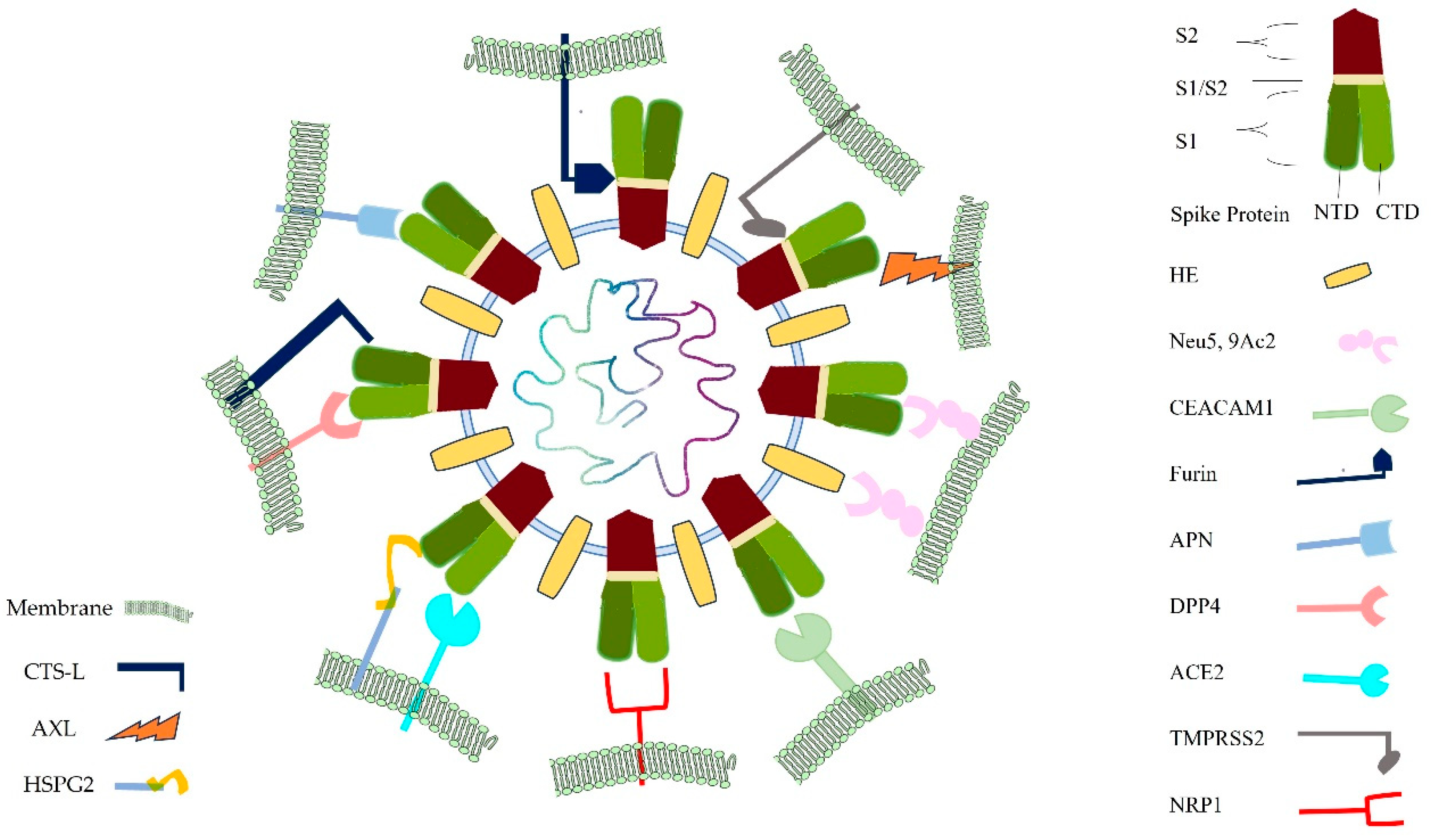
1. Introduction
Virus replication occurs in many consecutive steps to generate mature viral particles in the permissive cells. The virus always hijacks the cellular machinery and directs them to synthesize their viral proteins instead of cellular one’s [
1,
2]. The viral entry is a crucial step in any viral infection, including BCoV [
3]. The viral entry involves many host cells and viral proteins [
1]. The presence of viral-specific receptors is considered an important factor in the process of viral entry into the host cells. The host cell is called permissive for certain viruses if they express the specific viral receptors and provide suitable environments for virus replication, including any auxiliary receptors, transcription, and translation factors. Coronaviruses are enveloped viruses containing positive sense RNA genomes and belong to the order Nidovirales and are classified into four genera (α, β, λ, and δ) [
4]. The genus β-coronavirus includes five important human coronaviruses (the Sever Acute Respiratory Syndrome Coronavirus-1 (SARS-CoV-1), the Middel East respiratory syndrome coronavirus (MERS-CoV), the SARS-CoV-2, human coronavirus-HKU1 (HCoV-HKU1), and the human coronavirus-OC43 (HCoV-OC-43). This genus β-coronavirus includes some other important viruses affecting animals, particularly the BCoV and the equine coronavirus (ECoV) [
4]. The coronavirus’s genome size ranges from 27-31 kb in length and has a unique organization. The full-length genome is flanked with two untranslated regions at the 5′ and 3′ ends. Coronaviruses are characterized by the production of a subset of sub-genomic messenger RNA (mRNA) at their 3′ end [
5]. The 5′ end of the genome of most coronaviruses contains a large gene called Gene-1, which consists of two overlapping open reading frames (ORFs) with a ribosomal frameshifting between those two ORFs. However, the 3′ end of the genome is mainly occupied by the common structural proteins interspersed with some small accessory proteins. There are four major structural proteins in most coronaviruses, including the spike glycoprotein (S), the envelope (E), the membrane (M), and the nucleocapsid protein (N). Some members of the genus β-coronavirus, including BCoV and HCoV-OC43, have an additional structural protein called hemagglutinin esterase (HE); thus, their genome is a little larger in size compared to other coronaviruses (31 Kb) [
6]. The S glycoprotein is a key player in all coronavirus replication. There are several proteins, including some cellular receptors, co-receptors, and cellular enzymes are involved in the BCoV/host interaction. Both the BCoV spike (BCoV/S) and (BCoV-HE) proteins play important roles during BCoV replication and pathogenesis (6). BCoV/S has potential binding to the 5-N-acetyl-9-O-acetylneuraminic acid (Neu5,9Ac2), suggesting its possible roles as BCoV receptors [
7]. On the other hand, the BCoV-HE acts as a receptor-destroying enzyme during BCoV replication [
7]. However, there is a lack of comprehensive understanding of the interplay of the BCoV-S/BCoV-HE with the cellular receptors during BCoV replication. The availability of specific receptors is one of the main factors that make the target cells permissive to coronavirus (CoVs) infection. Each group of CoVs recognizes certain types of receptors and may require the presence of additional auxiliary receptors to facilitate virus attachment and downstream replication. The SARS-CoV-2 uses the angiotensin-converting enzyme 2 (ACE2) as the main receptor, and the chaperone GRP78 acts as an auxiliary receptor [
8,
9]. It was also shown that MERS-CoV utilizes the dipeptidyl peptidase-4 (DPP4) as receptors in humans and dormitory camels [
10,
11]. The amino-peptidase N (APN), also called cluster of differentiation -13 (CD13), acts as a major receptor for the transmissible gastroenteritis virus that causes enteric infections in pigs [
12]. The carcinoembryonic antigen cell adhesion molecule 1 (CEACAM-1) also acts as a major receptor to another coronavirus called murine hepatitis virus (MHV) [
13]. Although the presence of the CoVs receptors and co/receptors is important for the success of viral replication, most coronaviruses require the presence of host cell enzymes that help in the cleavage of the CoV-S and CoV-HE proteins to initiate the process of viral infection. Usually, these host cell enzymes are highly expressed in the target tissues, which play crucial roles during the CoVs entry to the host cells, particularly the mucosal surfaces of the respiratory and enteric tracts of the affected hosts. H
omology modeling is a well-established method that has been shown to produce quite accurate models for a protein sequence if an X-ray structure of a protein with a sufficient degree of sequence similarity is available [
14]
. The method is based on the fact that the structural conformation of a protein is more highly conserved than its amino acid sequence and that small or medium changes in sequence normally result in little variation in the 3D structures [
15]
. The quality of the model is directly linked to the identity between template and target sequences. As a rule, models built with over 50% sequence similarities are accurate enough for drug discovery applications [
16]
.
During the CoVs replication, various host cell proteases, including transmembrane serine protease 2 (TMPRSS2), furin, and cathepsin-L (CTS-L), are involved in processing the S protein. However, the precise order of protease cleavage and the interactions between these host proteases are not yet fully understood [
17]. It has been recently shown that the TMPRSS2 (transmembrane protease serine-2) is a serine protease that plays an important role in SARS-CoV-2 replication, particularly during viral entry to the host cells. The TMPRSS2 usually cleaves the viral spike glycoprotein, which enhances the interaction between the virus and the cellular receptors, thus facilitating its entry into the host cells [
18]. The Furin is another host cell enzyme that is a subtilisin-like proprotein convertase. Furin usually cleaves the target proteins at a polybasic amino acid sequence R-X-(K/R)-R (where R is arginine, K is lysine, and X can be any amino acid). The Furin cleavage to the SARS-CoV-2-S protein enhances the pathogenicity transmissibility and increases the virus infectivity in the target host [
19,
20]. Although the roles of the receptors mentioned above and enzymes were intensively studied in SARS-CoV2, there is a lack of knowledge about the roles of these receptors and enzymes in BCoV infection and the replication cycle. The main goals of the current study are (1) to use the in-silico prediction and docking tools to study the roles of these proteins and enzymes in BCoV replication. (2) to confirm the molecular docking results and in silico prediction of the short-listed potential receptors and host enzymes by drafting the host gene expression profiles during the BCoV replication of some bovine host cells. The results of this study highlighted some unknown aspects of BCoV tissue tropism and pathogenesis through the identification of some potential novel receptors and enzymes involved in BCoV entry and virus replication. It will also pave the way for the development of some novel vaccines and antiviral therapies for BCoV infection in cattle.
4. Discussion
Bovine coronavirus (BCoV) is one of the most common pathogens affecting cattle of all ages. BCoV possesses dual tissue tropism in cattle, particularly in the respiratory and enteric tracts. The process of BCoV entry to the target cells is a crucial step in the replication cycle of the virus. This process requires the orchestration between several viral and host cell proteins. The coronavirus spike glycoproteins contributed substantially to other viral infections and the molecular pathogenesis of coronaviruses, including BCoV. Meanwhile, among all coronaviruses, the hemagglutinin esterase enzyme is found in only BCoV and the human coronavirus-OC43 (HCoV-OC43). The viral receptors are among the key players in the viral entry and could fine-tune the viral tissue tropism. Some host cell proteases also enhance the coronavirus replication from entry to the release of the virus from the host cells. Previous studies have shown that the 9- O-acetylated sialic acids could potentially act as receptors for the BCoV. The tropism of coronaviruses is a complicated process that requires the availability of some factors from the viral side, including some attachment proteins, particularly the spike glycoprotein. This process also involves some cellular factors, including the receptors and other transcription and translation factors. It also requires some factors from the infected host, particularly the availability of host enzymes that help activate some essential proteins [
36,
37,
38]. BCoV possesses a dual tissue tropism in cattle. The virus mainly affects the digestive and respiratory tract of the affected animals; this pattern is called pneumoenteritis [
39]. The viral tropism primarily depends on the availability of specific viral receptors, some other transcription translation factors, and some host cell enzymes [
40]. Members of the family coronaviridae utilize many host cell receptors to attach to their target cells [
41].
Most coronaviruses require activation through the cleavage of their spike glycoprotein by some host cell proteases to facilitate the viral infection [
32,
33,
34,
42]. Although BCoV was discovered long ago, little is still known about viral tropism, especially the roles of the host cell receptors and the enzymes in fine-tuning the viral tissue tropism, pathogenesis, and replication [
35]. The current study aims to identify some novel BCoV receptors with higher affinities that bind to the S and HE proteins to initiate and facilitate viral infection and pathogenesis. The BCoV/S glycoprotein is the main viral protein involved in the process of viral attachment to the host cells [
43]. The BCoV/S protein consists of two subunits (S1 and S2): the N-terminal domain (NTD) and BCoV-S1-CTD are in the S1 subunit, whereas the fusion peptide (FP) and heptad repeat (HR) domains 1 and 2 are located in the S2 subunit of the BCoV/S glycoprotein. The BCoV/S protein usually attaches to the cell membrane by interacting with viral receptors on the surface of the target cells, initiating the viral infection. Spike protein S2 mediates the fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Also, it acts as a viral fusion peptide, which is unmasked following the S2 cleavage site occurring upon virus endocytosis [
44,
45]. The distal S1 subunit of the coronavirus spike protein is responsible for receptor binding. Either the S1-NTD or the S1-RBD at the C-terminal domain of the BCoV-S1 protein chain, or occasionally both, are involved in the binding to the host receptors [
46,
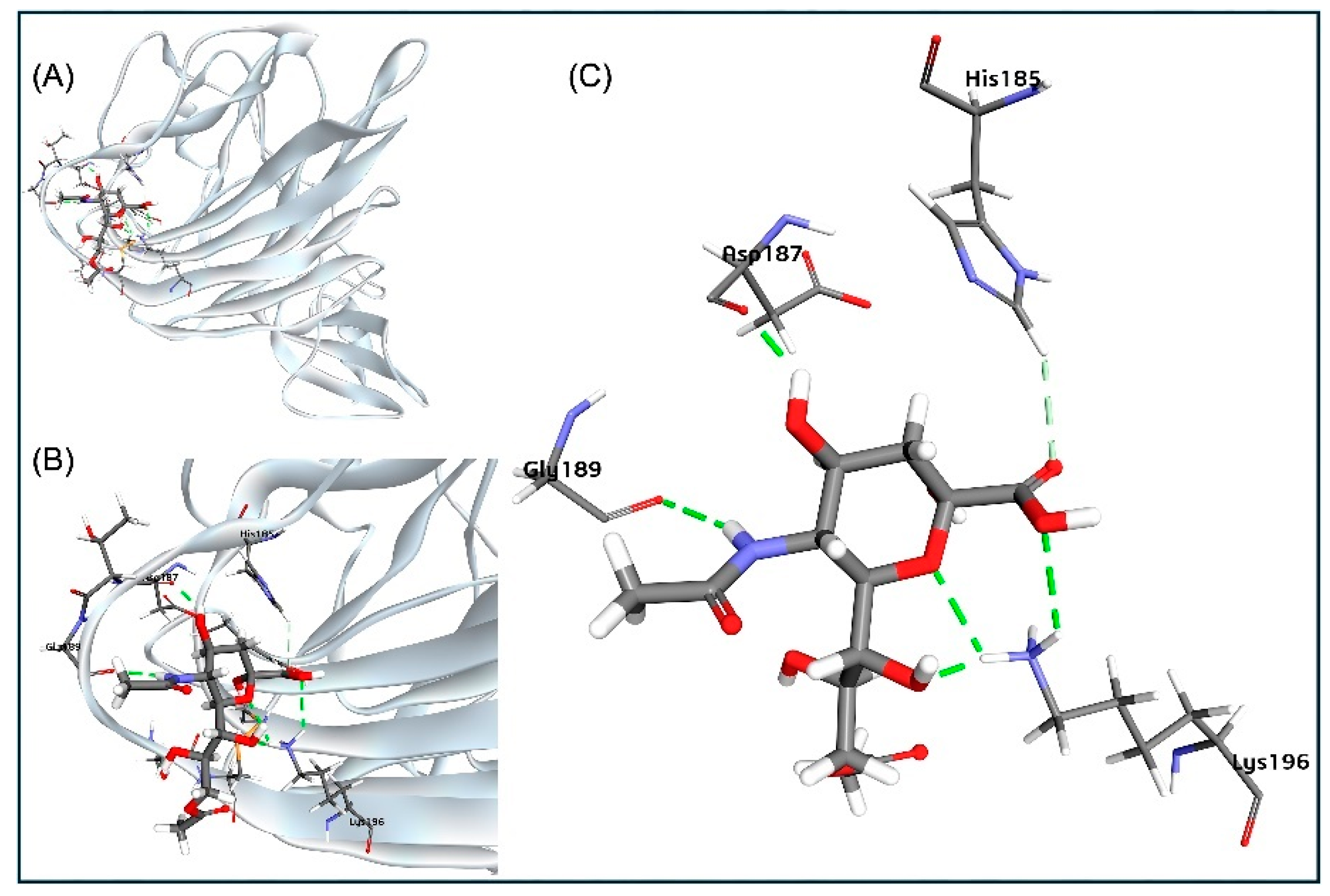
47]. The observed binding between Neu5,9Ac2 and the BCoV/S-NTD is particularly interesting because BCoV haemagglutinin esterase (HE) also utilizes this O-acylated sugar molecule as a substrate. While our study identified interacting residues like Asp-187, Gly-189, His-185, and Lys-196, another report [
34] proposed a slightly similar set of critical residues for Neu5,9Ac2 binding (Tyr-162, Glu-182, Trp-184, and His-185).
