1. Introduction
Meconopsis, a representative genus of the Papaveraceae family, has garnered attention due to its unique high-altitude ecological adaptability and diverse flower colors [
1]. Typically,
Meconopsis species grow in alpine environments at elevations ranging from 2,500 to 5,500 meters [
2,
3]. Globally, there are 79 species of
Meconopsis, with 58 species distributed in China, primarily in the Qinghai-Tibet Plateau and surrounding areas.
Meconopsis holds significant importance in traditional Tibetan medicine, where it is called "LvRongHao" in Chinese and is honored as one of the "Four Divine Herbs" [
4]. Its medicinal value is primarily seen in its ability to clear heat, detoxify, reduce inflammation, and alleviate pain. As such, it is widely used to treat various diseases such as pneumonia, hepatitis, headaches, and edema [
5].
Traditionally, blue-flowered
Meconopsis (
Meconopsis betonicifolia) has been valued for its medicinal properties despite its relatively low yield [
6]. In contrast, yellow-flowered
Meconopsis (
Meconopsis integrifolia), while more abundant, has not had its medicinal potential fully explored [
7]. This disparity highlights the need for a systematic comparison of the chemical compositions and potential pharmacological effects between blue and yellow
Meconopsis varieties, which could yield significant scientific and economic benefits.
The increasing scarcity of wild blue-flowered
Meconopsis and intensified conservation efforts have created an urgent demand for alternative sources [
8]. Yellow-flowered
Meconopsis, with its higher yield and wider geographical distribution, presents a promising alternative. However, to fully leverage its potential as a medicinal resource, comprehensive research is required to elucidate its chemical composition, pharmacological activity, and how these characteristics compare to its blue-flowered counterpart [
9]. This research is crucial not only for potentially expanding the medicinal applications of yellow-flowered
Meconopsis but also for addressing the sustainability concerns surrounding the more scarce blue variety.
Recent studies have revealed that
Meconopsis species contain a rich variety of bioactive substances, including amino acids, organic acids, cardiac glycosides, volatile oils, sugars, tannins, alkaloids, and coumarins [
10]. To better understand the chemical composition of blue and yellow
Meconopsis, we conducted a detailed analysis using liquid chromatography-mass spectrometry (LC-MS) techniques. Our results demonstrated a high degree of similarity in chemical composition between the two-color varieties, with over 70% of the detected compounds present in both. This finding provides a crucial foundation for exploring the medicinal potential of yellow-flowered
Meconopsis as a possible alternative to its blue-flowered counterpart.
To elucidate the potential medicinal applications of these shared compounds, we performed a systematic Gene Ontology (GO) and disease enrichment analysis. Our findings suggest that many compounds in
Meconopsis are potentially associated with multiple physiological systems, including the nervous, cardiovascular, and respiratory systems, as well as mental health and metabolic diseases. Notably, certain fatty acids such as arachidonic acid, γ-linolenic acid, and linoleic acid may be closely related to inflammatory conditions [
11,
12]. Furthermore, compounds like quercetin and arachidonic acid have demonstrated antimicrobial, anticancer, and anti-inflammatory activities.
Given the prominence of inflammation-related pathways in our enrichment analysis, we propose to further validate and elucidate the anti-inflammatory effects and mechanisms of
Meconopsis using the lipopolysaccharide (LPS)-induced RAW 264.7 macrophage model [
13,
14,
15]. This well-established in vitro model effectively simulates key processes of inflammatory responses, allowing for a comparative assessment of the anti-inflammatory potential of yellow and blue
Meconopsis.
To gain deeper insights into the molecular mechanisms underlying
Meconopsis' anti-inflammatory action, we will employ metabolomics analysis methods. As an essential branch of systems biology, metabolomics provides a global view of cellular metabolism, facilitating the identification of key metabolic pathways and molecular markers [
16]. This approach will not only validate our previous disease enrichment analysis results but may also reveal new targets and mechanisms of action.
This comprehensive exploration of yellow-flowered Meconopsis carries significant scientific, economic, and ecological implications. Firstly, confirming the superior anti-inflammatory effects of yellow-flowered Meconopsis compared to its blue-flowered counterpart would provide new options for the sustainable use of medicinal plant resources. Secondly, increased utilization of yellow-flowered Meconopsis could alleviate collection pressure on wild blue-flowered varieties, contributing to biodiversity conservation. Lastly, a deeper understanding of the chemical composition and pharmacological actions of yellow-flowered Meconopsis may reveal new bioactive compounds or pharmacological mechanisms, providing valuable leads for new drug development.
In conclusion, our study aims to comprehensively evaluate the anti-inflammatory potential of Meconopsis through a series of systematic experimental approaches. We will focus on comparing the medicinal efficacy of yellow and blue-flowered Meconopsis, using LC-MS technology for detailed chemical composition analysis, followed by target prediction and pathway analysis to reveal potential pharmacological mechanisms. The LPS-induced RAW 264.7 macrophage anti-inflammatory model, combined with metabolomics analysis, will allow for an in-depth comparison of the anti-inflammatory effects and mechanisms of action of the two Meconopsis extracts.
This comprehensive research approach will not only deepen our understanding of the pharmacological actions of Meconopsis but may also provide important clues for the development of new anti-inflammatory drugs. By comparing the medicinal efficacy of yellow and blue-flowered Meconopsis, we aim to provide a solid experimental foundation for the application of Meconopsis in the prevention and treatment of inflammation-related diseases. Simultaneously, this study will contribute valuable insights into the sustainable utilization of medicinal plant resources and biodiversity conservation.
2. Materials and Methods
2.1. Plant Material and Extraction
Meconopsis betonicifolia and Meconopsis integrifolia were obtained from Tibet Ganlu Tibetan Medicine Co., Ltd. and authenticated by Professor Xin Feng (Tibetan Medicine Institute, China Tibetan Research Center). The plants were dried, powdered separately, and stored under the accession numbers 2023112504 and 2023112505, respectively, at the Center for Molecular Metabolism, Nanjing University of Science and Technology.
For each plant species, 5 g of powder was subjected to triple extraction using 100 mL of 70% (v/v) ethanol. The extraction process was carried out under ultrasonic conditions (400 W, 4 kHz) for 30 minutes per cycle. The resulting extract solutions from all three cycles were combined for each species.
The merged solutions were then concentrated using a rotary evaporator under reduced pressure at 55°C to recover the ethanol solvent. Following rotary evaporation, the concentrated extracts were subjected to vacuum freeze-drying to eliminate any residual solvent, yielding the final solid phytochemical Meconopsis extracts (ME). The ME was then categorized by variety into MIE (M. integrifolia extract) and MBE (M. betonicifolia extract).
2.2. Cell Culture and ME Treatment
RAW 264.7 cells (obtained from the National Biomedical Laboratory Cell Repository, China) were used between passages 8 and 15. The cells were maintained in Dulbecco's Modified Eagle Medium (DMEM; Hyclone, Thermofisher, China) supplemented with 10% heat-inactivated fetal bovine serum (Gemini Bio, USA) and 1% penicillin-streptomycin antibiotics (Gibco, USA). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2 (Thermo Fisher Scientific, USA). The culture media were replaced daily.
For experimental procedures, RAW 264.7 cells were seeded into 96-well plates (NEST, China) at a density of 5 × 104 cells per well and incubated overnight. Stock solutions of MIE and MBE were prepared by dissolving the extracts in dimethyl sulfoxide (DMSO) at a concentration of 200 mg/mL.
To determine the non-toxic concentration range, cells were treated with MIE and MBE at concentrations ranging from 12.5 μg/mL to 3,200 μg/mL for 24 hours. Cell viability was assessed using the MTT assay to measure cellular metabolic activity. Based on the viability results, a concentration range of 31.25 μg/mL to 500 μg/mL was selected for subsequent experiments. Cells were treated with these concentrations of MIE and MBE for 24 hours. Following treatment, the media containing the extracts were removed, and fresh DMEM containing 1 μg/mL lipopolysaccharide (LPS) was added to the cells. The cells were then incubated for an additional 24 hours. Nitric oxide (NO) production, an indicator of inflammatory response, was quantified using a nitric oxide nitrate reductase assay kit (Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer's instructions.
2.3. Determination of mRNA Transcription Levels
Total RNA was extracted with Trizol reagent and 1 μg RNA was reversely transcribed into cDNA with all-in-one RT MasterMix (Takara). Synthesized cDNA was used as a template for qPCR with SYBR Green qPCR Master Mixes (Yeasen, China) on ABI 7300 Real-Time PCR Systems (Applied Biosystems, USA). The β-actin gene was selected as the internal reference gene. Samples were incubated at 95 ℃ for 3 min, followed by 40 cycles of amplification at 95 ℃ for 3 s and 60 ℃ for 30 s. The relative gene expression changes were quantified by the 2-ΔΔCt method. The primer sequences for the target genes were designed and synthesized by Tsingke Biotech (Beijing, China), as shown in Table 1.
2.4. Composition Identification and Cell Metabolomics Based on LC-MS
MIE and MBE were dissolved with 70% (v/v) ethanol at 1 mg/mL. After vortex and centrifuged the supernatant was obtained for LC-MS analysis.
