability confirmed by next generation sequencing, highlighting that the PCR assays provide an accurate, and cost-effective real-time alternative to the more expensive and time-consuming sequencing methods
The SARS-CoV-2 Omicron variant was first reported in South Africa and immediately noted as a variant of concern by the WHO on November 24, 2021 [
33]. Following this, evidence emerged that the variant was present in human samples collected in the Netherlands on November 19 [
34] and wastewater samples collected in New York on November 21[
20]. In this paper we present results that demonstrate the presence of Omicron in Oklahoma wastewater as early as November 11 and 19, lending support to other observations of the variant outside South Africa before November 24. This also provides evidence to back up the hypothesis that the Omicron variant was circulating worldwide before the first official notifications.
In addition to early detection of a SARS-CoV-2 variant of concern in Oklahoma, our results also highlight the significance and potential for using RT-qPCR assays as a simple, rapid, and cost-effective method of detecting the emergence and community transmission of novel SARS-CoV-2 variants. Because the PCR assay offers a realistic alternative for variant tracking, this finding is especially relevant for national surveillance institutes and laboratories who lack timely access to traditional sequencing equipment and expertise. Considering the longer processing times for genomic sequencing, our method also has the distinct advantage of providing real-time ‘early warning’ for healthcare providers needing updated information for control, treatment and prevention action.
Our results indicate that Omicron was introduced in Oklahoma neighborhoods from mid-November to early/late December 2021 and rapidly became dominant over Delta, corresponding to what was reported from other wastewater surveillance efforts across the US [
20,
35,
36]. We show that, until mid-late January 2022, increases in overall concentrations of SARS-CoV-2 in wastewater seemed largely driven by an increase in Omicron’s proportion. This lends further support to the observed high infectiousness and rapid spread of the variant in most areas across the world. By combining wastewater indicators with demographic data, we also demonstrate how the introduction of Omicron and its transition towards becoming the dominant variant was influenced by the ethnic composition of the area covered by the treatment plant or sewershed. Omicron was introduced later in African American and American Indian populations and among persons aged 65 or older, however once present, it rapidly became dominant. Interestingly, areas with higher household incomes experienced a more rapid introduction but slower spread of Omicron. The Delta variant disproportionately affected specific ethnic groups and lower-income populations [
37,
38], and these may have still experienced some cross-protective immunity from a Delta infection, causing the introduction of Omicron to be delayed. Following established transmission, dominance of Omicron would be rapid because of its high infectiousness. In ethnically diverse and low-income communities, the time to Omicron dominance was most likely further shortened because of reduced healthcare access and reduced access to – and use of – personal protective equipment [
39,
40,
41].
Our study presents relevant insights into the detection of a novel SARS-CoV-2 in wastewater and evidence for how this variant spread and became dominant across diverse population groups in Oklahoma. However, the results need to be interpreted with consideration of potential limitations. Firstly, primers may selectively amplify a single variant from a diverse pool, which could result in an unbalanced representation of the variants present. Secondly, our approach operates under the assumption that the prevalence of both Omicron and Delta variants mirrors the national trends observed at the time of the analysis. Recent evidence nonetheless suggests a significant deviation from this assumption, particularly in Oklahoma [
42]. Further, reports of ‘Long COVID’ are comparatively high in Oklahoma [
42], highlighting a continued presence and possible overabundance of older variants, including the Alpha variant which also presents the 69-70del mutation [
43,
44]. This finding implies that the viral landscape in Oklahoma may not align with national trends, potentially due to localized factors or the persistence of earlier variant strains.
Wastewater surveillance has proven to be a highly effective and useful tool for monitoring trends in SARS-CoV-2, including measuring community transmission dynamics of genetic variants. Our results from the State of Oklahoma confirm this with an added novel aspect of using allele-specific RT-qPCR as a timelier alternative to traditional genomic sequencing. To our knowledge, we also present the first results to show how the introduction and spread of Omicron differed significantly between areas depending on ethnic and socioeconomic composition. This evidence can ultimately form a crucial foundation for healthcare providers and decision-makers with respect to planning and implementing future variant-focused control and prevention measures.