BCoV/S and BCoV/HE proteins act synergistically and harmoniously to orchestrate the BCoV infection in the target cell [
7,
48]. The BCoV/S is mainly involved in the initial attachment of the virus to the host cells, while the BCoV/HE destroys the sialic acid in the cell’s surface, promoting the viral release from the host cells [
7].
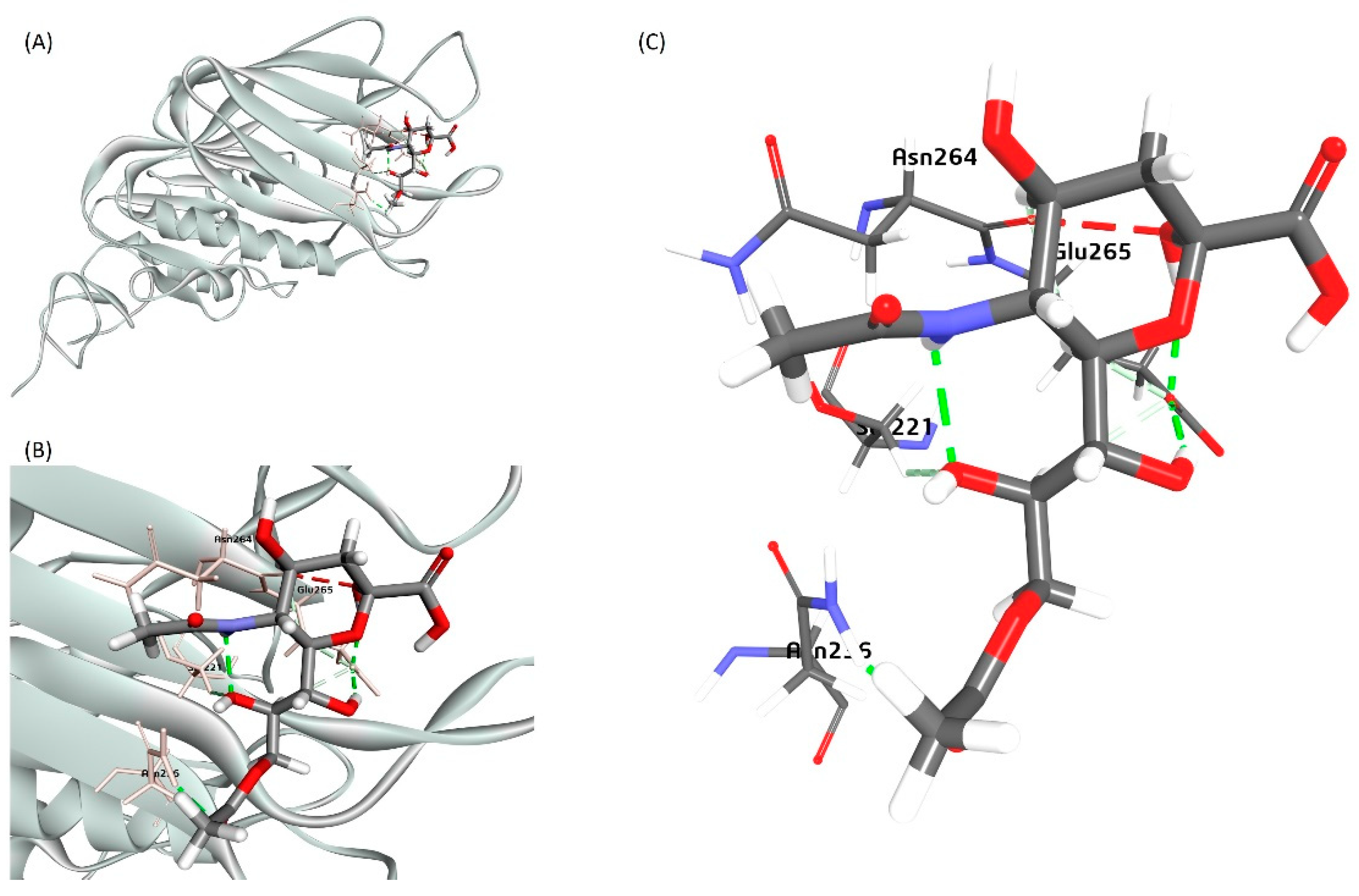
Most of the Betacoronaviruses, including the BCoV isolates, use the 9-O-acetyl-SA as receptors; however, during the evolution of these viruses, some isolates started to recognize other forms of sialic acid (the 4-O-acetyl-SA isoform) [
49]. It was recently shown that the HE gene of the SARS-CoV-2 recognizes and binds to the 9-O-acetyl-SA in contrast to the type-II HE protein of SARS-CoV-2 uses the other isoform; the 4-O-acetyl-SA [
49]. The ligand-interaction sites of the BCoV HE and the subset of coronavirus S glycoproteins evolved to recognize 9-O-acetyl-SA via hydrogen bonding [
50].
In the case of most Betacoronaviruses, the RBD of the CTD or domain of the spike glycoproteins showed the highest variability within S1 subunits across various members of the coronaviruses, including Betacoronaviruses. This phenomenon allows coronaviruses to bind to various types of host cell receptors [
46]. It has been proved that SARS-CoV-2 uses the ACE-2 as a valid receptor for the viral entry into the target host cells [
51]. However, there are no records about the potential roles of ACE2 as receptors for BCoV. In our study, based on the interaction between bovine ACE2 and BCoV /S interaction with its CTD, the bovine cell surface ACE2 could act as a putative receptor for BCoV/S (
Figure 17). This finding is based on the BCoV/S binding to the ACE2 receptor’s tendency to show lower interaction energy [
52]. This claim is also supported by the nature of the interaction energy, which is consistent with the structure of the virus receptor interface [
53]. Additionally, we demonstrated that the expression level of the bovine ACE2 is highly expressed in the MDBK following BCoV infection, whereas the ACE2 gene expression is downregulated in bovine endothelial cells (BEC) under the same infection conditions (
Figure 5D,E).
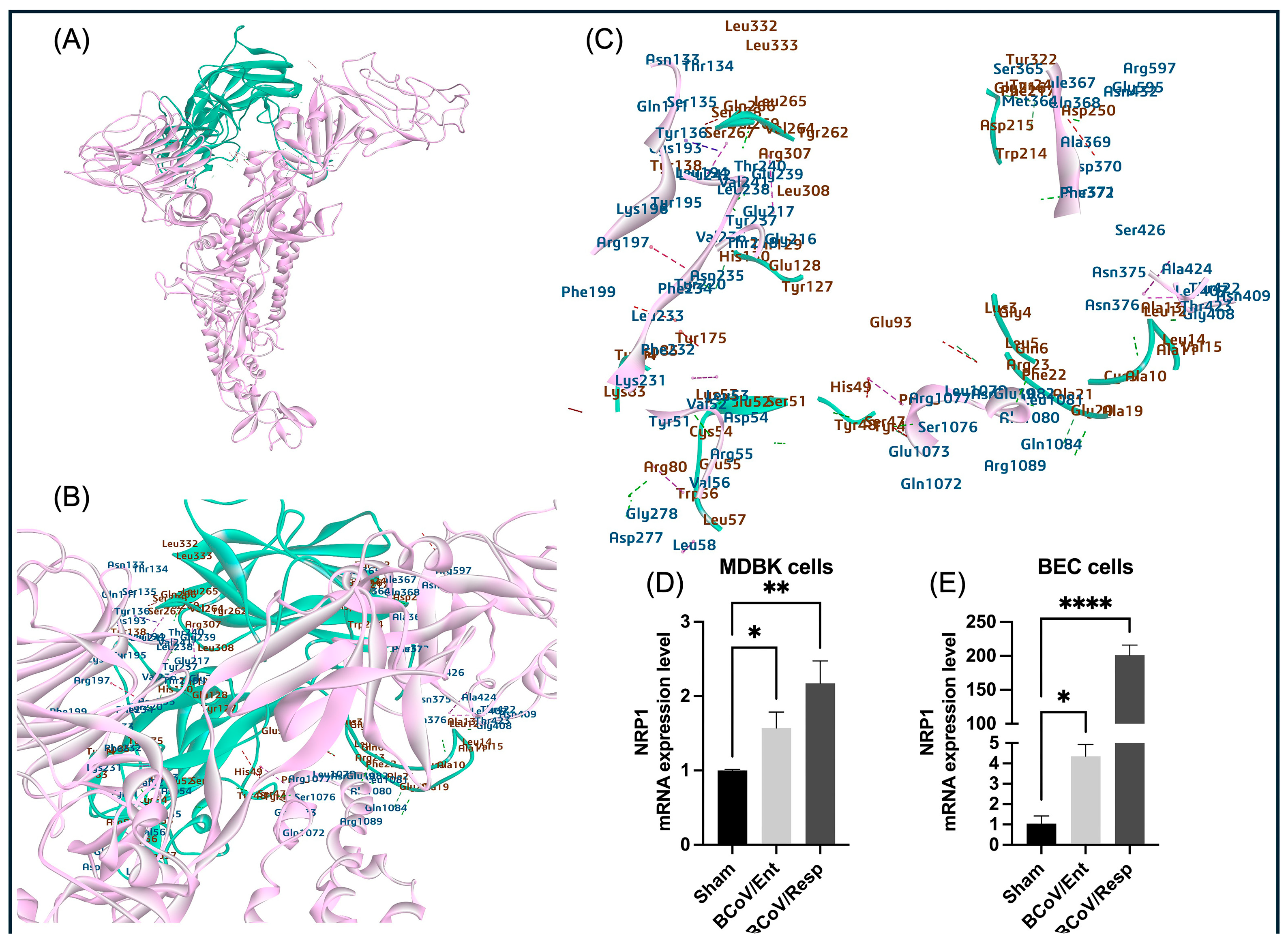
We also investigated the potential roles of Neuropilin-1 (NRP1) as a receptor for the BCoV. Our ZDock method result suggested the high interaction affinity (-22.99 kcal/mol) of NRP1 with BCoV/S-NTD and BCoV/S-CTD region (
Figure 5 and
Figure 16, and
Table 2). Furthermore, the gene expression studies in some bovine cell lines revealed a significant upregulation of the expression levels of the NRP1 following BCoV infection (
Figure 6D,E). This raises the possibility that BCoV could potentially use the NRP receptors for viral entry into the host cell. According to some previous reports, the higher expression of NRP1 will facilitate virus-host cell interactions, especially in cells that do not express other potential BCoV receptors [
54,
55]. Previous research indicates that Neuropilin-1 (NRP-1) exhibits stronger binding affinity to the CTD region of S1, and this interaction stabilizes the folded conformation of the S protein [
56]. Conversely, in the absence of NRP-1, the SARS-CoV-2 S protein tends to stretch and unfold.
It is well known that MHV utilizes the CEACAM1 protein for a dual function. The CEACAM-1 acts as a functional receptor for MHV in addition to enhancing the activation of the MHV-S glycoprotein by inducing some conformational changes, allowing the fusion of the virus with the target cells [
57]. However, there is no data about the potential roles of CEACAM1 in the BCoV replication. Our in-silico data showing low binding affinity interaction between BCoV/S and CEACAM1 suggested that CEACAM1 interacted with low affinity to the NTD of BCoV/S protein (
Figure 17). The gene expression analysis in BCoV-infected bovine cells revealed downregulation of CEACAM1 gene expression levels in the case of the BCov/Ent isolate infected cells, whereas a significant upregulation in the CEACAM1 gene expression levels was observed in the case of the BCoV/Resp infected cells (
Figure 2 and
Figure 7D,E). These findings indicate that CEACAM1 activation is specific to the BCoV/Resp isolate infection. The variations in the binding of coronaviruses to CEACAM1 can be directly attributed to the structural differences between their N-terminal domains, particularly those of BCoV and MHV. Unlike MHVs sugar-binding ancestors, contemporary MHVs utilize their NTDs for specific interaction with the host cell protein CEACAM1 [
58].
The aminopeptidase N (APN), a widely found enzyme on cell surfaces, is involved in various cellular processes like survival, migration, blood pressure regulation, and even virus uptake [
59]. Coronaviruses like the porcine delta coronavirus (PDCoV) can utilize APN as a receptor to enter cells, highlighting APN’s potential role as a viral infection gateway from different host species [
60]. Aminopeptidase N from porcine functions as a receptor for the enveloped RNA virus TGEV [
12]. This underscores the wide variety of membrane-bound proteins viruses exploit to infiltrate cells. The APN is a common receptor for the members of the alpha coronaviruses, particularly the HCoV-229E and the TGEV in pigs [
61,
62,
63]. Based on our protein-protein interaction (E_RDock), the interaction energy of the best pose of APN interaction with BCoV/S shows low-affinity interaction (-2.96 kcal/mol). This result confirms that the binding interaction of BCOV/S is not strong, and it shows a putative binding with the APN. The RBD of CTD from BCoV/S is involved in interaction with Bovine APN (
Figure 17). Meanwhile, the host APN gene was not expressed in MDBK cells following BCoV infection, but the limited expression of the APN in the BCoV infected BEC cells (
Figure 8D,E). This pattern of APN expression suggesting a subtile expression of APN in response to BCoV infection in the BEC.