RAW 264.7 cells were cultured into 60 mm culture dishes, and treated as described in
Section 2.2. with the concentrations at 200 and 500 μg/mL of MIE and MBE and divided into 6 groups with 7 dishes per group. Cells were washed with PBS twice at room temperature. 1mL pre-cooled 80% (v/v) methanol was added into each culture dish and cells were obtained with a cell scraper. After repeatedly freeze-thaw cycles, brief vortex, and ultrasound-treated at 0 ℃, broken cells were incubated at -20 ℃ for over 1 h and centrifuged at 16,000 g, 4 ℃ for 15 min. The supernatant was concentrated until dry under a vacuum. The dried cell extract was redissolved with 200 μL 50% (v/v) methanol. After vortex, ultrasound-treated and centrifuged the supernatant was obtained for LC-MS based metabolomics study.
LC-MS analysis was performed with TripleTOF 5600+ mass spectrometer coupled with SCIEX Exion LC system and controlled by Analyst TF 1.8 software (AB SCIEX, Canada). Atlantis PREMIER BEH C18 AX column (100×2.1 mm, 1.7 μm) with protection column (VanGuard FIT) was performed for chromatographic separation. The column temperature was set at 40 ℃ and the auto sampler temperature was set at 4 ℃. The liquid solvent system was composed of 0.1% (v/v) formic acid (phase A) and acetonitrile (phase B). The flow rate was set at 0.4 ml/min. The sample injection was 5 μL. The gradient elution program was set as 1% phase B at 0-1 min, 1-99 % phase B at 1-10 min, 99 % phase B at 10-13 min, 99-1 % phase B at 13-14 min and 1 % phase B at 14-17 min.
The electrospray ionization (ESI) spray voltage was +5,500(+)/-4,500(-) V. Nitrogen was used as the nebulizer gas (GS1) at 55 psi, turbo gas (GS2) at 55 psi, and curtain gas (CUR) at 35 psi. The source temperature was set at 550 ℃. Information-dependent acquisition (IDA) parameters were set as follows: declustering potential (DP) = +80(+)/-80(-) V, collision energy (CE) = +35±15(+)/-35±15(-) eV, intensity threshold ≥ 100 cps, ion tolerance = 50 mDa, exclude isotopes within 4 Da, 10 candidate ions monitored per cycle. In the TOF MS-IDA-MS/MS acquisition, the TOF MS scan was performed with a mass range from 50 to 1,000 m/z and an accumulation time of 0.10 s per spectrum. The product ion scan was performed with a mass range from 40 to 1,000 m/z and an accumulation time of 0.05 s per spectrum. Dynamic Background Subtraction (DBS) was enabled. Auto calibrate was performed per 5 samples for TOF MS and TOF MS/MS with a calibration delivery system (CDS). The data were analyzed using SCIEX OS 2.0 software.
2.5. Data Processing and Compound Identification
Raw LC-MS data files were converted to the open mzXML format using MSConvert (ProteoWizard, version 3.0.23114). The converted files were processed using MS-DIAL software (version 4.9.221218) for peak detection, alignment, and deconvolution. MS-DIAL parameters were optimized for our specific dataset, with key settings including minimum peak height for MS1 data, 1,500 counts; MS2 tolerance, 0.05 Da; and retention time tolerance, 0.3 min. A multi-faceted approach was employed for compound identification:
MS-DIAL's built-in libraries and algorithms were used for initial compound annotation. MS1 and MS2 spectra were matched against multiple databases including MassBank, GNPS, and Riken MSP. A minimum spectral similarity score of 70% was required for tentative identification.
In silico fragmentation tools, including CFM-ID 4.0, were utilized to predict MS2 spectra for compounds not found in spectral libraries. These predicted spectra were compared to experimental data to aid in structural elucidation.
Manual interpretation of MS2 spectra was conducted for compounds of particular interest that were not confidently identified through automated methods.
2.6. Comprehensive Metabolomics Analysis: From Multivariate Statistics to Pathway Enrichment and Network Visualization
Multivariate statistical analysis was performed using R package mixOmics. Principal component analysis (PCA) was initially used to visualize overall variation in the dataset and detect potential outliers. Partial least squares discriminant analysis (PLS-DA) was then applied to maximize separation between experimental groups. Metabolites contributing significantly to group separation were identified based on variable importance in projection (VIP) scores > 1.0.
We performed metabolite set enrichment analysis (MSEA) against the SMPDB pathway database to gain biological insight into metabolic differences between groups. Significantly enriched pathways were determined using a hypergeometric test with Benjamini-Hochberg false discovery rate correction (q < 0.05). The gene-metabolite interaction network and metabolite levels across samples were distributed in pathways.
We constructed a network visualization to further elucidate the relationships between differentially abundant metabolites and associated genes within the enriched pathways. This network graphically represented the connections between metabolites and genes, providing a comprehensive view of the molecular interactions underlying the observed metabolic differences. Additionally, we generated a heatmap to visualize the relative abundance of differentially abundant metabolites across sample groups within the enriched pathways. This heatmap allowed for the identification of metabolite patterns and trends specific to each experimental group, facilitating a more nuanced understanding of the metabolic shifts occurring under different conditions.
2.7. Gene Ontology (GO) Analysis Methodology
Target prediction for the identified compounds was performed using multiple computational approaches. The SwissTargetPrediction web server (
http://www.swisstargetprediction.ch/) was employed as the primary tool, utilizing a similarity-based approach to predict potential molecular targets. For each compound, the 2D structure represented by SMILES notation was input into the server. To enhance prediction accuracy and coverage, we also consulted the BindingDB database (
https://www.bindingdb.org/) with settings restricted to "Homo sapiens" and a minimum interaction score threshold of 0.700 to identify high-confidence target predictions.
To elucidate the biological functions of compounds, we performed Gene Ontology (GO) enrichment analysis on its predicted molecular targets. The protein targets were converted to their corresponding gene symbols using the UniProtKB database (
https://www.uniprot.org/). The GO analysis was conducted using the ShinyGO v0.76 graphical gene-set enrichment tool (
https://bioinformatics.sdstate.edu/go). The corresponding gene identifiers were then input into the ShinyGO tool. We set the significance threshold at p ≤ 0.05, with p-values adjusted using the Benjamini-Hochberg procedure to control for a false discovery rate. The GO analysis was performed across three main categories: biological process (BP), molecular function (MF), and cellular component (CC). An over-representation analysis was conducted using the DisGeNET database (
http://www.disgenet.org/) to disclose the target association with diseases.
3. Results
3.1. Evaluation of Safety and Efficacy: MIE and MBE's Impact on Cell Viability and NO Production in Inflammatory Conditions
MIE and MBE's cytotoxicity and anti-inflammatory effects were evaluated using LPS-stimulated RAW264.7 cells.
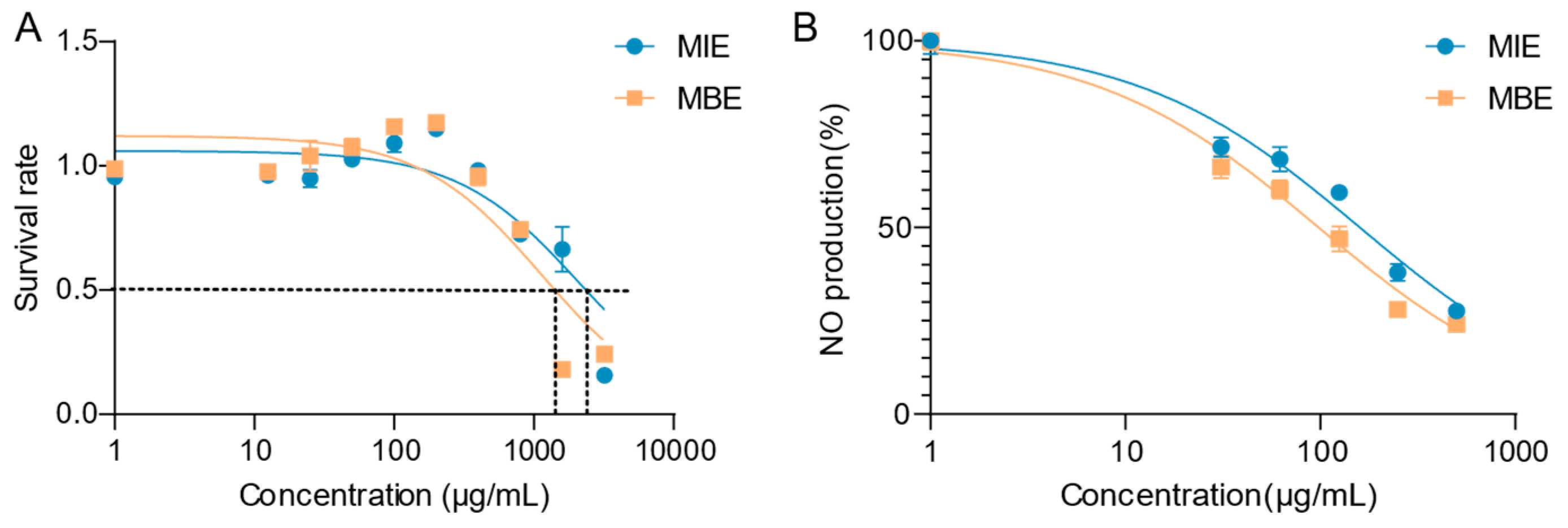
Figure 1A illustrates the cytotoxicity profiles of both extracts. MIE and MBE demonstrated minimal cytotoxicity towards RAW264.7 cells after 24 hours of treatment. The IC
50 values for MIE and MBE were determined to be 1,140 μg/mL and 2,105 μg/mL, respectively. Notably, at a concentration of 400 μg/mL, both MIE and MBE maintained over 90% cell viability, indicating their safety at therapeutically relevant concentrations.