Author contributions
Conceptualization, Jason Vogel, Bradley Stevenson and Katrin Kuhn; Data curation, Kristen Shelton, Gilson Sanchez, Gabriel Florea and Erin Jeffries; Formal analysis, Kristen Shelton, Gargi Deshpande, Gilson Sanchez and Katrin Kuhn; Funding acquisition, Jason Vogel, Bradley Stevenson and Katrin Kuhn; Investigation, A Caitlin Miller, Gabriel Florea, Erin Jeffries and Kara De Leon; Methodology, Kristen Shelton, Gilson Sanchez, A Caitlin Miller, Gabriel Florea, Erin Jeffries, Kara De Leon and Bradley Stevenson; Project administration, Jason Vogel, Bradley Stevenson and Katrin Kuhn; Supervision, Bradley Stevenson; Writing – original draft, Kristen Shelton, Gargi Deshpande, Jason Vogel, Erin Jeffries, Kara De Leon, Bradley Stevenson and Katrin Kuhn; Writing – review & editing, Kristen Shelton, Gargi Deshpande, Gilson Sanchez, A Caitlin Miller, Gabriel Florea, Kara De Leon, Bradley Stevenson and Katrin Kuhn.
Funding
The work was funded by the City of Oklahoma City through the Federal Coronavirus Aid, Relief and Economic Security (CARES) Act, the Presbyterian Health Foundation and the City of Tulsa through the Rockefeller Foundation.
Data statement
Measures of raw wastewater concentrations by location and date, including all corresponding positive and negative controls, concentrations of Omicron and Wild Type variants by location and date, are available upon request to the corresponding author.
Acknowledgements
We are grateful for the contributions Ms. Madelynn Henderson for assistance with sequencing results. We also acknowledge the support and collaboration from the City of Oklahoma City and the City of Tulsa, in particular the Oklahoma City County Health Department and the Tulsa City County Health Department. We acknowledge the continued support and knowledge sharing from the Rockefeller Foundation with thanks. We are grateful to the following members of the OU Wastewater Team for their individual contributions: James Cutler, Grant Graves, Jacobey King, Will Ruley, Halley Reeves, Emily Rhodes, and Ralph Tanner.
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- Koelle, K.; Martin, M.A.; Antia, R.; Lopman, B.; Dean, N.E. The Changing Epidemiology of SARS-CoV-2. Science 2022, 375, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. The Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/emergencies/what-we-do/tracking-SARS-CoV-2-variants (accessed on 5 January 2022).
- Increased Risk of SARS-CoV-2 Reinfection Associated with Emergence of the Omicron Variant in South Africa | medRxiv. Available online: https://www.medrxiv.org/content/10.1101/2021.11.11.21266068v2 (accessed on 5 January 2022).