The DPP4 is a cell surface protease that exhibits exopeptidase activity and is expressed on the surface of various cell types, including those found in the human airways [
64]. Researchers have compared the potential binding interactions between the receptor binding domain (RBD) of different spike variants of SARS-CoV-2 and DPP4 with the interactions observed in the experimentally determined structure of the MERS-CoV complex with DPP4 [
65]. Members of the Betacoronaviruses, especially the SARS-CoV-2 and the MERS-CoV, utilize the ACE2 and DPP4 as receptors, respectively [
66]. The pathogenicity of MERS-CoV is caused by the specific binding of its S1B domain or S1-CTD to the human DPP4 receptor [
67]. In our study, docking of the BCoV spike with the DPP4 shows low interaction affinity of the DPP4 towards the BCoV/S (E_R Dock score -6.23 kcal/mol). However, the binding of DPP4 with BCoV/S1-CTD region or S1B domain is correlated with MERS-CoV binding of its S1B domain or S1-CTD to the human DPP4 (
Figure 17). Additionally, the DPP4 gene expression level was noticed to be specific to the BCoV/Resp infected MDBK cells, while in the case of the BEC cells, it was only expressed in the BCoV/Ent infected cells (
Figure 9D,E). This indicates an inconsistent pattern of bovine DPP4 expression in response to BCoV infection. This finding suggests that DPP4 is not a strong candidate receptor for the BCoV.
The AXL is a member of the TAM tyrosine kinase receptor family and appears to be the most efficacious at mediating viral entry, given their prevalent use among enveloped viruses [
68]. The AXL receptor is found on the human cell membrane and contributes to the pathogenesis of SARS-CoV-2 infection [
69]. The AXL has been reported to bind to the NTD of the spike protein S1 subunit and could synergistically work with the ACE2 to facilitate SARS-CoV-2 entry into the target cells [
70]. In our study, the NTD of the BCoV/S is predicted to be involved in the interaction with Bovine AXL (
Figure 17). The AXL shows high binding affinity with NTD of the BCoV/S due to multiple non-covalent interactions, suggesting the possibility of its role in infection with its potential synergistic effect with other receptors such as ACE2 as shown in the context of the SARS-CoV-2 infection [
42]. The AXL gene expression profiles in the bovine cell lines infected with BCoV support our in-silico prediction and modeling. However, it indicates that the AXL gene expression level is upregulated specifically during the infection with the BCoV/Resp isolate (
Figure 2 and
Figure 10D,E). Therefore, this AXL could be a strong candidate receptor, especially in the case of BCoV/Resp infection.
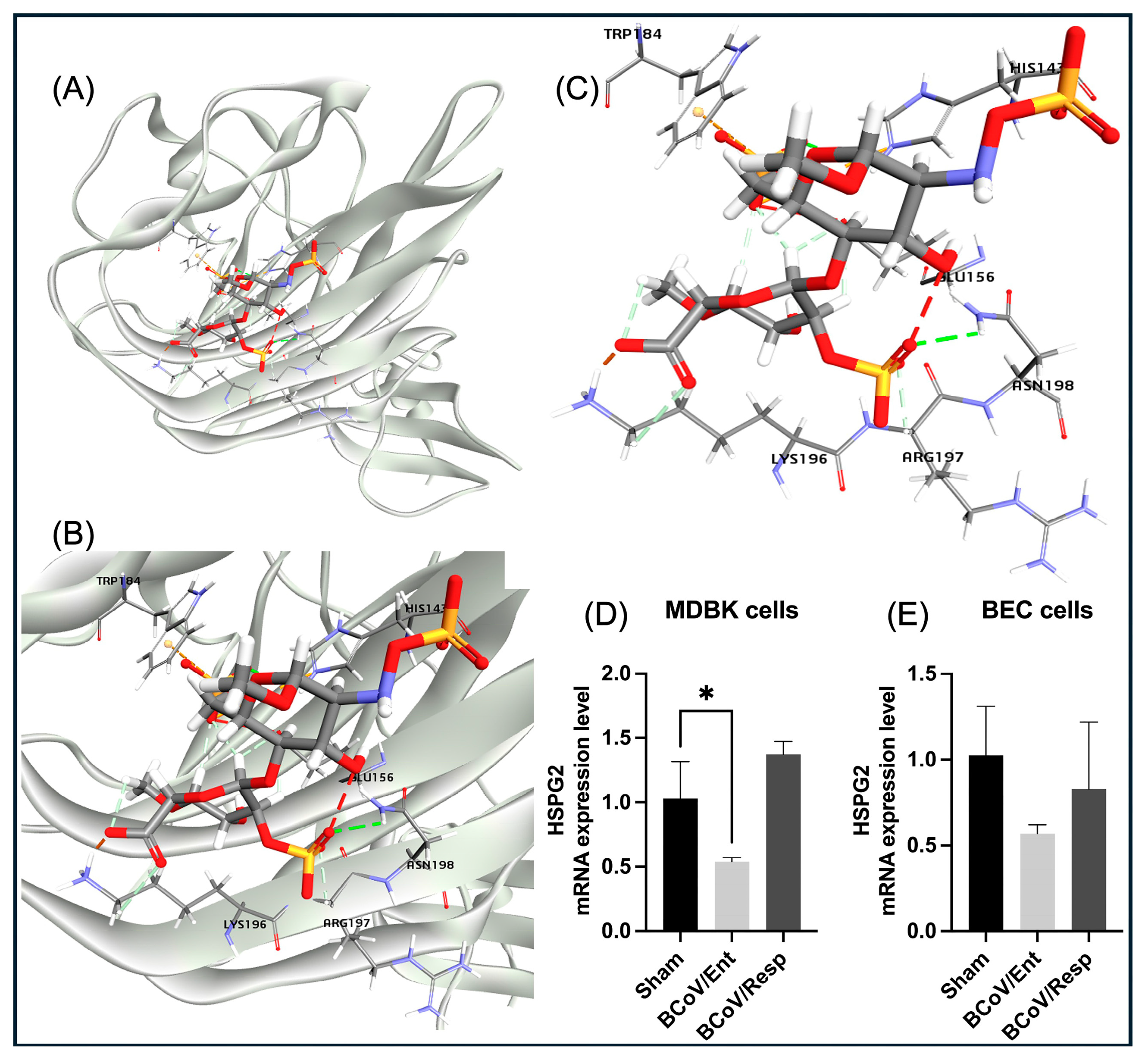
The Heparan sulfate proteoglycans of the HSPG2 play important roles during the entry of many viruses to the target cells [
71]. Glycan-binding may act as the initial step for cellular attachment, bringing the virus close to the epithelial cell membrane where it can interact with its protein receptor(s). The general mechanism appears conserved among coronaviruses (CoVs) such as MERS-CoV, SARS-CoV-1, and HCoV-NL63, which are related to SARS-CoV-2 [
71,
72]. The spike protein initially interacts with cell surface HS, resulting in an increase in the expression levels of the RBDs in the “up/open” conformation, which in turn leads to enhanced binding affinities to the bovine ACE2 receptors and subsequent stabilization of the interaction of the trimer and the virion with cells [
73]. The HS captures SARS-CoV-2 virions, increasing the local viral concentration and presentation of these virions to the ACE2 receptores and to the other proteinaceous receptors as well [
74]. Our in silico and molecualr docking results are showing the potential roles of the BCoV/S/NTD in the interaction with the HS of the HSPG2 (
Figure 17). The HS shows high binding energy with NTD of BCoV/S due to multiple non-covalent interactions such as hydrogen and pi-bonds, suggesting the possibility of its role in infection with its synergistic effect with other receptors such as ACE2. The BCoV/S-HS interaction could interact with its other cellular receptors, such as ACE2, may cause changes to the cell surface, which could lead to infection of the host cells. However, our results showed the downregulation of the HSPG2 gene expression, especially during the BCoV/Ent infection (
Figure 2 and
Figure 11D,E). Therefore, HS binding inhibition could also be a novel antiviral therapeutic for inhibiting BCoV infection in cattle.
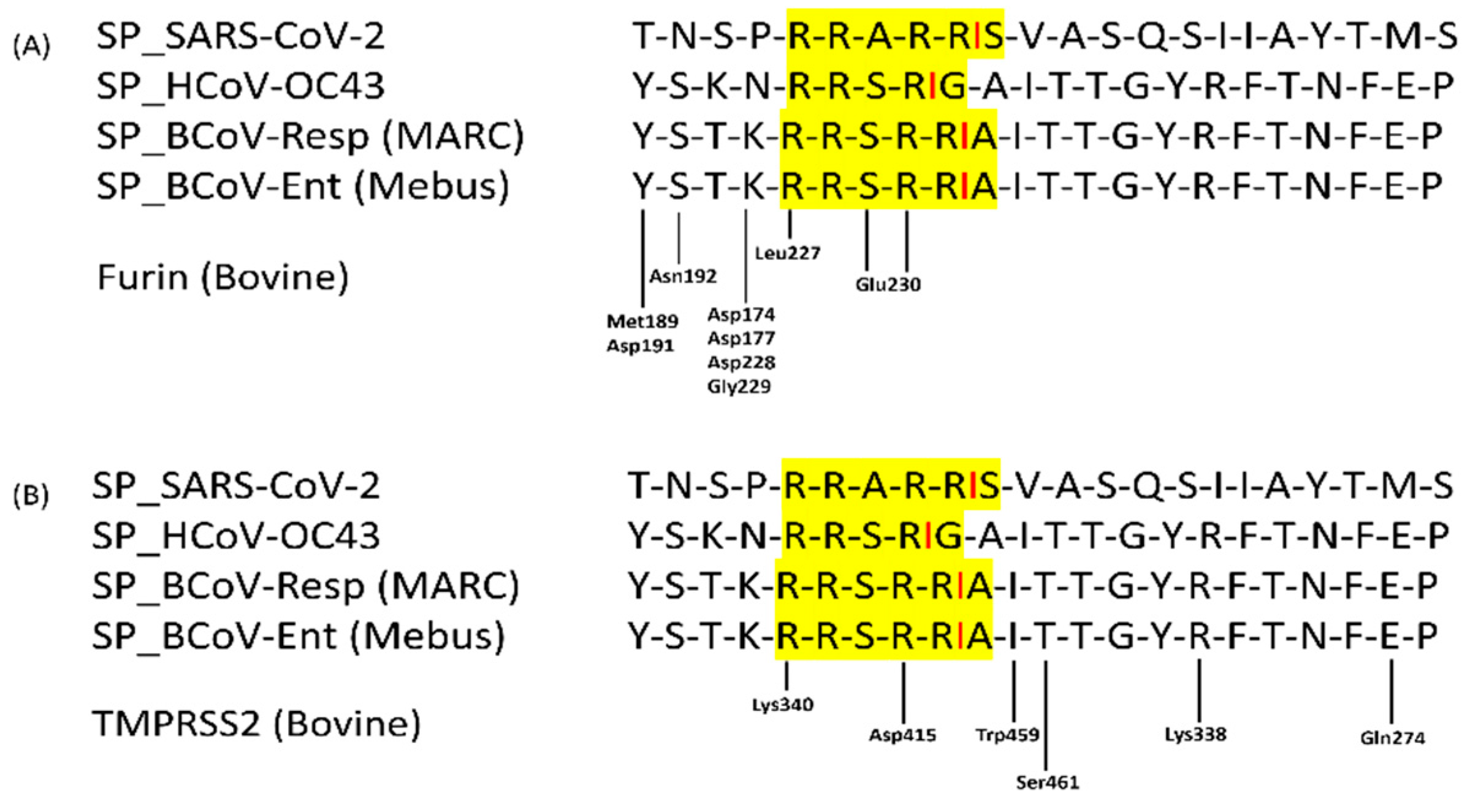
Previous studies on SARS-CoV-2, MERS-CoV, and other coronaviruses have shown that activation of the spike protein is often a complex process involving multiple cleavage events occurring at distinct sites and with the involvement of several host proteases [
75,
76]. For virus entry and infections, proteolytic cleavage is widely used to activate the fusion machinery of viral glycoproteins. Furin binding to RRSRR (Arginine-Arginine-Serine-Arginine-Arginine) site of BCoV spike protein, which is related to the conserved cleavage site of SARS-CoV-2 cleavable PRRAR|S residues at receptor-binding (S1) and fusion (S2) domains of the spike protein [
77]. Such a motif may allow Spikes to be cut into S1 and S2 by Furin and TMPRSS2-like proteases before maturity, which provides S1 with the flexibility to change the conformation to better fit the host receptor. The arginine residues at the SARS-CoV-2 spike protein catalytic site are popped out of the closed state of the S protein to form multiple non-covalent interactions with the furin [
78]. In our study, the BCoV/S interaction with the furin shows that furin binds near the spike protein S1/S2 cleavage RRSRR site specific for Furin and TMPRSS2 proteases (
Figure 17). Our gene expression data strongly supports the in-silico prediction of host cell Furin targeting the BCoV-S glycoprotein; it is also shows that host Furin is highly expressed during BCoV infection, with particular upregulation of the Furin expression observed in response to BCoV/Resp isolate infection in bovine cells (
Figure 2 and
Figure 13D,E). Our recent study shows the host cell miRNA-16a targeting the host cell furin, limiting the BCoV replication and enhancing the host immune response [
79]. The TMPRSS2 has been shown to proteolytically activate the S glycoprotein of many coronaviruses, including SARS-CoV-2, SARS-CoV-1, MERS-CoV, 229E, as well as influenza virus [
80,
81,
82]. The TMPRSS2 triggers HKU1-mediated cell-cell fusion and viral entry and binds with high affinity to both HKU1A and HKU1B RBDs [
83].
Our modeling results show that furin binds firmly with the S protein RRSRR site, which is conserved in BCoV as RRSRR at the S1/S2 site, whereas the TMPRSS2 cleaves the S protein in the lungs of SARS-CoV-2 infected person and promotes pathogenicity [
84]. SARS-CoV-2-S glycoprotein (S1/S2) is cleaved by furin protease, and subsequently, the TMPRSS2 mediates the cleavage and activation of the S2 region on the S2 protein [
85]. The TMPRSS2, which colocalizes with the ACE2 at the cell membrane, has been identified as the dominant proteolytic driver of the SARS-CoV-2/S protein activation that potentiates the viral infection in the aerodigestive tract [
86]. The TMPRSS2 protease can be opted as a potential therapeutic target for bovine host-specific viruses. In this study, TMPRSS2 binds near the S1/S2 junction RRSRR protease cleavage-specific region for furin and TMPRSS2 proteases. The in-vitro results also demonstrated high expression levels of the TMPRSS2especaiaaly during the BCoV/Ent isolate infection in the bovine cells (
Figure 14D,E). This finding strongly suggests the involvement of the TMPRSS2 in the entry of BCoV enteric isolate into the host cell.
The CTS-L is a member of the lysosomal cysteine protease family [
17]. The expression level of CTS-L mRNA is higher than ACE2, FURIN, and TMPRSS2 in human lung tissues during SARS-CoV-2 replication [
87]. In the context of SARS-COV-2, the S protein contains two highly conserved CTS-L cleavage sites (CS-1 and CS-2) among all known SARS-CoV-2 variants. CTS-L cleavage promoted S to adopt receptor-binding domain (RBD) “up” activated conformations, facilitating receptor-binding and membrane fusion [
17]. The highly conserved CS-2 site seems to be the most essential for the life cycle of SARS-CoVs, while CS-1 is likely to play an auxiliary role in virus infection. However, with the help of multiple sequence alignment, it has been observed that the BCoV/S has no conserved region for CTS-L cleavage site as the SARS-CoV-2 sequence of the S chain. On the basis of our protein-protein interaction result, cathepsin showed binding to BCoV/S in NTD and downstream of the S1/S2 region of CTD of the S chain (
Figure 17). However, our gene expression analysis of the host genes during BCoV infection in some bovine cells showed the down-regulation of the CST-L expression upon BCoV infection (
Figure 15D,E). To understand the catalytic role of bovine cathepsin towards BCoV/S glycoprotein, the interaction of BCoV/S with cathepsin could be further investigated in vitro.