Figure 1B depicts the effects of MIE and MBE on NO production in LPS-stimulated RAW264.7 cells. Both extracts exhibited a dose-dependent inhibition of NO production. MIE showed superior potency in reducing NO levels compared to MBE, with IC
50 values of 93.45 μg/mL and 152.1 μg/mL, respectively. This significant decrease in NO production suggests that both MIE and MBE possess strong anti-inflammatory properties, with MBE demonstrating greater efficacy.
The dose-response curves for both cytotoxicity and NO inhibition provide valuable insights into the therapeutic window of these extracts. MBE, in particular, shows promise as an anti-inflammatory agent, given its lower IC50 for NO inhibition and higher IC50 for cytotoxicity, indicating a favorable safety profile coupled with potent anti-inflammatory activity.
3.2. Dose-Dependent Anti-Oxidative and Anti-inflammatory Effects of MIE and MBE
We investigated the therapeutic potential of yellow (MIE) and blue (MBE)
Meconopsis species by examining their effects on oxidative stress markers (
Figure 2A) and inflammatory gene expression (
Figure 2B) across various dosages. Both
Meconopsis species demonstrated significant antioxidant and anti-inflammatory properties, with their effects generally increasing in a dose-dependent manner. The yellow and blue varieties showed similar trends in their impact on biochemical markers and gene expression, suggesting comparable therapeutic efficacy.
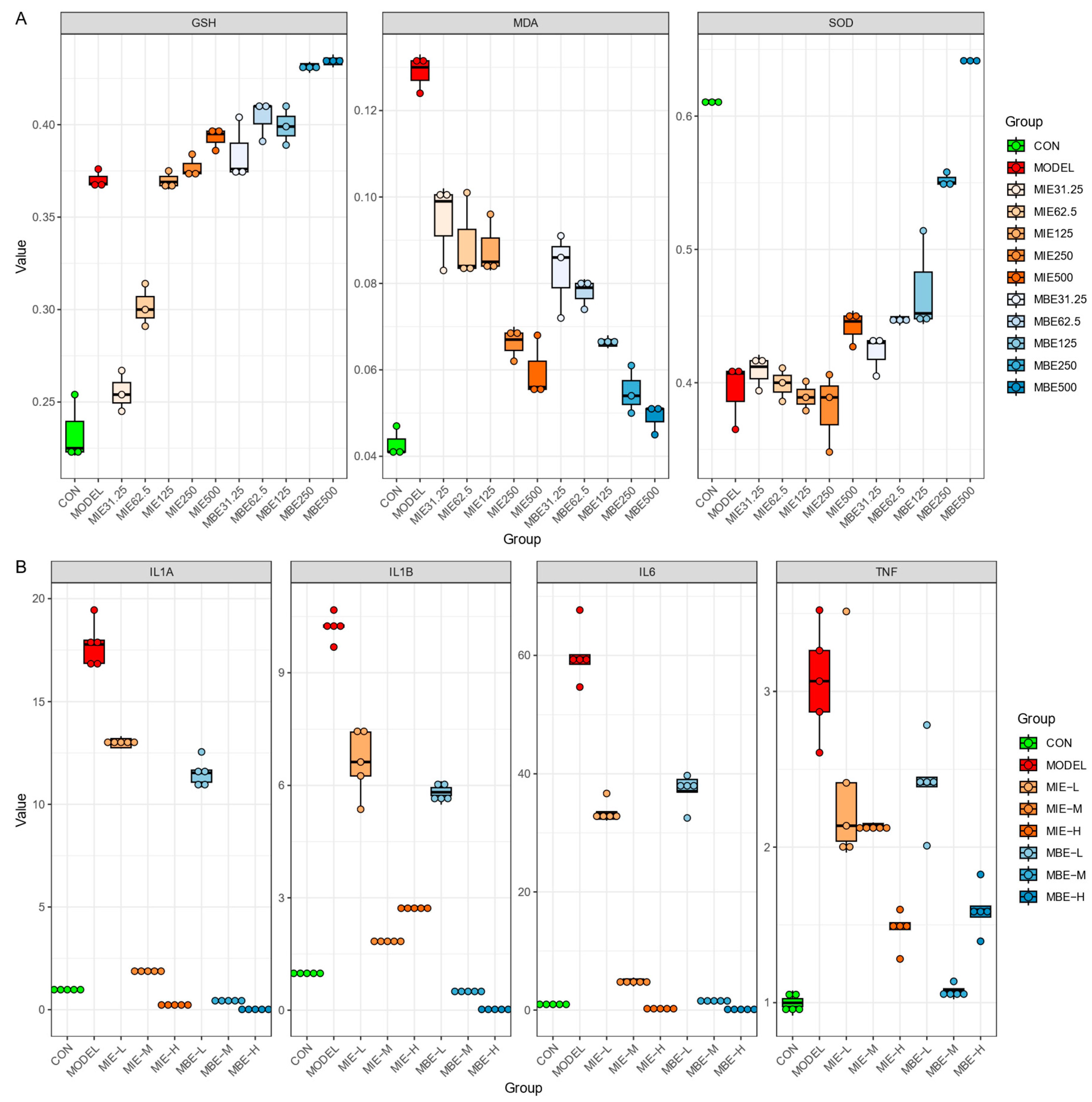
Both MIE and MBE treatments effectively enhanced the antioxidant defense system. Glutathione (GSH) levels increased dose-dependently, with the highest doses (500 ug/ml) of both species elevating GSH to levels comparable to or exceeding the control group. Concurrently, Superoxide Dismutase (SOD) activity, which was diminished in the model group, was restored by both species, approaching control levels at higher doses. The antioxidant effects were further evidenced by the reduction of Malondialdehyde (MDA) levels, a marker of oxidative stress. Both MIE and MBE lowered MDA concentrations dose-dependently, with higher doses exhibiting more pronounced effects. Notably, the yellow variety (MIE) showed a slightly more consistent dose-response relationship in MDA reduction compared to the blue variety (MBE), though the differences were not substantial.
The anti-inflammatory properties of both Meconopsis species were demonstrated through the suppression of key inflammatory genes: IL-1A, IL-1B, IL-6, and TNF. Both MIE and MBE effectively reduced the expression of these genes, with medium and high doses showing the most significant effects. IL-1B and IL-6 expression exhibited the most pronounced dose-dependent decrease for both species. The yellow variety (MBE) appeared to have a marginally stronger effect on IL-1B suppression at higher doses, while the blue variety (MBE) showed slightly more potent IL-6 suppression. However, these differences were subtle, and both species demonstrated robust anti-inflammatory action overall. TNF and IL-1A expression were also markedly reduced by both MIE and MBE treatments, particularly at medium and high doses. The effects on these genes were largely comparable between the two species, underscoring their similar anti-inflammatory potentials.
While both Meconopsis species exhibited strong therapeutic properties, some nuanced differences were observed. The yellow variety (MBE) showed a slightly more consistent dose-response relationship in certain parameters, such as MDA reduction and IL-1B suppression. Conversely, the blue variety (MBE) demonstrated marginally superior effects in IL-6 suppression at higher doses. These subtle variations notwithstanding, both species displayed remarkably similar overall efficacy in improving antioxidant status and reducing inflammatory gene expression. The results suggest that both yellow and blue Meconopsis possess significant and comparable therapeutic potential, particularly in conditions involving oxidative stress and inflammation.