- Kuhn, K.G.; Jarshaw, J.; Jeffries, E.; Adesigbin, K.; Maytubby, P.; Dundas, N.; Miller, A.C.; Rhodes, E.; Stevenson, B.; Vogel, J.; et al. Predicting COVID-19 Cases in Diverse Population Groups Using SARS-CoV-2 Wastewater Monitoring across Oklahoma City. Sci. Total Environ. 2021, 151431. [Google Scholar] [CrossRef]
- Ahmed, W.; Tscharke, B.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Choi, P.; Clarke, L.; Dwyer, J.; Edson, J.; Nguyen, T.M.H.; et al. SARS-CoV-2 RNA Monitoring in Wastewater as a Potential Early Warning System for COVID-19 Transmission in the Community: A Temporal Case Study. Sci. Total Environ. 2021, 761, 144216. [Google Scholar] [CrossRef]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Amman, F.; Markt, R.; Endler, L.; Hupfauf, S.; Agerer, B.; Schedl, A.; Richter, L.; Zechmeister, M.; Bicher, M.; Heiler, G.; et al. Viral Variant-Resolved Wastewater Surveillance of SARS-CoV-2 at National Scale. Nat. Biotechnol. 2022, 40, 1814–1822. [Google Scholar] [CrossRef]
- Vo, V.; Tillett, R.L.; Papp, K.; Shen, S.; Gu, R.; Gorzalski, A.; Siao, D.; Markland, R.; Chang, C.-L.; Baker, H.; et al. Use of Wastewater Surveillance for Early Detection of Alpha and Epsilon SARS-CoV-2 Variants of Concern and Estimation of Overall COVID-19 Infection Burden. Sci. Total Environ. 2022, 835, 155410. [Google Scholar] [CrossRef]
- Carcereny, A.; Garcia-Pedemonte, D.; Martínez-Velázquez, A.; Quer, J.; Garcia-Cehic, D.; Gregori, J.; Antón, A.; Andrés, C.; Pumarola, T.; Chacón-Villanueva, C.; et al. Dynamics of SARS-CoV-2 Alpha (B.1.1.7) Variant Spread: The Wastewater Surveillance Approach. Environ. Res. 2022, 208, 112720. [Google Scholar] [CrossRef]
- Lippi, G.; Adeli, K.; Plebani, M. Commercial Immunoassays for Detection of Anti-SARS-CoV-2 Spike and RBD Antibodies: Urgent Call for Validation against New and Highly Mutated Variants. Clin. Chem. Lab. Med. 2021. [Google Scholar] [CrossRef] [PubMed]
- Metzger, C.M.J.A.; Lienhard, R.; Seth-Smith, H.M.B.; Roloff, T.; Wegner, F.; Sieber, J.; Bel, M.; Greub, G.; Egli, A. PCR Performance in the SARS-CoV-2 Omicron Variant of Concern? Swiss Med. Wkly. 2021, 151, w30120. [Google Scholar] [CrossRef] [PubMed]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A.; et al. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 2021, 12, e02703–20. [Google Scholar] [CrossRef]
- Van Poelvoorde, L.A.E.; Delcourt, T.; Coucke, W.; Herman, P.; De Keersmaecker, S.C.J.; Saelens, X.; Roosens, N.H.C.; Vanneste, K. Strategy and Performance Evaluation of Low-Frequency Variant Calling for SARS-CoV-2 Using Targeted Deep Illumina Sequencing. Front. Microbiol. 2021, 12, 3073. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.L.; Gu, X.; Armas, F.; Chandra, F.; Chen, H.; Wu, F.; Leifels, M.; Xiao, A.; Chua, F.J.D.; Kwok, G.W.; et al. Quantitative SARS-CoV-2 Tracking of Variants Delta, Delta plus, Kappa and Beta in Wastewater by Allele-Specific RT-qPCR; 2021; p. 2021.08.03.21261298;
- Yaniv, K.; Ozer, E.; Shagan, M.; Lakkakula, S.; Plotkin, N.; Bhandarkar, N.S.; Kushmaro, A. Direct RT-qPCR Assay for SARS-CoV-2 Variants of Concern (Alpha, B.1.1.7 and Beta, B.1.351) Detection and Quantification in Wastewater. Env. Res 2021, 111653–111653.