Therefore, there is limited knowledge about the bovine-specific virus-host interactions that determine cellular entry of SARS-CoV-2. Viruses display considerable redundancy and flexibility because they can exploit weak multivalent interactions to enhance affinity. To date, studies of BCoV entry have focused almost entirely on ACE2 and Neu5,9Ac2. In this study, we aimed to see the interaction of bovine cell surface molecules as receptors for BCoV/S, including Nue5,9Ac2, NRP1, DPP4, CEACAM-1, APN ACE2, AXL, HS of HSPG2, TMPRSS2, and Furin shown in
Figure 17. The results from our study suggested that NRP1 and AXL showed greater binding affinity towards BCoV/S other than receptor Neu5,9Ac2 and HS. However, bovine HE also showed a stronger binding affinity with Neu5,9Ac2. Our in silico structural interaction data provide a blueprint for understanding the BCoV specificity for the different bovine receptors.
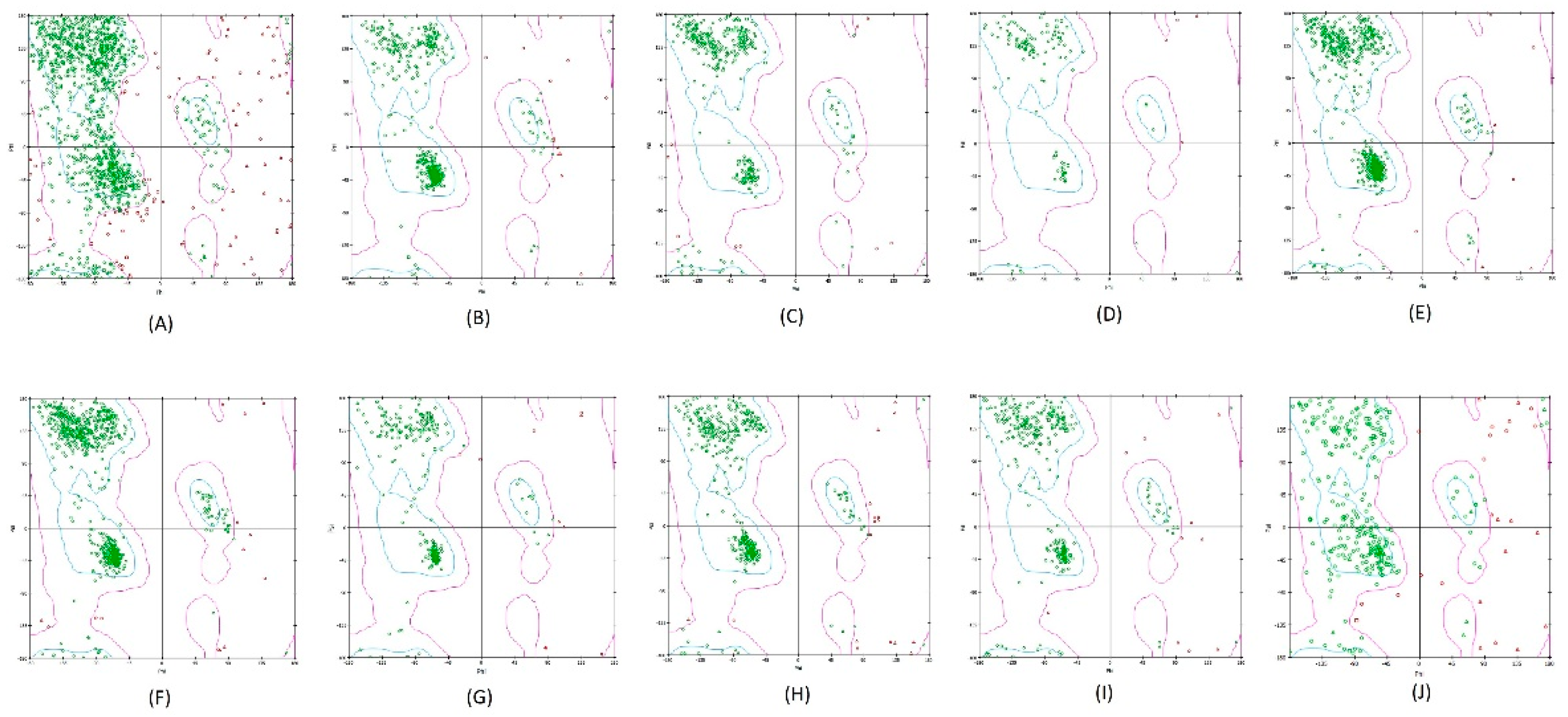
Figure 1.
The validity and stability of the homology-modeled protein structure were determined using the Ramachandran plot. (A) BCoV-Spike (B) Bovine ACE2 (C) Bovine NRP1 (D) Bovine CEACAM-1 (E) Bovine APN (F) Bovine DPP4 (G) Bovine AXL (H) Bovine Furin (I) Bovine TMPRSS2 (J) CTS-L.
Figure 1.
The validity and stability of the homology-modeled protein structure were determined using the Ramachandran plot. (A) BCoV-Spike (B) Bovine ACE2 (C) Bovine NRP1 (D) Bovine CEACAM-1 (E) Bovine APN (F) Bovine DPP4 (G) Bovine AXL (H) Bovine Furin (I) Bovine TMPRSS2 (J) CTS-L.
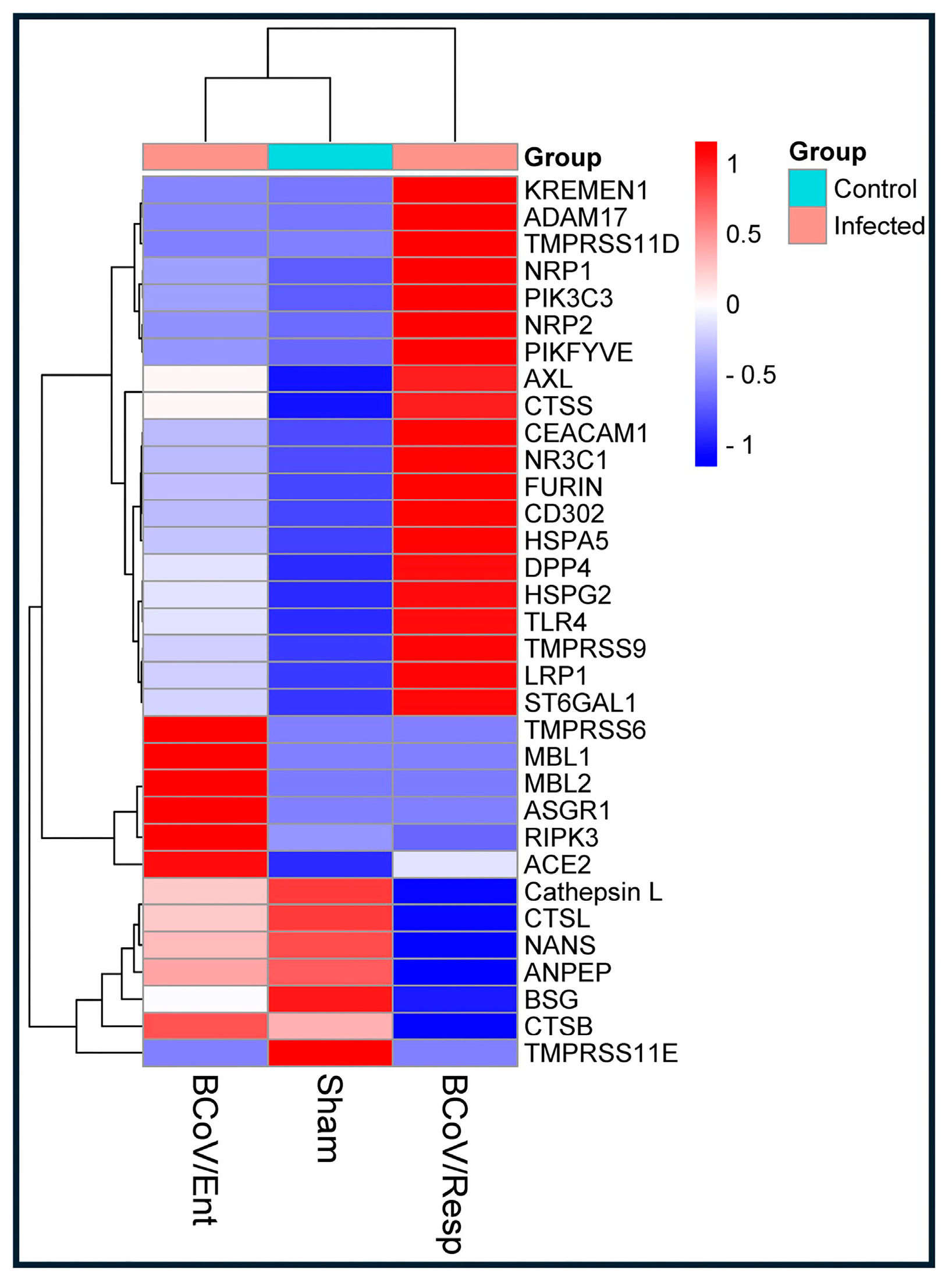
Figure 2.
The expression profiles of some known Coronavirus Receptors detected during the BCoV/Ent or BCoV/Resp Infection in Bovine Cell lines. Based on the NGS data analysis, The heatmap shows the differential expression profiles of some coronavirus-related receptors in the control (sham), BCoV/Ent, and BCoV/Resp infected groups of BEC cells.
Figure 2.
The expression profiles of some known Coronavirus Receptors detected during the BCoV/Ent or BCoV/Resp Infection in Bovine Cell lines. Based on the NGS data analysis, The heatmap shows the differential expression profiles of some coronavirus-related receptors in the control (sham), BCoV/Ent, and BCoV/Resp infected groups of BEC cells.
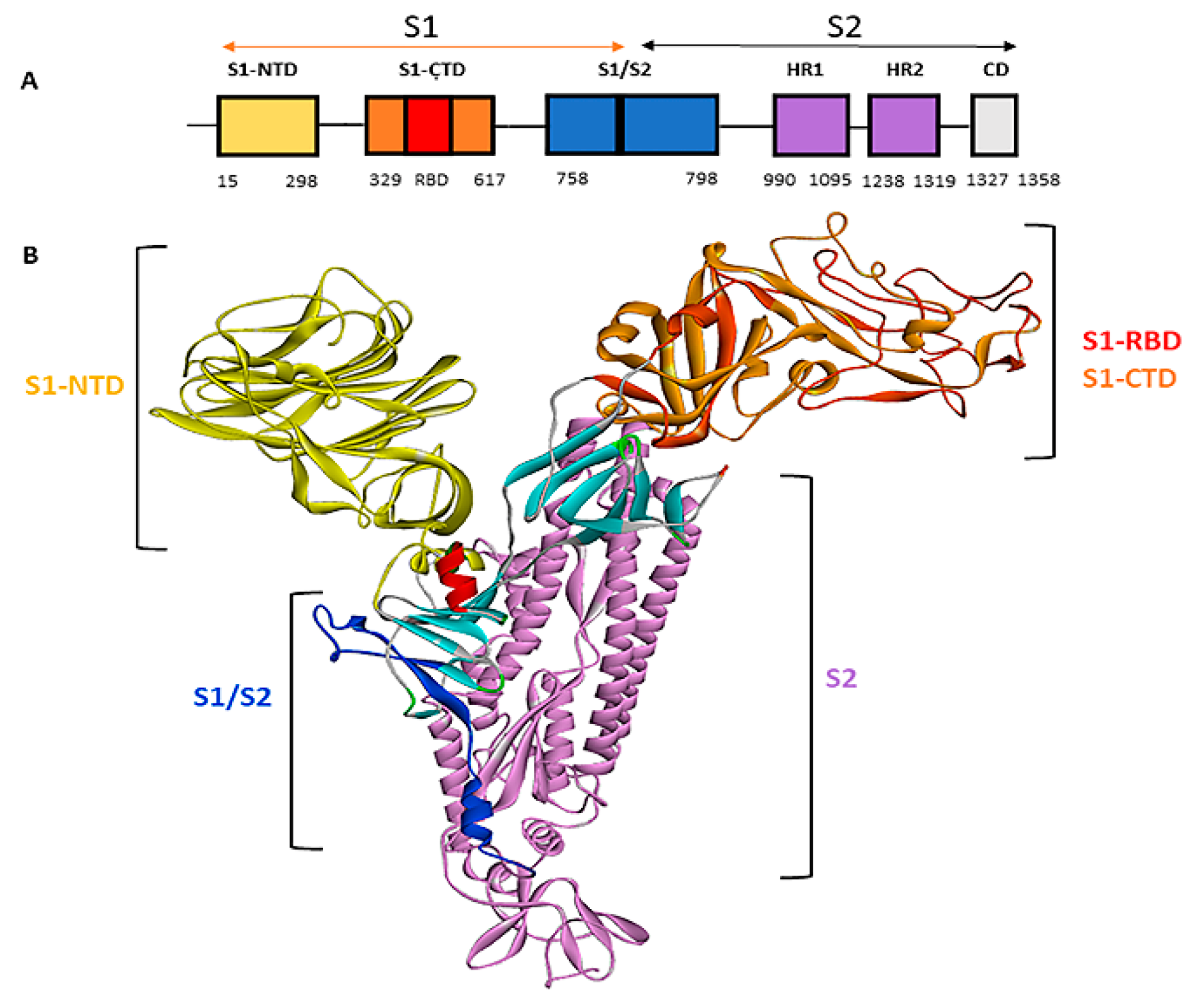
Figure 3.
Schematic representation of the BCoV/S Protein: S1 and S2 Chains (A) The listed domain boundaries are mostly defined as S-NTD, N-terminal Domain; RBD, Receptor Binding Domain; S1-CTD, C-Terminal domain; S2-HR1, Heptad Repeat 1; S2-HR2, Heptad Repeat 2; S2-CD, Cytoplasm Domain. (B) Schematic drawing of the three-dimensional structure of BCoV/S protein showing different domains on S1 and S2 Chains.
Figure 3.
Schematic representation of the BCoV/S Protein: S1 and S2 Chains (A) The listed domain boundaries are mostly defined as S-NTD, N-terminal Domain; RBD, Receptor Binding Domain; S1-CTD, C-Terminal domain; S2-HR1, Heptad Repeat 1; S2-HR2, Heptad Repeat 2; S2-CD, Cytoplasm Domain. (B) Schematic drawing of the three-dimensional structure of BCoV/S protein showing different domains on S1 and S2 Chains.
Figure 4.
Sialic acid (Neu5,9Ac2) as the receptor for BCoV Spike protein at the NTD site. (A) Neu5,9Ac2 binding to the N-terminal domain (NTD) of the BCoV Spike protein is illustrated. (B) and (C) Detailed views of the interactions between Neu5,9Ac2 and the specific amino acid residues at the NTD site of the BCoV Spike protein: His185, Asp187, Gly189, and Lys196.