3.3. Comparative Metabolomic Profiling of Yellow and Blue Meconopsis Species: MIE and MBE Shared about 70% Composition
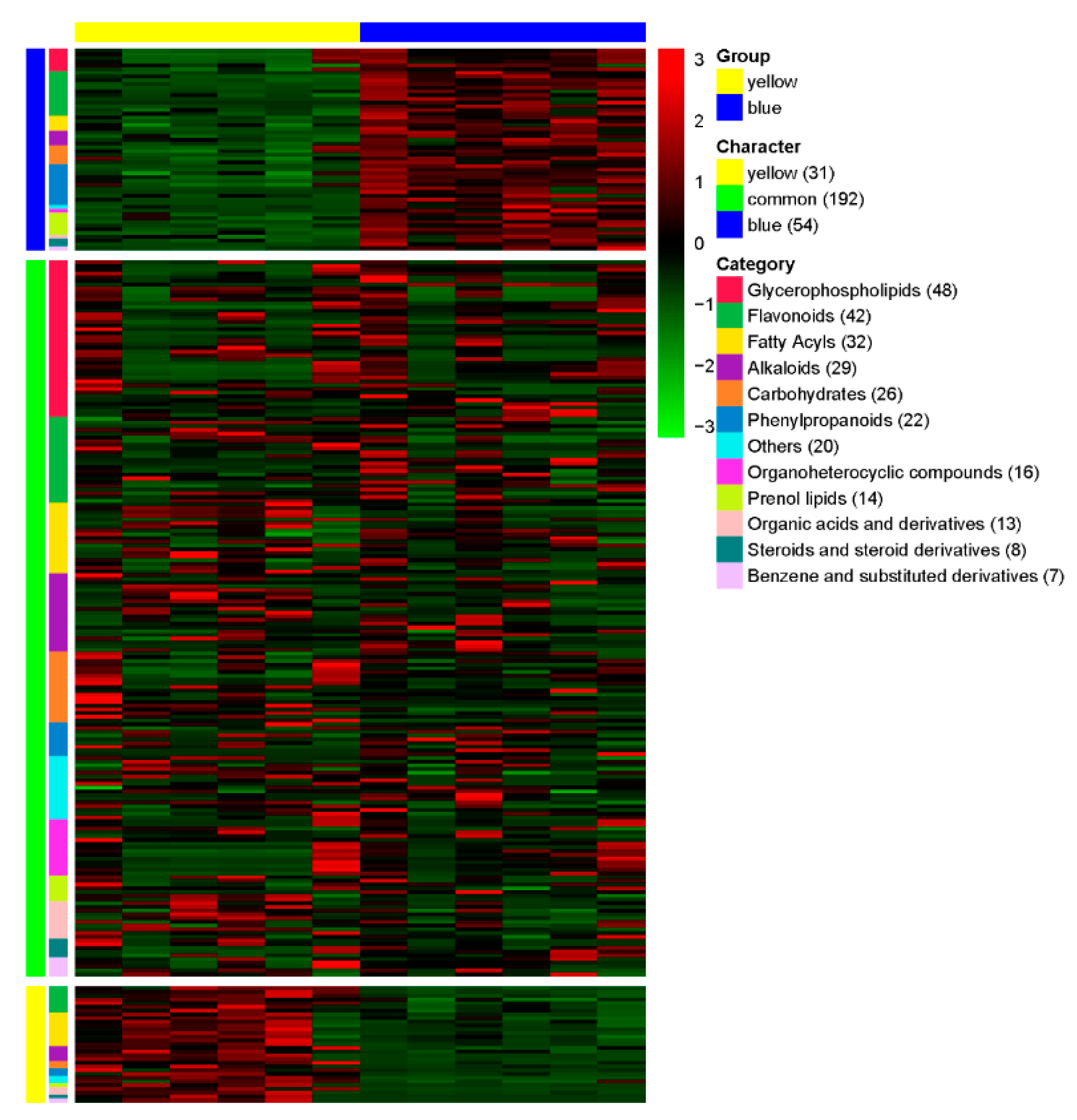
LC-MS analysis was performed on two
Meconopsis species, one with yellow flowers and another with blue flowers, to identify and compare their chemical compositions. The heat map visualization in
Figure 3 illustrates the relative abundance of compounds across categories for both species, with colors ranging from dark blue (lower abundance) to dark red (higher abundance). This representation allows for a quick visual comparison of the chemical profiles between the yellow and blue-flowered
Meconopsis species. A total of 277 compounds were detected and categorized based on their relative abundance in each species (
Table S1). The compounds were classified into 12 major categories: Glycerophospholipids, Flavonoids, Fatty Acyls, Alkaloids, Carbohydrates, Phenylpropanoids, Organoheterocyclic compounds, Prenol lipids, Organic acids and derivatives, Steroids and steroid derivatives, Benzene and substituted derivatives, and Others. Of the 277 compounds, 192 were found to be common between the yellow and blue flower species, showing no significant difference in their relative abundance. These common compounds represent shared characteristics between the two species. The yellow-flowered species exhibited 31 compounds that were significantly more abundant compared to the blue-flowered species. These compounds can be considered characteristic of the yellow-flowered
Meconopsis. Conversely, the blue-flowered species showed 54 compounds with significantly higher abundance compared to the yellow-flowered species, representing the characteristic compounds of the blue-flowered
Meconopsis. The distribution of compounds across categories varied, with Glycerophospholipids being the most represented category (48 compounds), followed by Flavonoids (42 compounds), and Fatty Acyls (32 compounds). Alkaloids and Carbohydrates were also well-represented with 29 and 26 compounds, respectively. The least represented categories were Steroids and steroid derivatives (8 compounds) and Benzene and substituted derivatives (7 compounds).
3.4. Molecular Insights into Meconopsis: GO Enrichment and Disease Network Analysis of Common Feature Targets
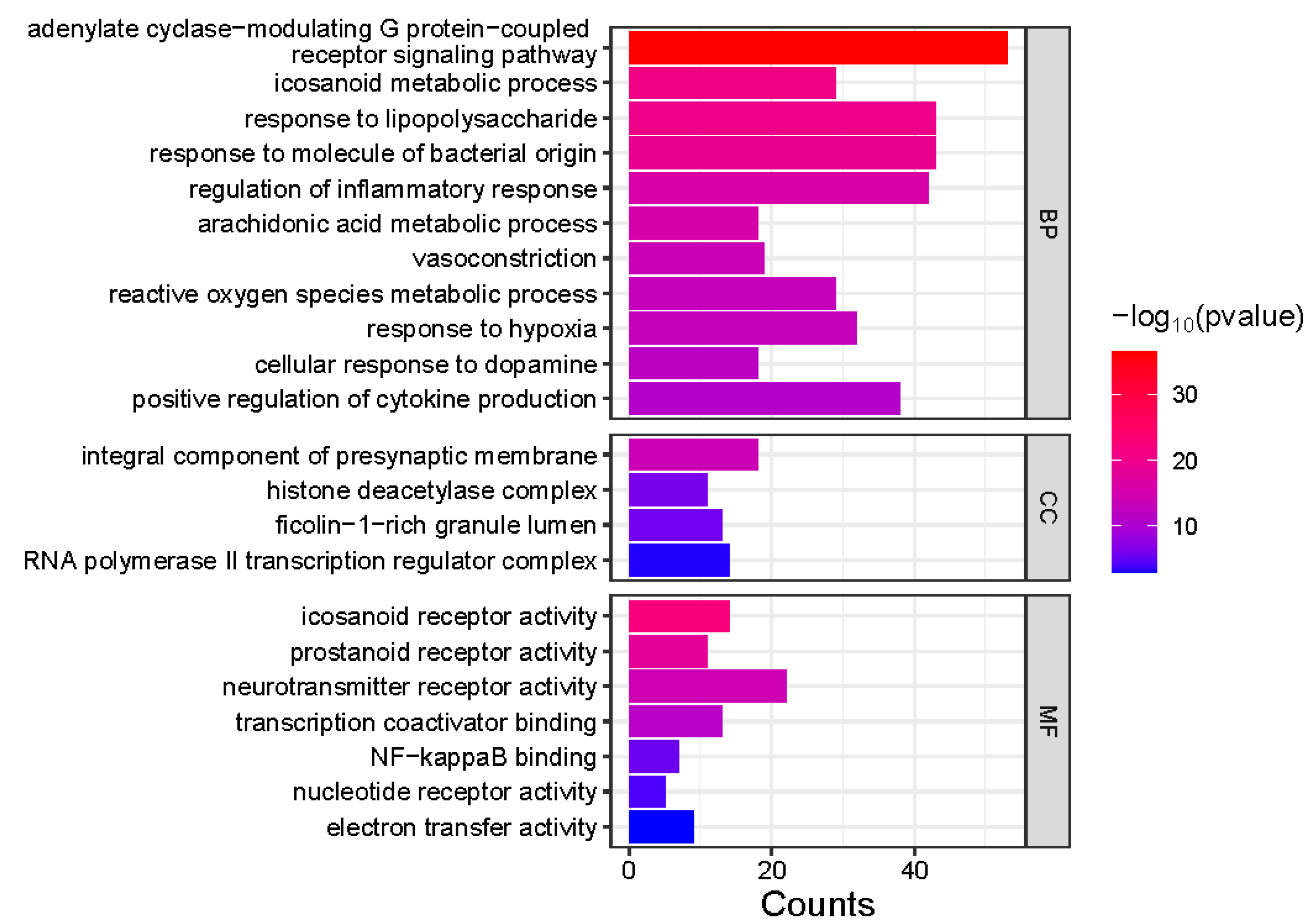