- Graber, T.E.; Mercier, É.; Bhatnagar, K.; Fuzzen, M.; D’Aoust, P.M.; Hoang, H.-D.; Tian, X.; Towhid, S.T.; Plaza-Diaz, J.; Eid, W.; et al. Near Real-Time Determination of B.1.1.7 in Proportion to Total SARS-CoV-2 Viral Load in Wastewater Using an Allele-Specific Primer Extension PCR Strategy. Water Res. 2021, 205, 117681. [Google Scholar] [CrossRef]
- Kirby, A.E. Notes from the Field: Early Evidence of the SARS-CoV-2 B.1.1.529 (Omicron) Variant in Community Wastewater — United States, November–December 2021. MMWR Morb. Mortal. Wkly. Rep. 2022, 71. [Google Scholar] [CrossRef]
- Kumblathan, T.; Liu, Y.; Pang, X.; Hrudey, S.E.; Le, X.C.; Li, X.-F. Quantification and Differentiation of SARS-CoV-2 Variants in Wastewater for Surveillance. Environ. Health Wash. DC 2023, 1, 203–213. [Google Scholar] [CrossRef]
- Lee, W.L.; Armas, F.; Guarneri, F.; Gu, X.; Formenti, N.; Wu, F.; Chandra, F.; Parisio, G.; Chen, H.; Xiao, A.; et al. Rapid Displacement of SARS-CoV-2 Variant Delta by Omicron Revealed by Allele-Specific PCR in Wastewater. Water Res. 2022, 221, 118809. [Google Scholar] [CrossRef]
- Sutcliffe, S.G.; Kraemer, S.A.; Ellmen, I.; Knapp, J.J.; Overton, A.K.; Nash, D.; Nissimov, J.I.; Charles, T.C.; Dreifuss, D.; Topolsky, I.; et al. Tracking SARS-CoV-2 Variants of Concern in Wastewater: An Assessment of Nine Computational Tools Using Simulated Genomic Data. Microb. Genomics 2024, 10, 001249. [Google Scholar] [CrossRef]
- Xu, X.; Deng, Y.; Ding, J.; Shi, X.; Zheng, X.; Wang, D.; Yang, Y.; Liu, L.; Wang, C.; Li, S.; et al. Refining Detection Methods for Emerging SARS-CoV-2 Mutants in Wastewater: A Case Study on the Omicron Variants. Sci. Total Environ. 2023, 904, 166215. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhang, J.; Xiao, A.; Gu, X.; Lee, W.L.; Armas, F.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N.; et al. SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases. mSystems 2020, 5, e00614–20. [Google Scholar] [CrossRef] [PubMed]
- Oberacker, P.; Stepper, P.; Bond, D.M.; Höhn, S.; Focken, J.; Meyer, V.; Schelle, L.; Sugrue, V.J.; Jeunen, G.-J.; Moser, T.; et al. Bio-On-Magnetic-Beads (BOMB): Open Platform for High-Throughput Nucleic Acid Extraction and Manipulation. PLOS Biol. 2019, 17, e3000107. [Google Scholar] [CrossRef] [PubMed]
- CDC Labs. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html (accessed on 13 May 2021).
- TaqMan SARS-CoV-2 Mutation Panel - US Available online: //www.thermofisher.com/us/en/home/clinical/clinical-genomics/pathogen-detection-solutions/real-time-pcr-research-solutions-sars-cov-2/mutation-panel.html (accessed on 4 February 2022).
- Synthetic Viral Controls | Twist Bioscience. Available online: https://www.twistbioscience.com/products/ngs/synthetic-viral-controls?mkt_tok=OTQzLVVHRC0wMDMAAAGDSPZFUrAXrIszxxl3qTp5_-oQVnERkn_K6exdhXre-wXUbQv8Q-OLaEkboH5_RgqTTz3fOBK9clN_sFYm4aWDEJVq2YF22EJ6f7fr9XIGtg&tab=sars-cov-2-controls (accessed on 23 March 2022).
- Kayikcioglu, T.; Amirzadegan, J.; Rand, H.; Tesfaldet, B.; Timme, R.E.; Pettengill, J.B. Performance of Methods for SARS-CoV-2 Variant Detection and Abundance Estimation within Mixed Population Samples. PeerJ 2023, 11, e14596. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Levy, J.I.; De Hoff, P.; Humphrey, G.; Birmingham, A.; Jepsen, K.; Farmer, S.; Tubb, H.M.; Valles, T.; Tribelhorn, C.E.; et al. Wastewater Sequencing Reveals Early Cryptic SARS-CoV-2 Variant Transmission. Nature 2022, 609, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Stata: Software for Statistics and Data Science. Available online: https://www.stata.com/ (accessed on 4 February 2022).
- Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 4 February 2022).
- Ingraham, N.E.; Ingbar, D.H. The Omicron Variant of SARS-CoV-2: Understanding the Known and Living with Unknowns. Clin. Transl. Med. 2021, 11, e685. [Google Scholar] [CrossRef]
- Boehm, A.B.; Hughes, B.; Wolfe, M.K.; White, B.J.; Duong, D.; Chan-Herur, V. Regional Replacement of SARS-CoV-2 Variant Omicron BA.1 with BA.2 as Observed through Wastewater Surveillance. Environ. Sci. Technol. Lett. 2022, 9, 575–580. [Google Scholar] [CrossRef]
- Smith, M.F.; Holland, S.C.; Lee, M.B.; Hu, J.C.; Pham, N.C.; Sullins, R.A.; Holland, L.A.; Mu, T.; Thomas, A.W.; Fitch, R.; et al. Baseline Sequencing Surveillance of Public Clinical Testing, Hospitals, and Community Wastewater Reveals Rapid Emergence of SARS-CoV-2 Omicron Variant of Concern in Arizona, USA. mBio 2023, 14, e0310122. [Google Scholar] [CrossRef]
- Malden, D.E. Distribution of SARS-CoV-2 Variants in a Large Integrated Health Care System — California, March–July 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70. [Google Scholar] [CrossRef]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital Admission and Emergency Care Attendance Risk for SARS-CoV-2 Delta (B.1.617.2) Compared with Alpha (B.1.1.7) Variants of Concern: A Cohort Study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Chopra, J.; Abiakam, N.; Kim, H.; Metcalf, C.; Worsley, P.; Cheong, Y. The Influence of Gender and Ethnicity on Facemasks and Respiratory Protective Equipment Fit: A Systematic Review and Meta-Analysis. BMJ Glob. Health 2021, 6, e005537. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Drew, D.A.; Graham, M.S.; Joshi, A.D.; Guo, C.-G.; Ma, W.; Mehta, R.S.; Warner, E.T.; Sikavi, D.R.; Lo, C.-H.; et al. Risk of COVID-19 among Front-Line Health-Care Workers and the General Community: A Prospective Cohort Study. Lancet Public Health 2020, 5, e475–e483. [Google Scholar] [CrossRef] [PubMed]
- Louis-Jean, J.; Cenat, K.; Njoku, C.V.; Angelo, J.; Sanon, D. Coronavirus (COVID-19) and Racial Disparities: A Perspective Analysis. J. Racial Ethn. Health Disparities 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Blanchflower, D.G.; Bryson, A. Long COVID in the United States. PLOS ONE 2023, 18, e0292672. [Google Scholar] [CrossRef]
- Peterson, S.W.; Lidder, R.; Daigle, J.; Wonitowy, Q.; Dueck, C.; Nagasawa, A.; Mulvey, M.R.; Mangat, C.S. RT-qPCR Detection of SARS-CoV-2 Mutations S 69-70 Del, S N501Y and N D3L Associated with Variants of Concern in Canadian Wastewater Samples. Sci. Total Environ. 2022, 810, 151283. [Google Scholar] [CrossRef]
- Gregory, D.A.; Trujillo, M.; Rushford, C.; Flury, A.; Kannoly, S.; San, K.M.; Lyfoung, D.; Wiseman, R.W.; Bromert, K.; Zhou, M.-Y.; et al. Genetic Diversity and Evolutionary Convergence of Cryptic SARS-CoV-2 Lineages Detected Via Wastewater Sequencing. medRxiv 2022, 2022.06.03.22275961. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).