Figure 4.
Sialic acid (Neu5,9Ac2) as the receptor for BCoV Spike protein at the NTD site. (A) Neu5,9Ac2 binding to the N-terminal domain (NTD) of the BCoV Spike protein is illustrated. (B) and (C) Detailed views of the interactions between Neu5,9Ac2 and the specific amino acid residues at the NTD site of the BCoV Spike protein: His185, Asp187, Gly189, and Lys196.
Figure 5.
The proposed Model for the BCoV/S glycoprotein interaction with the bovine ACE2 and the confirmation of the ACE2 expression profile in BCoV-infected cells (A) The BCoV Spike protein interacting with ACE2, showing possible binding interactions at the receptor-binding domain (RBD) of the C-terminal domain (CTD) of the S1 protein chain. (B) and (C) Detailed views of the protein-protein interaction interface, highlighting the amino acid residues involved in the interaction between ACE2 and the N-terminal domain (NTD) of the BCoV Spike protein. ACE2 interaction residues: Ser19, Thr20, Thr21, Glu23, Gln24, Thr27, Glu30, Lys81, Thr82, and other residues. BCoV Spike NTD interaction residues: Tyr661, Asp662, Ser663, Gly665, Asn666, Ser611, Thr425, Arg419, Ala527, and other residues. (D) Compared to the sham, the ACE2 expression levels in the MDBK cells infected with BCoV/Ent or BCoV/Resp isolates were analyzed by qRT-PCR. (E) Compared to the sham, the ACE2 mRNA expression levels in BEC cells infected with BCoV/Ent or BCoV/Resp isolates were analyzed by qRT-PCR. Cells were infected with 1 MOI of either BCoV/Ent or BCoV/Resp isolate for 72 hpi and used for qRT-PCR analysis.
Figure 5.
The proposed Model for the BCoV/S glycoprotein interaction with the bovine ACE2 and the confirmation of the ACE2 expression profile in BCoV-infected cells (A) The BCoV Spike protein interacting with ACE2, showing possible binding interactions at the receptor-binding domain (RBD) of the C-terminal domain (CTD) of the S1 protein chain. (B) and (C) Detailed views of the protein-protein interaction interface, highlighting the amino acid residues involved in the interaction between ACE2 and the N-terminal domain (NTD) of the BCoV Spike protein. ACE2 interaction residues: Ser19, Thr20, Thr21, Glu23, Gln24, Thr27, Glu30, Lys81, Thr82, and other residues. BCoV Spike NTD interaction residues: Tyr661, Asp662, Ser663, Gly665, Asn666, Ser611, Thr425, Arg419, Ala527, and other residues. (D) Compared to the sham, the ACE2 expression levels in the MDBK cells infected with BCoV/Ent or BCoV/Resp isolates were analyzed by qRT-PCR. (E) Compared to the sham, the ACE2 mRNA expression levels in BEC cells infected with BCoV/Ent or BCoV/Resp isolates were analyzed by qRT-PCR. Cells were infected with 1 MOI of either BCoV/Ent or BCoV/Resp isolate for 72 hpi and used for qRT-PCR analysis.
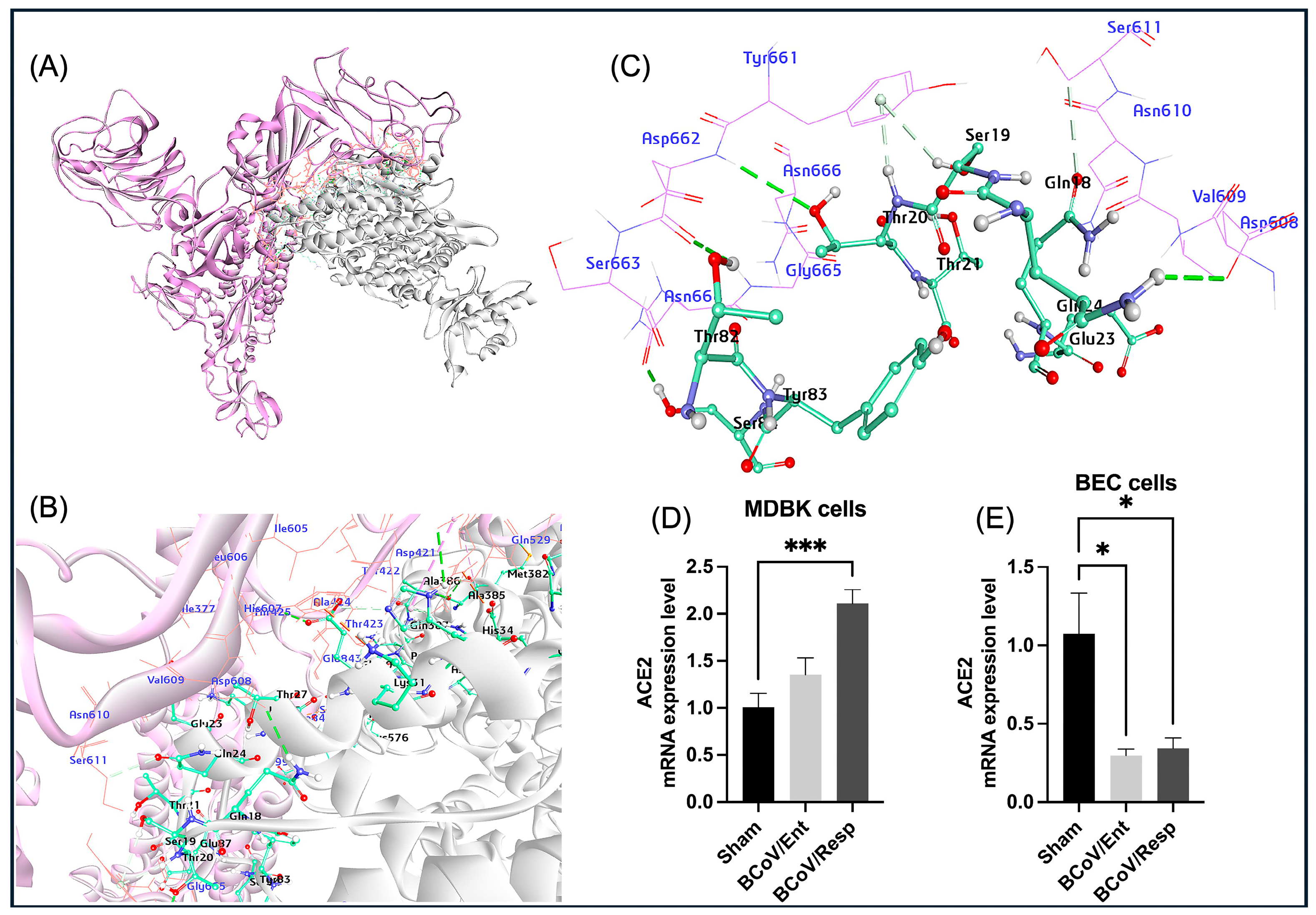
Figure 6.
The proposed model for the interaction between the bovine NRP1 and the BCoV/S glycoprotein and confirming the bovine NPR1 during BCoV infection in MDBK and BEC. (A) NRP1 interactions with the C-terminal domain (CTD) and N-terminal domain (NTD) of the S1 protein, illustrating possible binding conformations. (B) and (C) Specific NRP1 residues (Leu322, Leu333, Val269, Arg307, Tyr138, Gln266) and their binding affinities to amino acid residues in the NTD and CTD of the S1 protein. (D) The NRP1 expression levels in the MDBK cells infected with the BCoV/Ent or BCoV/Resp isolates, compared to the sham, were analyzed by the qRT-PCR. (E) The NRP1 expression levels in the BEC cells infected with the BCoV/Ent or the BCoV/Resp isolates, compared to the sham, were analyzed by the qRT-PCR. Both cell lines were infected with (MOI =1) of either BCoV/Ent or BCoV/Resp isolates for 72 hpi and used for qRT-PCR analysis.
Figure 6.
The proposed model for the interaction between the bovine NRP1 and the BCoV/S glycoprotein and confirming the bovine NPR1 during BCoV infection in MDBK and BEC. (A) NRP1 interactions with the C-terminal domain (CTD) and N-terminal domain (NTD) of the S1 protein, illustrating possible binding conformations. (B) and (C) Specific NRP1 residues (Leu322, Leu333, Val269, Arg307, Tyr138, Gln266) and their binding affinities to amino acid residues in the NTD and CTD of the S1 protein. (D) The NRP1 expression levels in the MDBK cells infected with the BCoV/Ent or BCoV/Resp isolates, compared to the sham, were analyzed by the qRT-PCR. (E) The NRP1 expression levels in the BEC cells infected with the BCoV/Ent or the BCoV/Resp isolates, compared to the sham, were analyzed by the qRT-PCR. Both cell lines were infected with (MOI =1) of either BCoV/Ent or BCoV/Resp isolates for 72 hpi and used for qRT-PCR analysis.
Figure 7.
The proposed model for the interaction between the bovine CEACAM-1 protein and the BCoV-S glycoprotein, mapping the interactive amino acid residues and confirming the bovine CECAM-1 expression during BCoV infection in some bovine cells by the qRT-PCR. (A) The bovine CEACAM-1 protein interacts with the BCoV/S-NTD. (B) and (C) CEACAM-1 binds through LEU233, PHE232, VAL229, Phe232, THR145, ASP148, and other amino acids with NTD of spike glycoprotein with residues ARG143, ASN170, THR171, ASN178, LYS196, LYS196 with hydrogen and Pi bond interactions. (D) Compared to the sham, the bovine CEACAM-1 gene expression levels in the MDBK cells infected with either the BCoV/Ent or BCoV/Resp isolates were analyzed by qRT-PCR. (E) Compared to the sham, the bovine CEACAM-1 gene expression levels in the BEC cells infected with either the BCoV/Ent or the BCoV/Resp isolates were analyzed by qRT-PCR. Both cell lines were infected with (MOI =1) either BCoV/Ent or BCoV/Resp isolates for 72 hpi and used for the qRT-PCR analysis.
Figure 7.
The proposed model for the interaction between the bovine CEACAM-1 protein and the BCoV-S glycoprotein, mapping the interactive amino acid residues and confirming the bovine CECAM-1 expression during BCoV infection in some bovine cells by the qRT-PCR. (A) The bovine CEACAM-1 protein interacts with the BCoV/S-NTD. (B) and (C) CEACAM-1 binds through LEU233, PHE232, VAL229, Phe232, THR145, ASP148, and other amino acids with NTD of spike glycoprotein with residues ARG143, ASN170, THR171, ASN178, LYS196, LYS196 with hydrogen and Pi bond interactions. (D) Compared to the sham, the bovine CEACAM-1 gene expression levels in the MDBK cells infected with either the BCoV/Ent or BCoV/Resp isolates were analyzed by qRT-PCR. (E) Compared to the sham, the bovine CEACAM-1 gene expression levels in the BEC cells infected with either the BCoV/Ent or the BCoV/Resp isolates were analyzed by qRT-PCR. Both cell lines were infected with (MOI =1) either BCoV/Ent or BCoV/Resp isolates for 72 hpi and used for the qRT-PCR analysis.
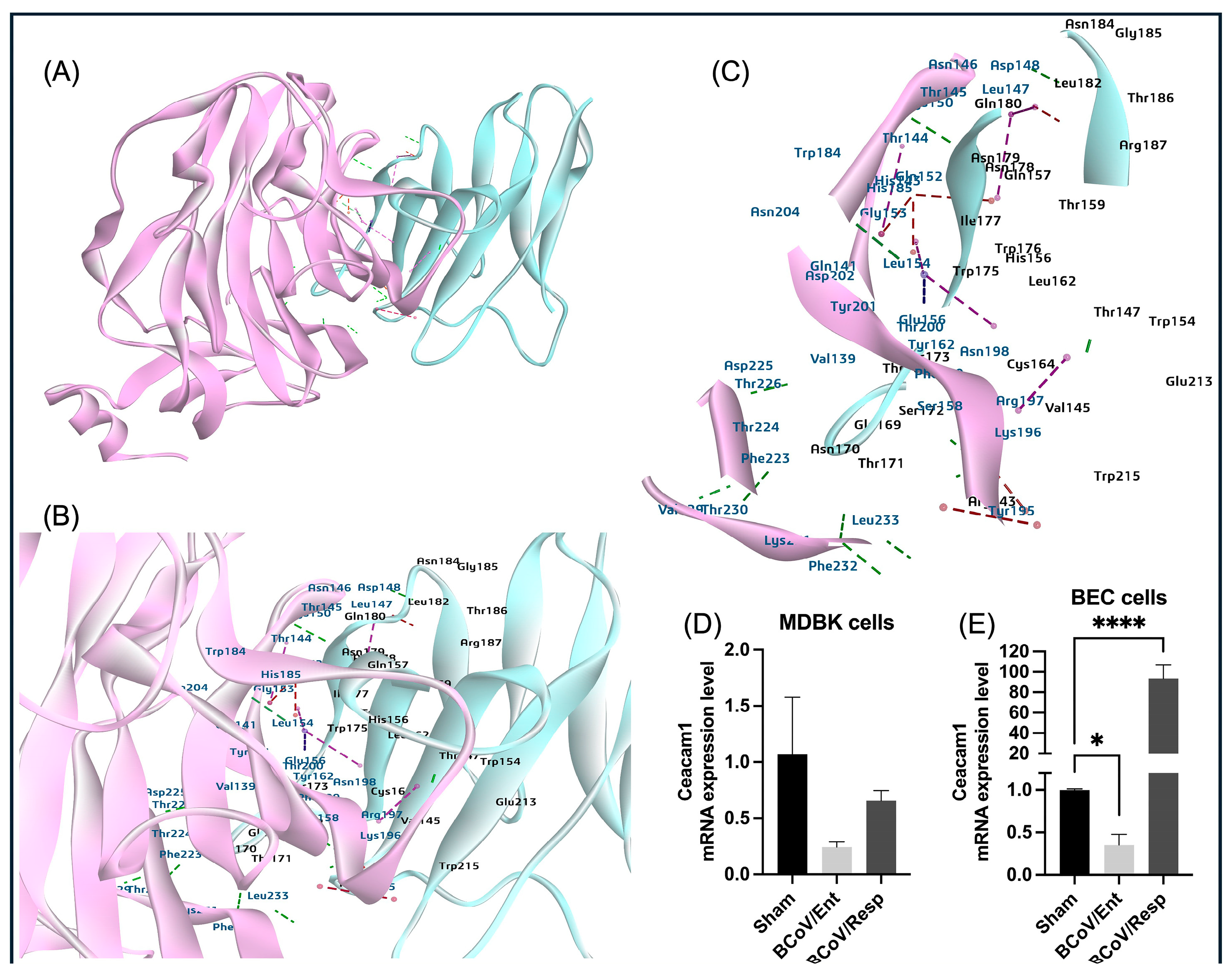
Figure 8.