Gene Ontology (GO) enrichment analysis was performed on 413 genes corresponding to the common feature targets of both
Meconopsis species (
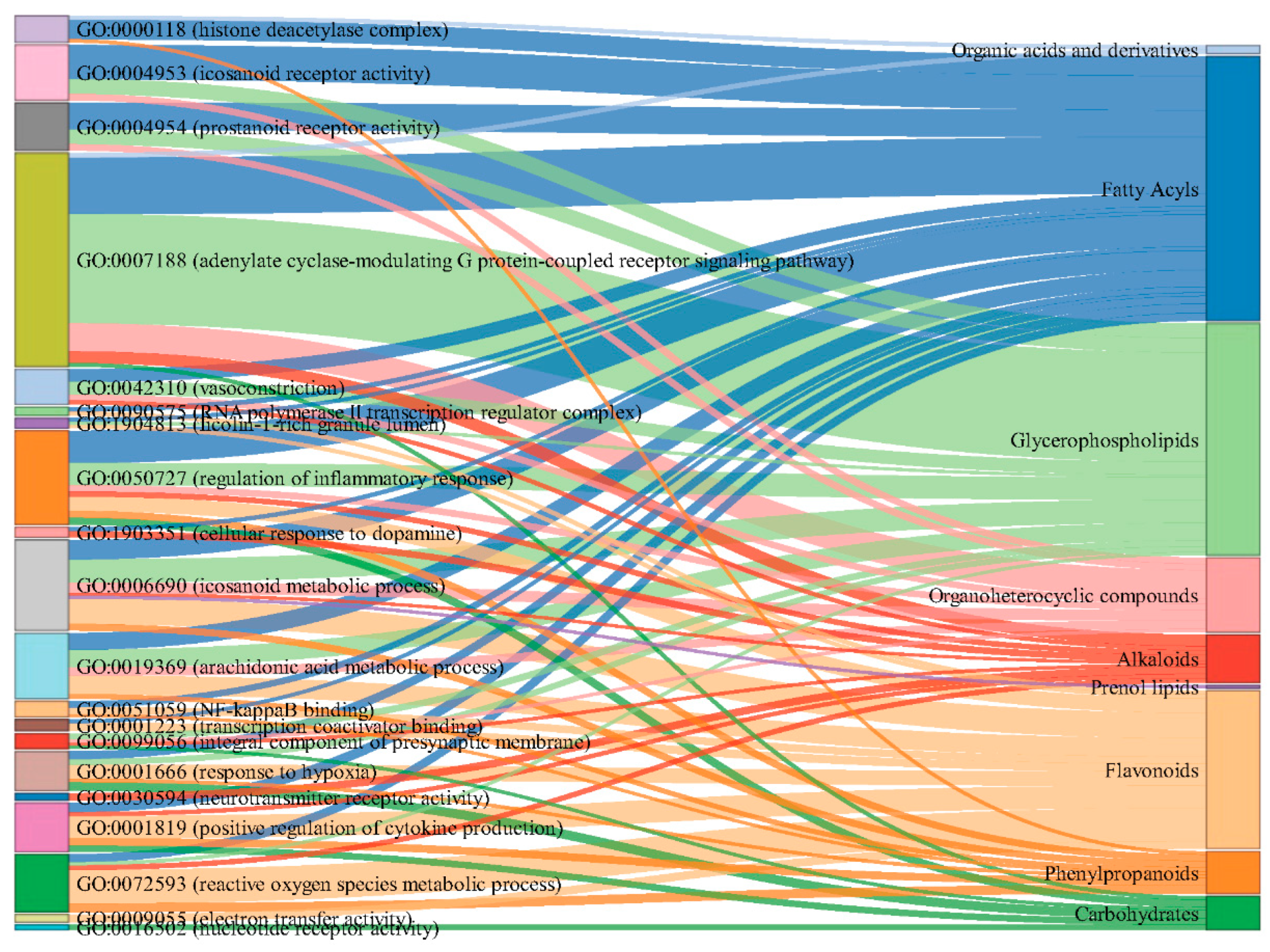
Figure 4). The analysis revealed significant enrichment in three main categories: Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). In the BP category, several processes were notably enriched, including positive regulation of cytokine production, cellular response to dopamine, response to hypoxia, and reactive oxygen species metabolic processes. Other significant processes included vasoconstriction, arachidonic acid metabolism, regulation of inflammatory response, and response to bacterial molecules and lipopolysaccharides. The CC category highlighted enrichment in RNA polymerase II transcription regulator complex, ficolin-1-rich granule lumen, histone deacetylase complex, and integral components of the presynaptic membrane. For MF, the analysis showed significant enrichment in electron transfer activity, nucleotide receptor activity, NF-kappaB binding, transcription coactivator binding, neurotransmitter receptor activity, and prostanoid and icosanoid receptor activities. The association of component types with the enriched GO terms was visualized as a Sankey diagram in
Figure 5.
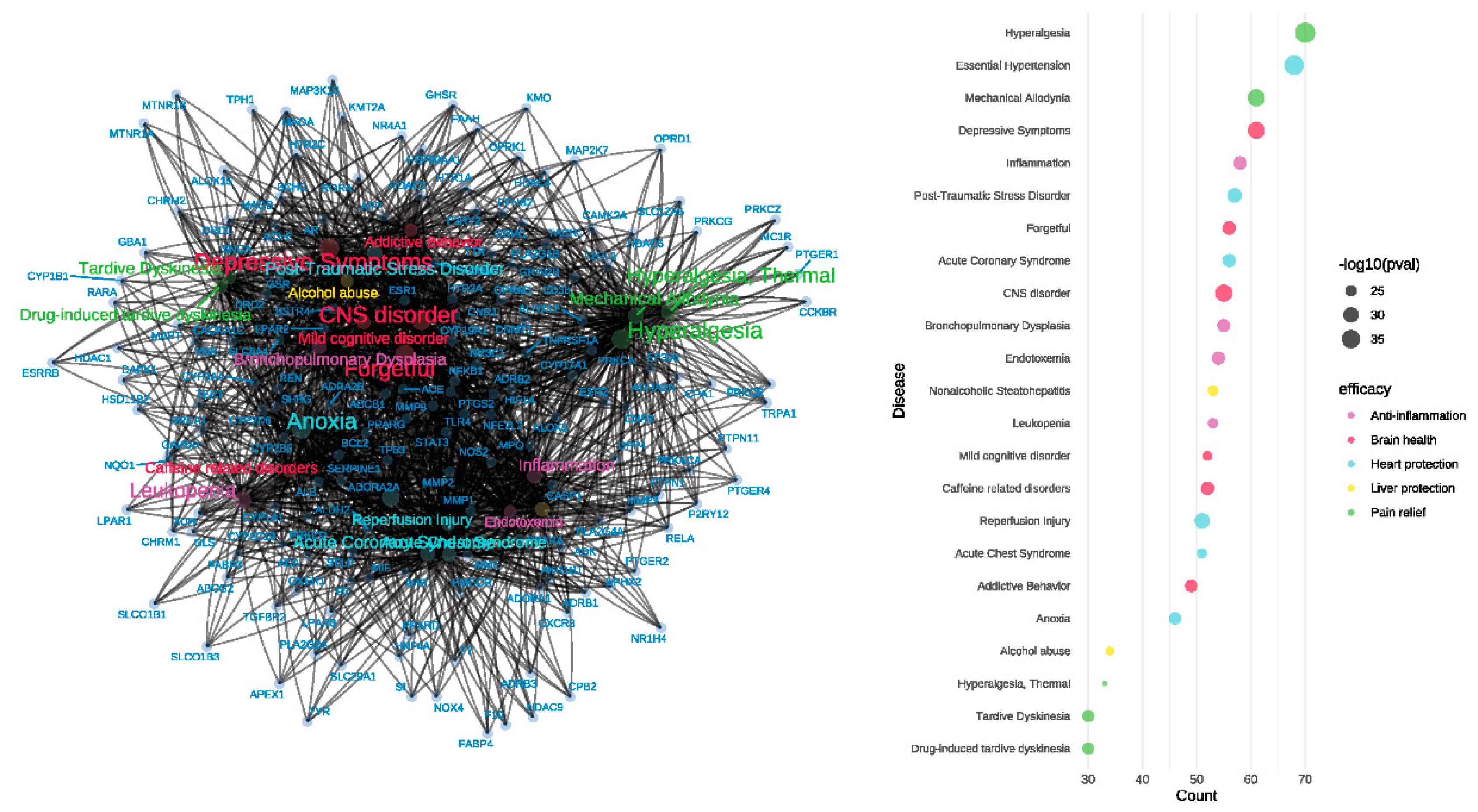
The gene-disease network analysis (
Figure 6) of the 413 common feature target genes revealed enrichment in various diseases, indicating the potential therapeutic efficacy of
Meconopsis. The diseases were categorized based on their corresponding efficacy, represented by different colors in the bubble plot. The size of the bubbles in the plot correlates with the adjusted p-values, with larger bubbles representing higher enrichment significance for the corresponding diseases. Significant enrichment in hyperalgesia, mechanical allodynia, and thermal hyperalgesia, suggesting potent analgesic properties. Enrichment in essential hypertension, acute coronary syndrome, and reperfusion injury, indicating cardiovascular protective effects. Enrichment in CNS disorders, depressive symptoms, mild cognitive disorders, and addictive behaviors, pointing to potential neuroprotective and psychiatric benefits. Significant enrichment in inflammation and bronchopulmonary dysplasia, suggesting anti-inflammatory properties. Enrichment in nonalcoholic steatohepatitis and alcohol abuse, indicating potential hepatoprotective effects.