The proposed model for the interaction between the BCoV/S glycoprotein and the bovine aminopeptidase N (APN) protein and the conformation of the APN expression levels during the BCoV infection in some bovine cell lines. (A) The APN interactions with BCoV/S1 RBD and CTD (B) (C) BCoV/S1 chain CTD residues binding to the bovine APN through the residues (TYR412, PHE416, HIS478, CYS524, ILE512, GLY511, THR514, CYS515, PRO516, THR518, ALA517 and GLY518 to amino acid residues ASN495, ALA555, ASP565, GLY575, ASP576, ARG614 and other amino acid residues of APN by hydrogen bond, Pi bond and salt bridge interactions). (D) The APN gene expression in the MDBK cells infected with either the BCoV/Ent or the BCoV/Resp isolates, compared to the sham, was analyzed by the qRT-PCR. (E) The APN gene expression levels in BEC cells infected with BCoV/Ent or BCoV/Resp isolates, compared to the sham, were analyzed by the qRT-PCR. Both cell lines were infected with 1 MOI of either BCoV/Ent or BCoV/Resp isolates for 72 hpi and used for qRT-PCR analysis.
Figure 8.
The proposed model for the interaction between the BCoV/S glycoprotein and the bovine aminopeptidase N (APN) protein and the conformation of the APN expression levels during the BCoV infection in some bovine cell lines. (A) The APN interactions with BCoV/S1 RBD and CTD (B) (C) BCoV/S1 chain CTD residues binding to the bovine APN through the residues (TYR412, PHE416, HIS478, CYS524, ILE512, GLY511, THR514, CYS515, PRO516, THR518, ALA517 and GLY518 to amino acid residues ASN495, ALA555, ASP565, GLY575, ASP576, ARG614 and other amino acid residues of APN by hydrogen bond, Pi bond and salt bridge interactions). (D) The APN gene expression in the MDBK cells infected with either the BCoV/Ent or the BCoV/Resp isolates, compared to the sham, was analyzed by the qRT-PCR. (E) The APN gene expression levels in BEC cells infected with BCoV/Ent or BCoV/Resp isolates, compared to the sham, were analyzed by the qRT-PCR. Both cell lines were infected with 1 MOI of either BCoV/Ent or BCoV/Resp isolates for 72 hpi and used for qRT-PCR analysis.
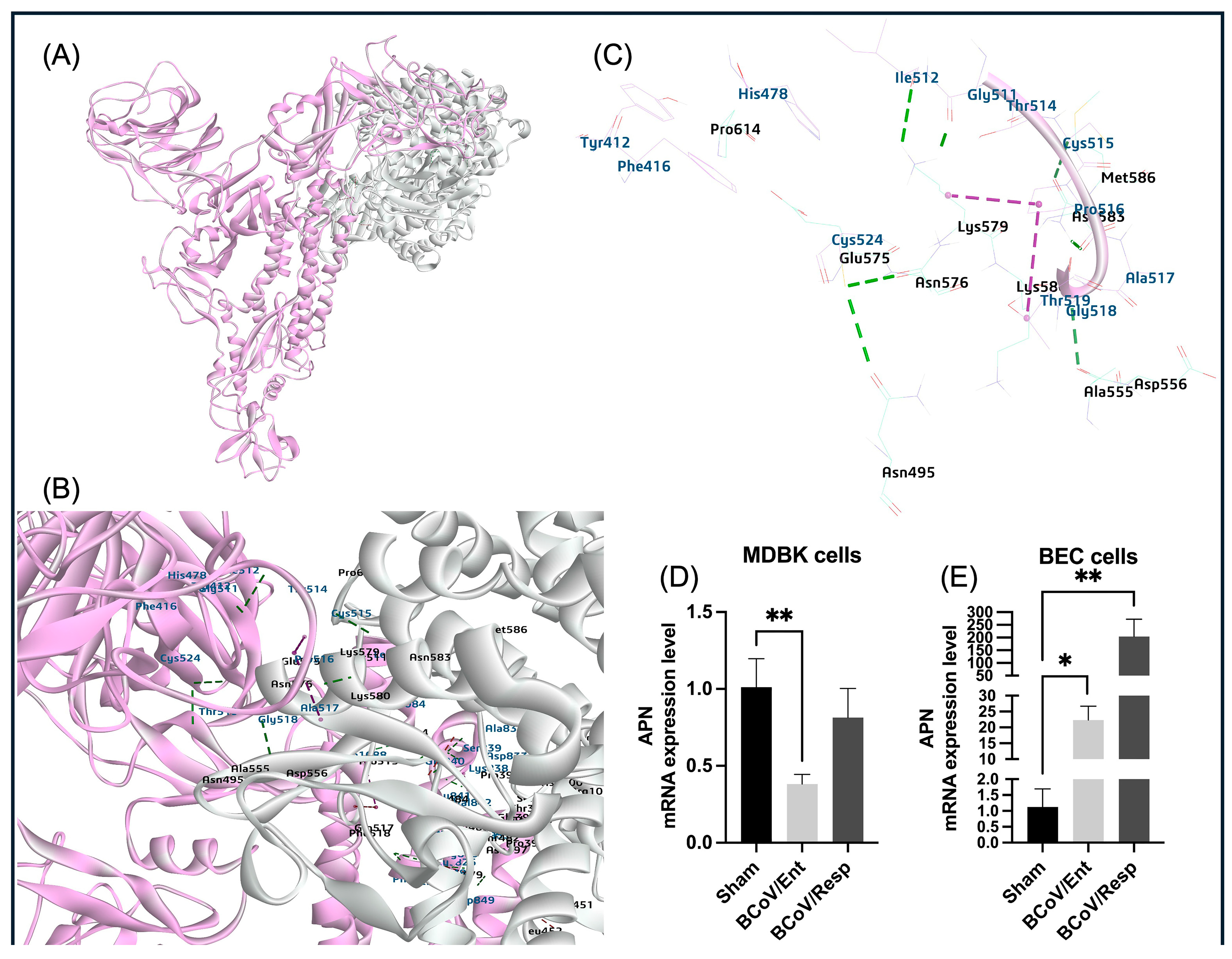
Figure 9.
The proposed model for the interaction between the bovine DPP4 proteins and the BCoV/S glycoprotein and the conformation of the DPP4 expression levels during BCOV infection in some bovine cell lines. (A) The DPP4 protein interacted with the BCoV/S1-NTD protein. (B) and (C) The BCoV/ S1 chain NTD residues bind to the DPP4 were mapped as (TRP184, HIS185, TRP186, ASP187, THR188, GLY189, LYS196) which were the core sites for Neu5, 9Ac2 binding to BCoV/S-NTD. The binding amino acid residues from the DPP4 with the NTD site are (TYR43, VAL441 ARG552 GLY475, LEU476, LYS-512, PHE515, LYS520, HIS532, PHE533, LYS535, MET615, and some other amino acid residues by hydrogen bond, Pi bond and salt bridge interactions). (D) The DPP4 gene expression levels in the MDBK cells infected with BCoV/Ent or BCoV/Resp isolates, compared to the sham, were analyzed by the qRT-PCR. (E) Compared to the sham, the DPP4 gene expression levels in the BEC cells infected with BCoV/Ent or BCoV/Resp isolate were analyzed by the qRT-PCR. Both cell lines were infected with (MOI=1) of either the BCoV/Ent or the BCoV/Resp isolates for 72 hpi and used for the qRT-PCR analysis.
Figure 9.
The proposed model for the interaction between the bovine DPP4 proteins and the BCoV/S glycoprotein and the conformation of the DPP4 expression levels during BCOV infection in some bovine cell lines. (A) The DPP4 protein interacted with the BCoV/S1-NTD protein. (B) and (C) The BCoV/ S1 chain NTD residues bind to the DPP4 were mapped as (TRP184, HIS185, TRP186, ASP187, THR188, GLY189, LYS196) which were the core sites for Neu5, 9Ac2 binding to BCoV/S-NTD. The binding amino acid residues from the DPP4 with the NTD site are (TYR43, VAL441 ARG552 GLY475, LEU476, LYS-512, PHE515, LYS520, HIS532, PHE533, LYS535, MET615, and some other amino acid residues by hydrogen bond, Pi bond and salt bridge interactions). (D) The DPP4 gene expression levels in the MDBK cells infected with BCoV/Ent or BCoV/Resp isolates, compared to the sham, were analyzed by the qRT-PCR. (E) Compared to the sham, the DPP4 gene expression levels in the BEC cells infected with BCoV/Ent or BCoV/Resp isolate were analyzed by the qRT-PCR. Both cell lines were infected with (MOI=1) of either the BCoV/Ent or the BCoV/Resp isolates for 72 hpi and used for the qRT-PCR analysis.
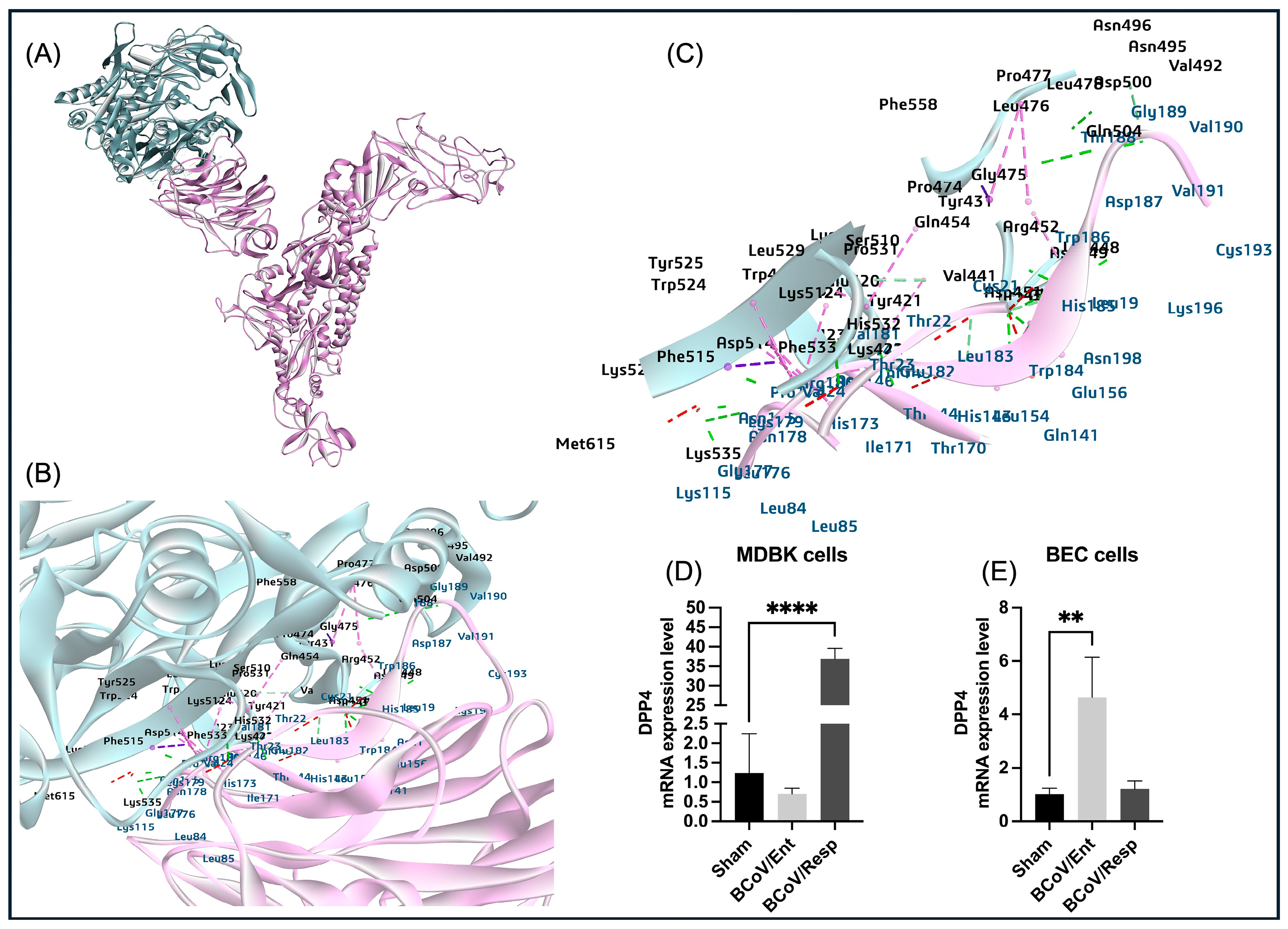
Figure 10.
The proposed model for the interaction between the bovine AXL protein and the BCoV/S glycoprotein and the AXL protein expression levels conformation in the BCoV-infected bovine cells. (A) The AXL protein interacts with the BCoV/S1 protein NTD. (B) and (C) The BCoV/ S1 chain N-terminal domain (NTD) residues binding to the AXL through the amino acids (TYR237, VAL191, ASN198, CYS160, HIS185, GLU182, VAL181, THR188, TYR136, GLN141, GLN161, CYS193, LYS196, GLY189, GLU156 and GLY239). The binding amino acid residues of the AXL to the BCoV/S-NTD sites are (LYS524, SER583, LYS665, TYR697, ASN698, ARG703, ILE707, GLU579, CYS587, LYS694, LYS695, and ARG706). The binding of these residues occurs though the hydrogen bond and Pi bond interactions. (D) The AXL gene expression levels in the MDBK cells infected with BCoV/Ent or BCoV/Resp isolates, compared to the sham, analyzed by the qRT-PCR. (E) The AXL gene expression levels in the BEC cells infected with BCoV/Ent or BCoV/Resp isolates, compared to the sham, analyzed by the qRT-PCR. Both bovine cell lines were infected with either BCoV/Ent or BCoV/Resp isolates (MOI=1) for 72 hpi and used for qRT-PCR analysis.
Figure 10.
The proposed model for the interaction between the bovine AXL protein and the BCoV/S glycoprotein and the AXL protein expression levels conformation in the BCoV-infected bovine cells. (A) The AXL protein interacts with the BCoV/S1 protein NTD. (B) and (C) The BCoV/ S1 chain N-terminal domain (NTD) residues binding to the AXL through the amino acids (TYR237, VAL191, ASN198, CYS160, HIS185, GLU182, VAL181, THR188, TYR136, GLN141, GLN161, CYS193, LYS196, GLY189, GLU156 and GLY239). The binding amino acid residues of the AXL to the BCoV/S-NTD sites are (LYS524, SER583, LYS665, TYR697, ASN698, ARG703, ILE707, GLU579, CYS587, LYS694, LYS695, and ARG706). The binding of these residues occurs though the hydrogen bond and Pi bond interactions. (D) The AXL gene expression levels in the MDBK cells infected with BCoV/Ent or BCoV/Resp isolates, compared to the sham, analyzed by the qRT-PCR. (E) The AXL gene expression levels in the BEC cells infected with BCoV/Ent or BCoV/Resp isolates, compared to the sham, analyzed by the qRT-PCR. Both bovine cell lines were infected with either BCoV/Ent or BCoV/Resp isolates (MOI=1) for 72 hpi and used for qRT-PCR analysis.
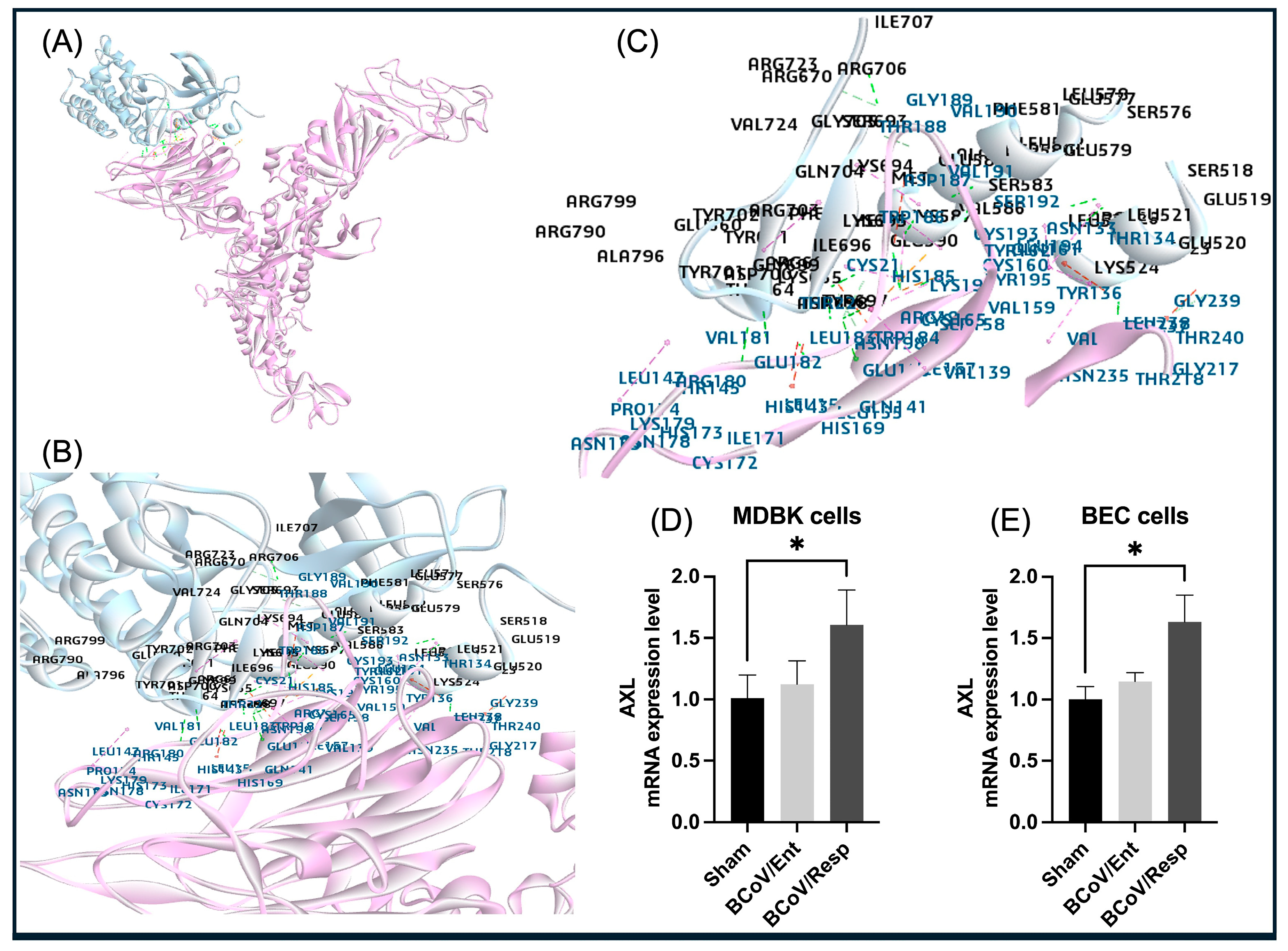
Figure 11.
The proposed model for the bovine heparan sulfate (HS) of HSPG2 interaction with the BCoV/S protein and the conformation of the HSPG2 expression levels during BCoV infection in some bovine cells. (A) The model for the binding of the HS with the BCoV/S protein receptor. (B) and (C) showing ligand binding to the interacted amino acid residues- (TRP184, HIS143, GLU156, ASN198, ARG197, and LYS192) of the BCoV/S protein. (D) Bovine HSPG2 gene expression levels in the MDBK cells infected with either the BCoV/Ent or BCoV/Resp isolates, compared to the sham, analyzed by the qRT-PCR. (E) The bovine HSPG2 gene expression in the BEC cells infected with either the BCoV/Ent or BCoV/Resp isolates, compared to the sham, was analyzed by the qRT-PCR. Both cell lines were infected with either BCoV/Ent or BCoV/Resp isolates (MOI=1) for 72 hpi and used for qRT-PCR analysis.
Figure 11.
The proposed model for the bovine heparan sulfate (HS) of HSPG2 interaction with the BCoV/S protein and the conformation of the HSPG2 expression levels during BCoV infection in some bovine cells. (A) The model for the binding of the HS with the BCoV/S protein receptor. (B) and (C) showing ligand binding to the interacted amino acid residues- (TRP184, HIS143, GLU156, ASN198, ARG197, and LYS192) of the BCoV/S protein. (D) Bovine HSPG2 gene expression levels in the MDBK cells infected with either the BCoV/Ent or BCoV/Resp isolates, compared to the sham, analyzed by the qRT-PCR. (E) The bovine HSPG2 gene expression in the BEC cells infected with either the BCoV/Ent or BCoV/Resp isolates, compared to the sham, was analyzed by the qRT-PCR. Both cell lines were infected with either BCoV/Ent or BCoV/Resp isolates (MOI=1) for 72 hpi and used for qRT-PCR analysis.
Figure 12.
The multiple sequence alignment of the putative furin cleavage site of the BCoV-S glycoprotein compared to the validated SARS-CoV-2 furin cleavage site and other members of the Betacoronavirus. (A) Mapping the bovine furin protease binding residues near its cleavage (RRSRR) at the S1/S2 site of the S protein of the BCoV. (B) Mapping the TMPRSS2 protease binding residues near its cleavage (RRSRR) at the BCoV/S1/S2 cleavage sites. The yellow highlighted sequences show the polybasic residues and their potential specific cleavage site for furin and TMPRSS2 proteases.
Figure 12.
The multiple sequence alignment of the putative furin cleavage site of the BCoV-S glycoprotein compared to the validated SARS-CoV-2 furin cleavage site and other members of the Betacoronavirus. (A) Mapping the bovine furin protease binding residues near its cleavage (RRSRR) at the S1/S2 site of the S protein of the BCoV. (B) Mapping the TMPRSS2 protease binding residues near its cleavage (RRSRR) at the BCoV/S1/S2 cleavage sites. The yellow highlighted sequences show the polybasic residues and their potential specific cleavage site for furin and TMPRSS2 proteases.
Figure 13.
The proposed model of the bovine furin protease interactions with BCoV/S cleavage site and the conformation of the furin expression levels in some bovine cell lines infected with BCoV. (A) Mapping the bovine furin protease interaction at the furin-specific recognition sites (RRSRR region) at the BCoV/S1/S2 junction. (B) and (C) Identifcation of the furin residues (Glu230, Leu227, Asp228, Asp174, Asp177, Asn192, Met189) that have strong binding affinity to the BCoV/S protein amino acid residues including (Val758, Asp759, Tyr760 Ser761, Thr762 Lys763, Arg764, Arg765, Ser766, Arg767) at the RRSRR|A (S1/S2) cleavage sites. (D) The bovine Furin gene expression levels in the MDBK cells infected with either the BCoV/Ent or BCoV/Resp isolates, compared to the sham-infected cells, data were analyzed by the qRT-PCR. (E) The bovine Furin gene expression levels in the BEC cells infected with either the BCoV/Ent or BCoV/Resp isolates were compared to those of the sham-infected cells, and the qRT-PCR analyzed data. Both cell lines were infected with either BCoV/Ent or BCoV/Resp isolates (MOI=1) for 72 hpi and used for the qRT-PCR analysis.
Figure 13.
The proposed model of the bovine furin protease interactions with BCoV/S cleavage site and the conformation of the furin expression levels in some bovine cell lines infected with BCoV. (A) Mapping the bovine furin protease interaction at the furin-specific recognition sites (RRSRR region) at the BCoV/S1/S2 junction. (B) and (C) Identifcation of the furin residues (Glu230, Leu227, Asp228, Asp174, Asp177, Asn192, Met189) that have strong binding affinity to the BCoV/S protein amino acid residues including (Val758, Asp759, Tyr760 Ser761, Thr762 Lys763, Arg764, Arg765, Ser766, Arg767) at the RRSRR|A (S1/S2) cleavage sites. (D) The bovine Furin gene expression levels in the MDBK cells infected with either the BCoV/Ent or BCoV/Resp isolates, compared to the sham-infected cells, data were analyzed by the qRT-PCR. (E) The bovine Furin gene expression levels in the BEC cells infected with either the BCoV/Ent or BCoV/Resp isolates were compared to those of the sham-infected cells, and the qRT-PCR analyzed data. Both cell lines were infected with either BCoV/Ent or BCoV/Resp isolates (MOI=1) for 72 hpi and used for the qRT-PCR analysis.
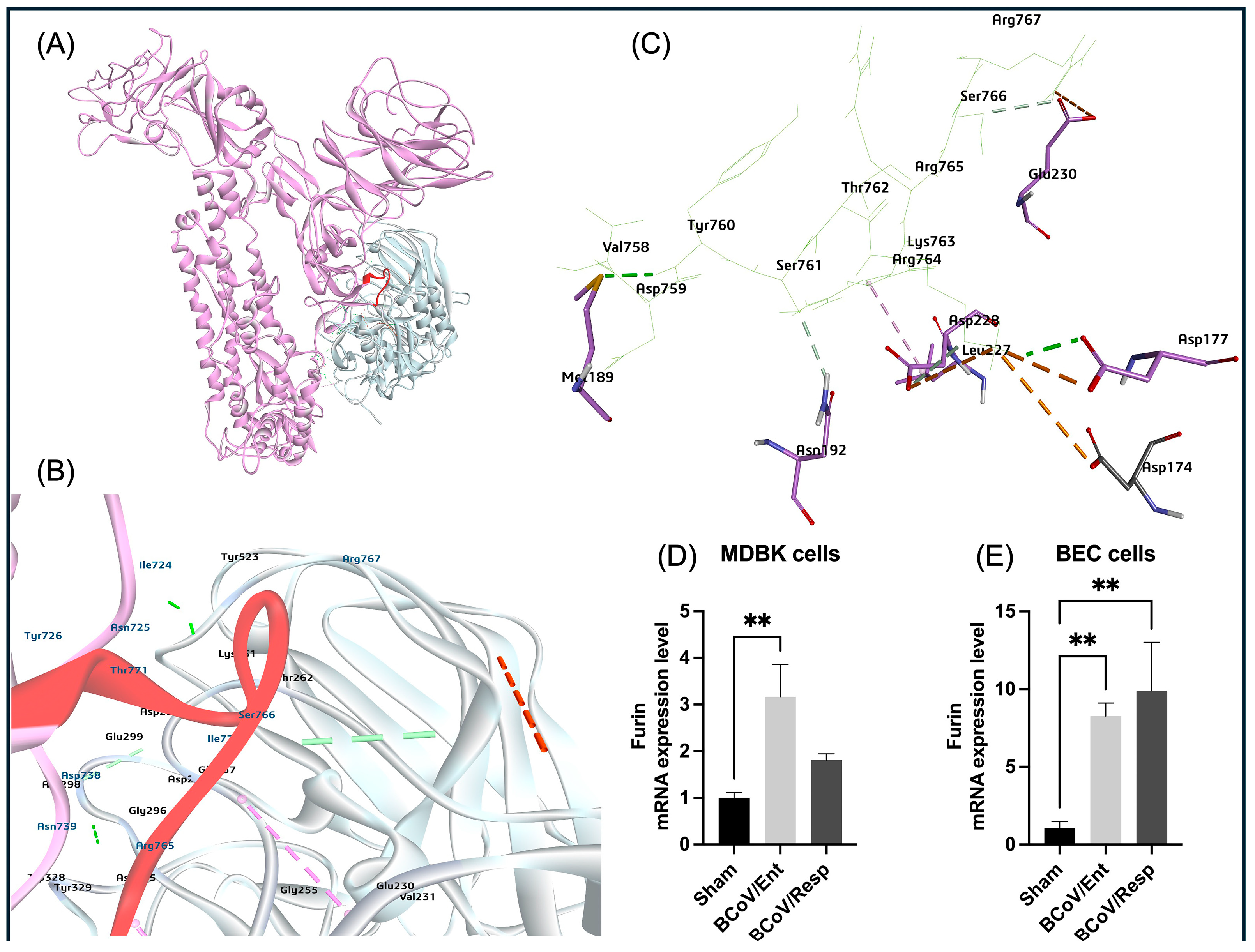
Figure 14.
The proposed putative model of the bovine TMPRSS2 interaction with BCoV/S glycoprotein and the confirmation of the TMPRSS2 gene expression levels during BCoV infection in some bovine cell lines. (A) The proposed model represents the TMPRSS2 serin protease binding upstream and downstream of the BCoV/S protein polybasic cleavage site RRSRR|A. (B) and (C) Mapping the TMPRSS2 residues (Val278, His294, Thr339, Asp415, Asn416, Trp459, Gly460, Ser461, Gly462 binding affinity to the BCoV/S1/S2 site (Asp738, Ser740, Thr741, Ser742, Ser743, Ser761, Arg764, Thr762, Lys763, Arg767, Thr771 and Ile779). (D) The Bovine TMPRSS2 gene expression levels in the MDBK cells infected with either the BCoV/Ent or BCoV/Resp isolates, compared to the sham, data was analyzed by the qRT-PCR. (E) The Bovine TMPRSS2 gene expression in the BEC cells infected with either the BCoV/Ent or the BCoV/Resp isolates, compared to the sham, data was analyzed by the qRT-PCR. Both cell lines were infected with either BCoV/Ent or BCoV/Resp isolates (MOI=1) for 72 hpi.
Figure 14.
The proposed putative model of the bovine TMPRSS2 interaction with BCoV/S glycoprotein and the confirmation of the TMPRSS2 gene expression levels during BCoV infection in some bovine cell lines. (A) The proposed model represents the TMPRSS2 serin protease binding upstream and downstream of the BCoV/S protein polybasic cleavage site RRSRR|A. (B) and (C) Mapping the TMPRSS2 residues (Val278, His294, Thr339, Asp415, Asn416, Trp459, Gly460, Ser461, Gly462 binding affinity to the BCoV/S1/S2 site (Asp738, Ser740, Thr741, Ser742, Ser743, Ser761, Arg764, Thr762, Lys763, Arg767, Thr771 and Ile779). (D) The Bovine TMPRSS2 gene expression levels in the MDBK cells infected with either the BCoV/Ent or BCoV/Resp isolates, compared to the sham, data was analyzed by the qRT-PCR. (E) The Bovine TMPRSS2 gene expression in the BEC cells infected with either the BCoV/Ent or the BCoV/Resp isolates, compared to the sham, data was analyzed by the qRT-PCR. Both cell lines were infected with either BCoV/Ent or BCoV/Resp isolates (MOI=1) for 72 hpi.