3.5. Metabolomic Profiling Reveals Anti-inflammatory Effects of Meconopsis Species in LPS-Stimulated RAW264.7 Cells
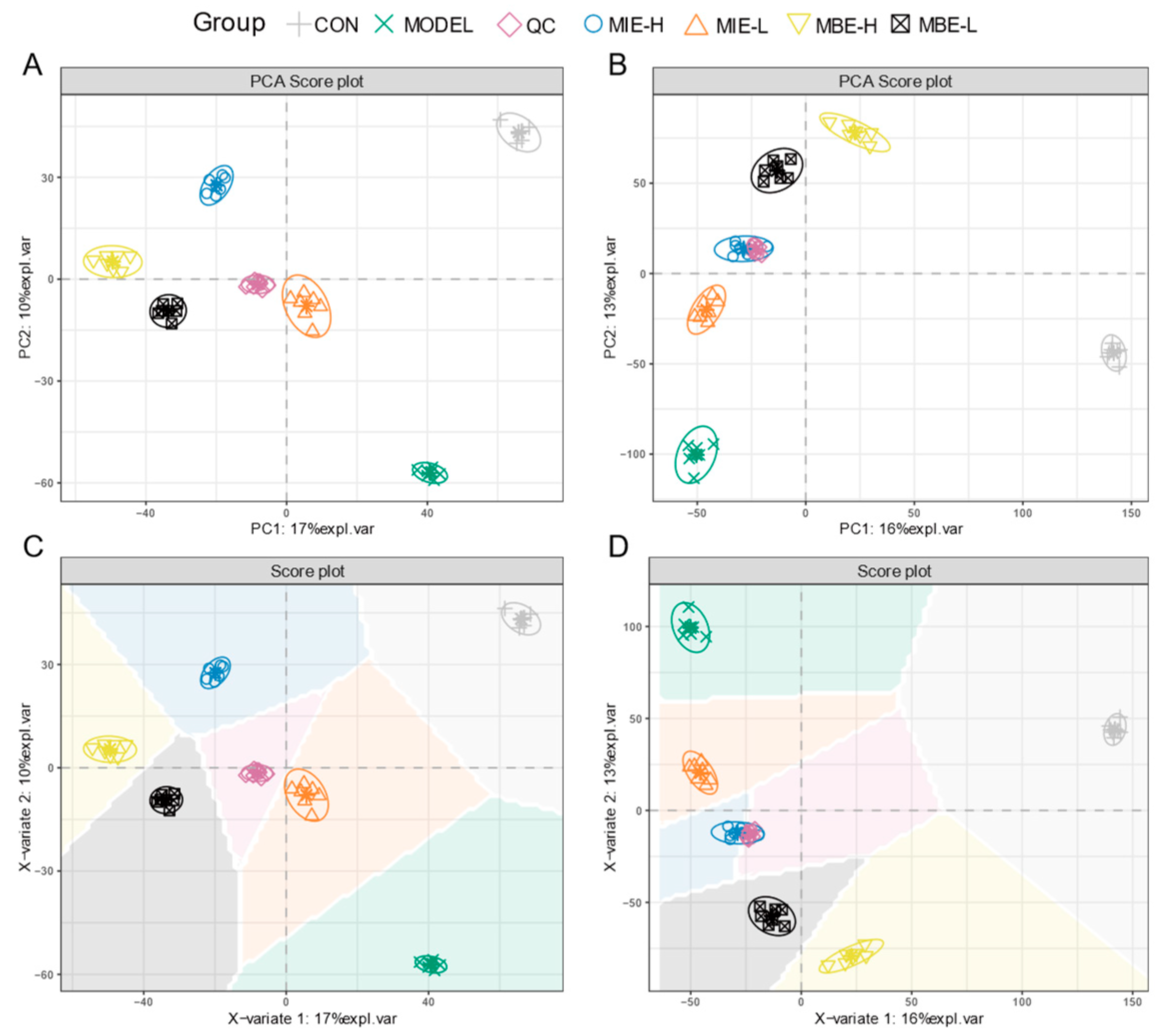
Multivariate statistical analysis of the metabolomic data revealed distinct metabolic profiles among the experimental groups. The PCA and PLS-DA score plots (
Figure 7) for both positive (A, C) and negative (B, D) ion modes demonstrated clear separation between the control (C), model (M), and treatment groups. The clustering of QC samples indicated the reliability and reproducibility of the metabolomic analysis. The PCA and PLS-DA plots showed that the yellow-flowered species treatment groups (MIE-L and MIE-H) were more closely clustered to the control group compared to the blue-flowered species treatment groups (MBE-L and MBE-H), suggesting that yellow-flowered species had a more pronounced effect in normalizing the LPS-induced metabolic perturbations. This observation aligns with the experimental findings that yellow-flowered species exhibited superior anti-inflammatory effects compared to blue-flowered species.
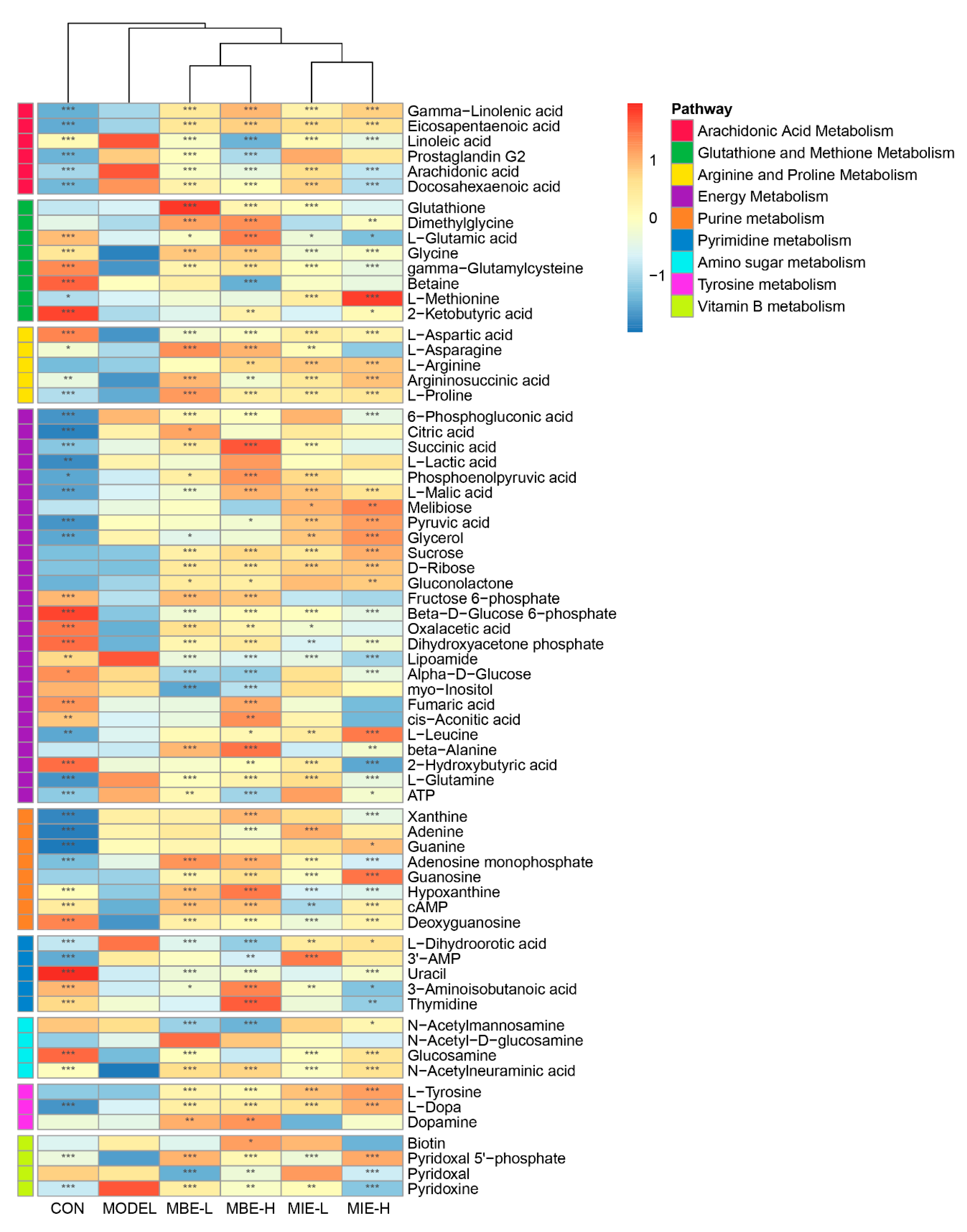
The heatmap (
Figure 8) of differentially abundant metabolites across various pathways provided further insights into the metabolic changes induced by LPS stimulation and the subsequent effects of
Meconopsis treatments. Nine key pathways were identified: Arachidonic Acid Metabolism, Glutathione and Methionine Metabolism, Arginine and Proline Metabolism, Energy Metabolism, Purine Metabolism, Pyrimidine Metabolism, Amino Sugar Metabolism, Tyrosine Metabolism, and Vitamin B Metabolism. Notably, the heatmap revealed that the LPS-stimulated model group (M) showed distinct metabolite patterns compared to the control group (C), particularly in pathways related to inflammation and energy metabolism. Both
Meconopsis species appeared to modulate these LPS-induced changes, with yellow-flowered
Meconopsis species demonstrating a more pronounced effect in restoring metabolite levels closer to those observed in the control group.
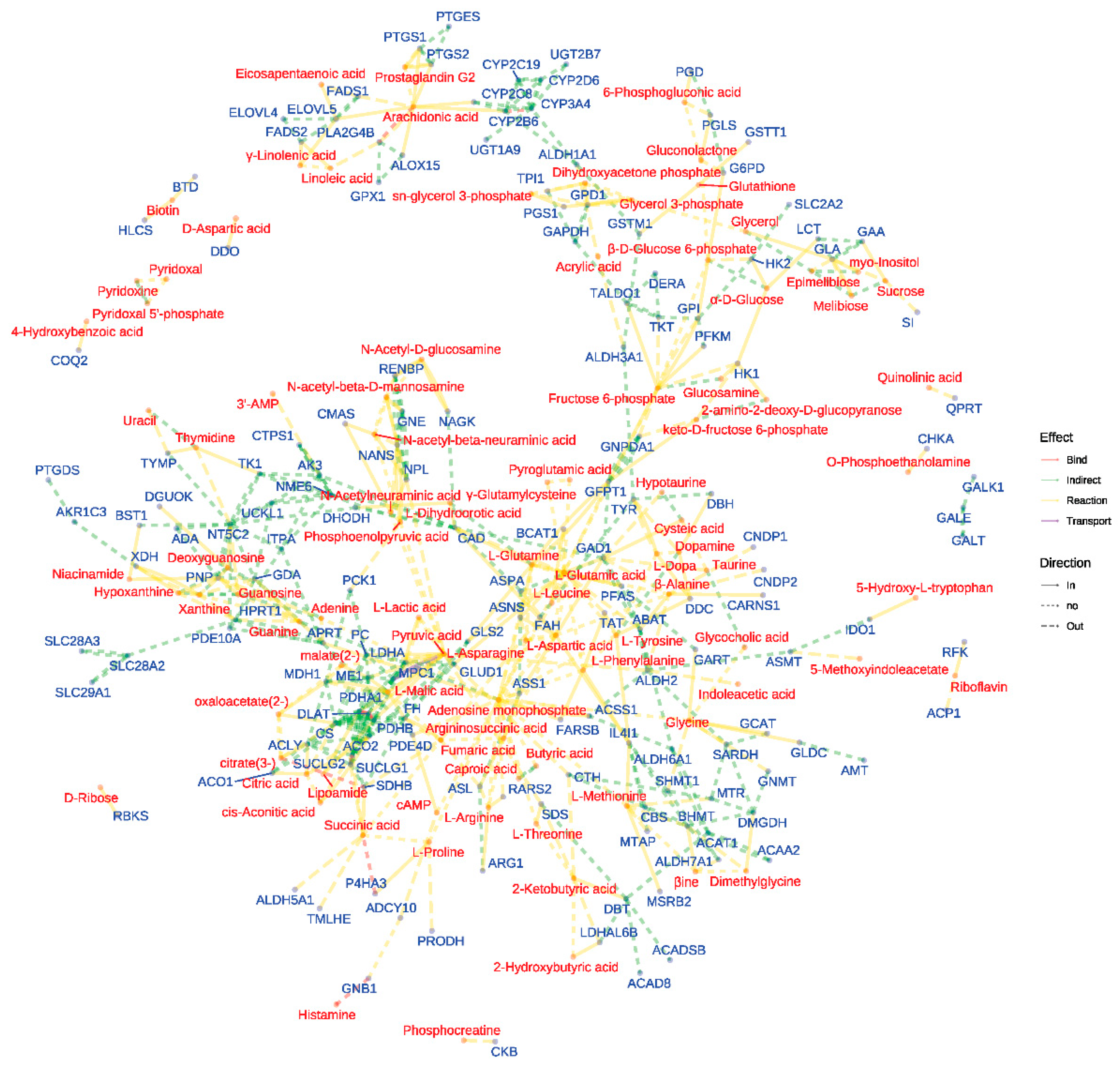
The network visualization (
Figure 9) of metabolite-gene interactions within enriched pathways provided a comprehensive view of the molecular mechanisms underlying the observed anti-inflammatory effects. This network highlighted key nodes and connections between differentially abundant metabolites and their associated genes, offering potential targets for further investigation into the anti-inflammatory actions of the two
Meconopsis species.
4. Discussion
This study comprehensively evaluated and compared the anti-inflammatory properties of yellow- and blue-flowered Meconopsis species using multiple complementary approaches. Firstly, LC-MS metabolomic profiling revealed that the two species shared around 70% of chemical constituents, establishing a material basis for their functional interchangeability.
Gene Ontology and disease enrichment analysis of the common constituents' putative targets indicated significant associations with processes and conditions relevant to the traditional uses documented for Meconopsis, validating their therapeutic potential. Notably, enriched terms were linked to inflammation, providing rationale for subsequent in vitro evaluation.
The cell-based assays using LPS-stimulated RAW 264.7 macrophages demonstrated that both MIE and MBE possessed dose-dependent anti-oxidative and anti-inflammatory activities, as evidenced by their effects on oxidative stress markers, inflammatory gene expression and NO production. MIE generally exhibited superior efficacy.
To gain deeper mechanistic insights, untargeted metabolomics was employed. Distinct metabolic phenotypes were observed between treatment groups and the inflammatory model. Both species modulated several metabolic pathways disrupted by LPS stimulation.
4.1. Arachidonic Acid Metabolism
LPS stimulation in macrophages significantly upregulates the levels of arachidonic acid and prostaglandin G2 compared to the unstimulated control group. Both MIE and MBE demonstrate a dose-dependent ability to reduce the elevated levels of arachidonic acid post-LPS stimulation, with MBE showing superior efficacy. Additionally, MBE exhibits a dose-dependent reduction in prostaglandin G2 levels, an effect not observed with MIE. The "Arachidonic Acid Metabolism" pathway is activated in LPS-stimulated macrophages and pivotal for the generation of inflammatory mediators [
17].
In the arachidonic acid metabolism pathway, arachidonic acid is liberated from cell membranes by the action of PLA2G4B and subsequently converted to prostaglandin G2 by the enzymes PTGS1/PTGS2 [
18]. It can also be transformed into various eicosanoids, including leukotrienes, through the actions of ALOX15 or cytochrome P450 enzymes [
19]. The observed trend of decreasing linoleic acid, a precursor to arachidonic acid, in treated groups, albeit less pronounced than that of arachidonic acid itself, suggests that the treatments may be influencing arachidonic acid production upstream in the pathway.
Gamma-Linolenic acid, positioned as an intermediate between Linoleic acid and Arachidonic acid, shows a consistent trend with eicosapentaenoic acid (EPA). EPA, renowned for its potent anti-inflammatory properties, is synthesized from linolenic acid through the catalytic action of fatty acid desaturase 2 and a series of elongase enzymes. Docosahexaenoic acid (DHA), synthesized from EPA via the activity of fatty acid desaturase 1 and additional elongase enzymes, also plays a significant role in modulating inflammation [
20].
The reduction in prostaglandin G2 levels coupled with the increased levels of gamma-linolenic acid and EPA in the drug treatment groups indicates that MIE and MBE may exert their anti-inflammatory effects by downregulating arachidonic acid metabolism. This might achieved by coordinately inhibiting the activity of PTGS1/PTGS2 and associated enzymes, thereby reducing the production of inflammatory eicosanoids and redirecting arachidonic acid towards the formation of less potent mediators. This orchestrated modulation of the arachidonic acid pathway presents a more sophisticated therapeutic approach than the inhibition of a solitary enzyme.
4.2. Glutathione and Methionine Metabolism
An enhancement in glutathione levels in both MIE and MBE-treated cells, more pronounced in the MBE groups, points to an increased cellular antioxidant capacity [
21]. Glutathione is a tripeptide composed of glutamic acid, cysteine, and glycine. Dimethylglycine (DMG) can be generated from choline metabolism to betaine [
22]. DMG can further metabolize to glycine via transamination, indicating its role as both a precursor and intermediate in glycine metabolism [
23]. Methionine undergoes transmethylation to form S-adenosylmethionine (SAM), which participates in various methylation reactions including glutathione synthesis [
24]. Methionine metabolism provides a source of sulfur to promote glutathione synthesis. While both herbs showed a trend towards increasing L-methionine levels, only MBE reached statistical significance, indicating distinct mechanisms of action on methionine metabolism, with MBE potentially promoting methionine salvage pathways. L-glutamic acid contributes to glutathione synthesis through the catalytic action of glutathione synthetase. Interestingly, both MIE and MBE exhibit a higher increase in glutathione levels at lower doses compared to higher doses, suggesting that lower doses may channel more L-glutamic acid into glutathione synthesis. Beyond its antioxidant role, glutathione is implicated in the conjugation of leukotriene C4 with reduced glutathione to form leukotriene C4. Elevated glutathione levels may further contribute to the anti-inflammatory effects by potentially curbing leukotriene production, given that glutathione is involved in the conjugation of LTA4 to form the pro-inflammatory mediator LTC4 [
25].