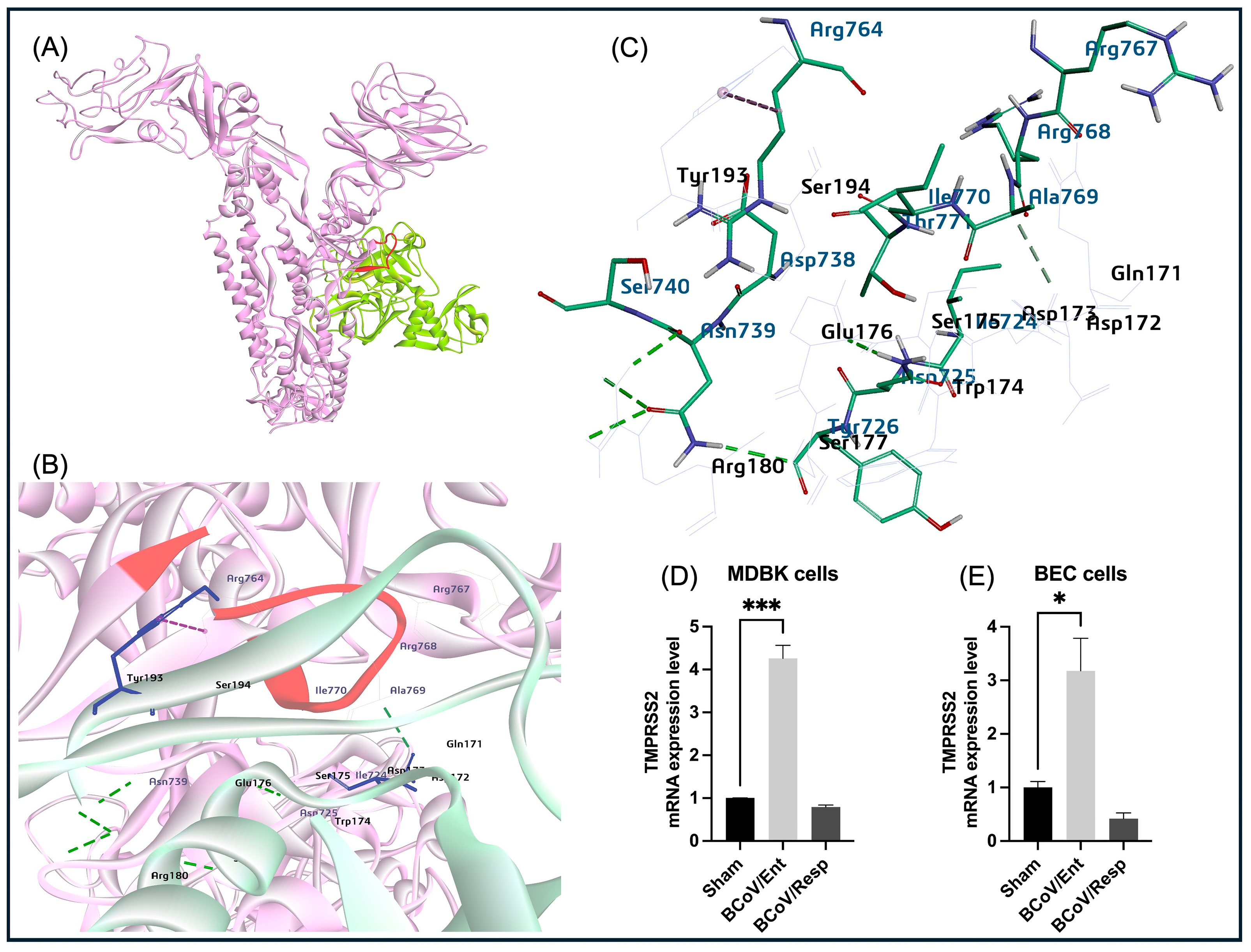
Figure 15.
The proposed model for the putative interaction between the bovine CTS-L protein and the BCoV-S glycoprotein and the confirmation of the CTS-L gene expression levels in some bovine cells infected with the BCoV. (A) The proposed model represents the CTS-L protease binding sites at the NTD and the CTD of the S1 chain from BCoV/S. (B) and (C) Mapping the interaction interface of the CTS-L residues (Ile36, Met291, Thr38, Ala33, Ser35, Ile36, Gl72, Arg768, Pro723, Gln722, Phe713) binding affinity to the BCoV/S1 chain residues (Trp154, Phe145, Lys267, Thr275, Asp326, Gly190, Phe186, Tyr159, Asp326, Glu142, Thr143 and Pro149). (D) The CTS-L gene expression levels in the MDBK cells infected with either the BCoV/Ent or the BCoV/Resp isolates, compared to the sham-infected cells, data was analyzed by the qRT-PCR. (E) The bovine CTS-L gene expression levels in the BEC cells infected with the BCoV/Ent or the BCoV/Resp isolates, compared to the sham-infected cells, data was analyzed by the qRT-PCR. Both cell lines were infected with the BCoV/Ent or the BCoV/Resp isolates (MOI=1) for 72 hpi.
Figure 15.
The proposed model for the putative interaction between the bovine CTS-L protein and the BCoV-S glycoprotein and the confirmation of the CTS-L gene expression levels in some bovine cells infected with the BCoV. (A) The proposed model represents the CTS-L protease binding sites at the NTD and the CTD of the S1 chain from BCoV/S. (B) and (C) Mapping the interaction interface of the CTS-L residues (Ile36, Met291, Thr38, Ala33, Ser35, Ile36, Gl72, Arg768, Pro723, Gln722, Phe713) binding affinity to the BCoV/S1 chain residues (Trp154, Phe145, Lys267, Thr275, Asp326, Gly190, Phe186, Tyr159, Asp326, Glu142, Thr143 and Pro149). (D) The CTS-L gene expression levels in the MDBK cells infected with either the BCoV/Ent or the BCoV/Resp isolates, compared to the sham-infected cells, data was analyzed by the qRT-PCR. (E) The bovine CTS-L gene expression levels in the BEC cells infected with the BCoV/Ent or the BCoV/Resp isolates, compared to the sham-infected cells, data was analyzed by the qRT-PCR. Both cell lines were infected with the BCoV/Ent or the BCoV/Resp isolates (MOI=1) for 72 hpi.
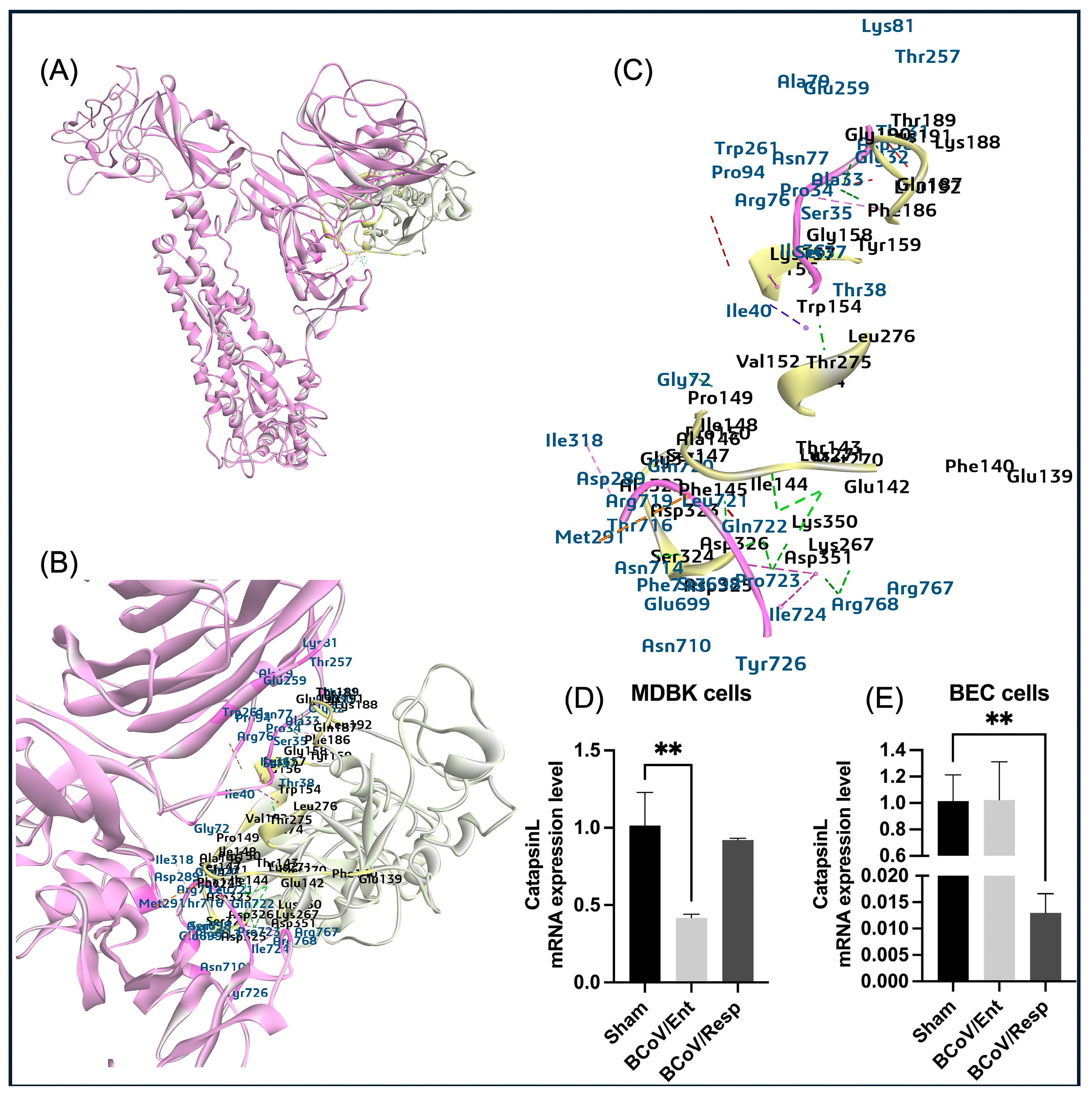
Figure 16.
A proposed bovine sialic acid (Neu5,9Ac2) interaction model with the BCoV/HE protein. (A) The model of the binding of Neu5,9Ac2 as a Ligand for the BCoV/HE protein receptor. (B) and (C) The ligand binding to the mapped amino acid residues- (Asn-264, Glu-265, Ser-221, and Asn-236) of the BCoV/HE protein.
Figure 16.
A proposed bovine sialic acid (Neu5,9Ac2) interaction model with the BCoV/HE protein. (A) The model of the binding of Neu5,9Ac2 as a Ligand for the BCoV/HE protein receptor. (B) and (C) The ligand binding to the mapped amino acid residues- (Asn-264, Glu-265, Ser-221, and Asn-236) of the BCoV/HE protein.
Figure 17.
Schematic representation showing the interaction between the BCoV/S and BCoV/HE proteins with the potential cellular receptors, including host cell proteases (Furin, CTS-L, and TMPRSS2). BCoV/S is predicted to interact with most of the potential host cell receptors through their NTD and CTD. The NTD is the N-terminal domain present in the BCoV/S1 protein. The CTD is the C-terminal domain of the BCoV/S1 chain.
Figure 17.
Schematic representation showing the interaction between the BCoV/S and BCoV/HE proteins with the potential cellular receptors, including host cell proteases (Furin, CTS-L, and TMPRSS2). BCoV/S is predicted to interact with most of the potential host cell receptors through their NTD and CTD. The NTD is the N-terminal domain present in the BCoV/S1 protein. The CTD is the C-terminal domain of the BCoV/S1 chain.
Table 1.
List of the oligonucleotides used for the amplification of the gene expression profiles of some potential BCoV receptors and host cell protases.
Table 1.
List of the oligonucleotides used for the amplification of the gene expression profiles of some potential BCoV receptors and host cell protases.
| Bovine Genes |
Forward Primer (5′ to 3′) |
Reverse Primer (5′ to 3′) |
| ACE2 |
GCTGTCGGGGAAATCATGTC |
TCTCTCGCTTCATCTCCCAC |
| Furin |
CGAGAAGAACCACCCAGACT |
CTACGCCACAGACACCATTG |
| TMPRSS2 |
CCTTCTTAGCAGCCCAGAGT |
CATCTTCAAGGGAGGCCAGA |
| NRP1 |
CCAGAAGCCAGAGGAGTACG |
GCCTTTTCCGATTTCACCCT |
| CEACAM1 |
TTCTTCTGCTTGCCCACAAC |
TCCTTTGTAACGAGCAGGGT |
| DPP4 |
AGAGACGCAGACCATGAAGA |
TCGGCTAGAGTGTAGGTTCTG |
| AXL |
TCTCAGATGCGGGATGGTAC |
AGCTCAGGTTGAAGGGAGTG |
| APN |
AGAGTGGGACTTTGCTTGGA |
TGGCAATGCTGGTAATGGTG |
| HSPG2 |
GTTGTCAGCGTGGTGTTCAT |
GAGAGGTGACGTAGGAGGC |
| Cathepsin-L |
CTTCGATTCCTCCATCCGTG |
TCTATGAAGCCACCGTGACA |
| β-actin |
CAAGTACCCCATTGAGCACG |
GTCATCTTCTCACGGTTGGC |
Table 2.
Homology Model with best DOPE Score and stability verification Using the Verify-3D protein, BIOVIA Discovery Studio software.
Table 2.
Homology Model with best DOPE Score and stability verification Using the Verify-3D protein, BIOVIA Discovery Studio software.
| Homology model |
Dope Score |
Verify Score |
Verify Expected High Score |
Verify Expected Low Score |
| BCoV-Spike |
-135770 |
476.141 |
561.414 |
252.636 |
| ACE2 |
-94345 |
308.35 |
358.11 |
161.153 |
| Furin |
-53696 |
227.99 |
216.48 |
97.4159 |
| TMPRSS2 |
-34507 |
142.08 |
152.573 |
68.6579 |
| Cathepsin-L |
-33161 |
132.18 |
144.31 |
64.94 |
| NRP1 |
-25920 |
125.32 |
159.001 |
71.5506 |
| DPP4 |
-90256 |
308.11 |
333.627 |
150.132 |
| APN |
-118318 |
385.36 |
415.465 |
186.959 |
| CEACAM-1 |
-10424 |
32.41 |
58.3052 |
26.2373 |
| AXL |
-35499 |
135.46 |
137.42 |
61.84 |
Table 3.
Summary of the results of the prediction of the Energetics and bonding interactions between the BCoV/ S glycoprotein with some potential BCoV host cell receptors.
Table 3.
Summary of the results of the prediction of the Energetics and bonding interactions between the BCoV/ S glycoprotein with some potential BCoV host cell receptors.
Complex
(Best Pose) |
ZDock score |
E_R Dock Score |
Total hydrogen bonds |
Total Pi bonds |
Salt Bridge |
| BCoV/S-ACE2 |
22.58 |
-12.26 |
7 |
28 |
0 |
| BCoV/S-NRP1 |
26 |
-21.22 |
29 |
9 |
0 |
| BCoV/S-CEACAM-1 |
16.8 |
-6.29 |
12 |
9 |
1 |
| BCoV/S-APN |
17.6 |
-2.95 |
19 |
2 |
1 |
| BCoV/S-DPP4 |
22.44 |
-6.23 |
20 |
9 |
1 |
| BCoV/S-AXL |
17.56 |
-13.68 |
21 |
9 |
0 |
| BCoV/S-Furin |
16.6 |
-13.14 |
30 |
1 |
0 |
| BCoV/S-TMPRSS2 |
20 |
0.641 |
15 |
6 |
0 |
| BCoV/S-CTS-L |
19.84 |
-16.45 |
15 |
2 |
0 |