4.3. Arginine and Proline Metabolism
In the Arginine and Proline Metabolism pathway, Arginine and Proline are pivotal metabolites. Argininosuccinic acid serves as a precursor for L-arginine synthesis, while argininosuccinic acid itself is synthesized from L-aspartic acid and citrulline [
26]. Both herbal varieties significantly elevate the levels of L-aspartic acid, argininosuccinic acid, and L-arginine, with MBE showing a more pronounced effect.
This finding is particularly relevant as arginine, a product of argininosuccinic acid metabolism via argininosuccinate lyase (ASL), serves as a precursor for nitric oxide (NO) synthesis, a potent anti-microbial and inflammatory mediator [
27,
28]. The observed changes suggest that MBE may exert its anti-inflammatory effects primarily by interfering with NO production, possibly through the modulation of arginine availability or alterations in the activity of enzymes such as argininosuccinate synthase 1 (ASS1) and ASL [
29].
L-asparagine and L-aspartic acid are interconvertible. Interestingly, MIE only moderately increased L-aspartic acid levels compared to the model group, with concentrations lower than both the control and MBE groups. However, MIE significantly elevated L-asparagine content, indicating an enhanced conversion of L-aspartic acid to L-asparagine. This is noteworthy because asparagine has been shown to differentially regulate mRNA expression of Toll-like receptor 4 (TLR4) and nucleotide-binding oligomerization domain (NOD) signaling pathways and their negative regulators [
30]. During early lipopolysaccharide (LPS) challenge, asparagine downregulates the mRNA expression of genes related to these signaling pathways, while in late-stage challenge, it upregulates their expression, potentially mitigating LPS-induced liver injury [
31].
Both varieties significantly increase proline levels, with MIE showing a more pronounced effect. Proline metabolism is a crucial pathway in regulating macrophage polarization. Studies have shown that M1-type macrophages exhibit enhanced proline oxidative metabolism, while M2-type macrophages display increased proline synthetic metabolism. Thus, proline metabolism plays a vital role in modulating the M1/M2 macrophage polarization balance [
32,
33].
These results suggest that MIE exerts its anti-inflammatory effects primarily by promoting the conversion of L-aspartic acid to L-asparagine and enhancing proline synthesis metabolism. This mechanism likely influences macrophage polarization towards an anti-inflammatory M2 phenotype. Conversely, MBE appears to achieve its anti-inflammatory effects mainly by inhibiting NO synthesis through modulation of arginine metabolism.
4.4. Energy Metabolism
Compared to the normal group, the LPS-stimulated model group exhibited a sharp decrease in fumaric acid, cis-aconitic acid, and malic acid, coupled with a dramatic increase in succinic acid and citric acid. This pattern suggests a potential disruption in TCA cycle flux in response to inflammatory stimuli. High doses of MBE significantly elevated levels of fumaric acid, cis-aconitic acid, succinic acid, and malic acid, supporting the idea of enhanced TCA cycle activity [
32,
34]. These changes indicate that MBE may effectively promote a shift towards oxidative metabolism, potentially countering the inflammatory disruption of the TCA cycle.
The model group showed markedly reduced levels of fructose 6-phosphate and beta-D-glucose 6-phosphate, alongside significantly elevated levels of L-lactic acid and pyruvic acid. This metabolic shift indicates an activation of macrophages by LPS stimulation, leading to enhanced glycolysis [
35]. The increased consumption of glycolytic intermediates and accumulation of end products (lactate and pyruvate) suggest that activated macrophages increase glucose uptake and glycolytic rates to meet heightened energy demands [
36]. Interestingly, both MIE and MBE treatments led to a dramatic increase in phosphoenolpyruvic acid (PEP) levels compared to both model and normal groups, with MBE demonstrating a stronger effect. As PEP is a regulatory point in the glycolytic pathway, its accumulation can inhibit early glycolytic steps, thus modulating the overall metabolic flux [
37]. MBE treatment also reduced pyruvic acid levels, further indicating its ability to modulate glycolysis. These changes suggest that both treatments, particularly MBE, can inhibit the inflammation-induced enhancement of glycolysis.
In the model group, lipoamide levels increased sharply compared to the normal group. Following treatment with both varieties, lipoamide levels decreased significantly, falling below both model and normal group levels. A decrease in lipoamide indicates increased utilization of this cofactor, potentially suggesting higher activity of pyruvate dehydrogenase (PDH), which catalyzes the conversion of pyruvic acid to acetyl-CoA [
38]. PDH activity is inhibited by LPS and this increased utilization by treatment could support oxidative phosphorylation. Levels of leucine were significantly higher in the treatment groups compared to the normal and model groups in a dose-dependent manner. Leucine is a branched-chain amino acid (BCAA), which undergoes transamination during metabolism in the body to release ammonia and the corresponding keto acids [
39]. These keto acids can further be converted to pyruvic acid, and then through transamination, pyruvic acid accepts the amino group from glutamic acid to generate alanine, a process that helps regulate nitrogen balance in the body. which is greatly disrupted by LPS. Alanine levels were also significantly higher in the treatment groups compared to the normal and model groups in a dose-dependent manner, with an opposite trend to pyruvic acid, indicating that MBE significantly promoted the oxidation of BCAAs. Oxidation of branched-chain amino acids can generate a large amount of ATP to provide energy for cells, and its oxidative product pyruvic acid is also an important substrate entering the TCA cycle to produce more ATP and NADH, thereby increasing energy supply to cells [
40].
A sharp increase in 6-phosphogluconic acid in the model group points to an upregulation of the Pentose Phosphate Pathway (PPP), which is crucial for providing NADPH for NOX2-dependent ROS production in M1 macrophages and generating ribose-5-phosphate for nucleotide synthesis [
41]. Both MIE and MBE treatments significantly reduced 6-phosphogluconic acid levels to a point between normal and model group levels, suggesting partial suppression of inflammation-induced PPP activation. Notably, treatment with both herbs led to a marked increase in gluconolactone levels compared to normal and model groups. This implies potential inhibition of 6-phosphogluconate dehydrogenase (G6PD), the enzyme converting 6-phosphogluconic acid to ribulose-5-phosphate in the PPP. These changes could contribute to the anti-inflammatory effect by reducing NADPH production for ROS generation [
42].
4.5. Other Pathways
LPS stimulation significantly altered metabolites involved in purine metabolism, pyrimidine metabolism, amino sugar metabolism, tyrosine metabolism, and vitamin B metabolism. Purines and pyrimidines are essential for DNA and RNA synthesis [
43,
44]. Metabolites like adenosine monophosphate are involved in the synthesis of cAMP and may be influenced by the treatments, affecting inflammatory processes [
45]. Both MIE and MBE showed changes in these metabolites, suggesting an influence on purine and pyrimidine metabolism. Levels of xanthine and hypoxanthine were significantly higher in the high-dose MBE group compared to other groups following treatment, further indicating that MIE has a greater ability to enhance antioxidant capacity compared to MBE. Metabolites in the vitamin B series function as coenzymes in biochemical reactions and participate in various biochemical processes [
46,
47]. Amino sugar metabolism is involved in the synthesis of amino sugars, which are important components of glycosaminoglycans and other polysaccharides. These molecules play a role in cell-cell interactions and signal transduction, relevant to immune cell activation and inflammatory responses. The modulation of this pathway by MIE and MBE may contribute to their anti-inflammatory effects by affecting the synthesis and function of these important biomolecules. The differential effects on dopamine might be related to the superior anti-inflammatory action of MIE. Dopamine has been shown to have immunomodulatory effects, potentially contributing to the anti-inflammatory response.
In conclusion, the metabolic changes induced by Meconopsis species point to a multifaceted mechanism of action. MIE appears to modulate central metabolic pathways, including glycolysis, the TCA cycle, and the PPP, while also enhancing antioxidant capacity. These effects collectively contribute to its anti-inflammatory action, potentially by reprogramming macrophage metabolism towards a less inflammatory state. The ability of MBE to more potently inhibit glycolysis, enhance TCA cycle activity, modulate the PPP, and boost antioxidant capacity compared to MBE suggests it may be a more effective anti-inflammatory agent. Further research into the specific components of MIE and their direct effects on these metabolic pathways would provide more detailed insights into its anti-inflammatory mechanism.
5. Conclusions
This study comprehensively evaluated the anti-inflammatory potential of Meconopsis species, focusing on the differences between yellow-flowered M. integrifolia and blue-flowered M. betonicifolia. Our findings demonstrate significant anti-inflammatory properties in both species, with M. integrifolia consistently exhibiting superior effects across multiple inflammatory markers and pathways.
Both Meconopsis varieties significantly reduced pro-inflammatory cytokine production in LPS-stimulated RAW 264.7 cells. However, yellow-flowered Meconopsis showed a more potent inhibition of NO production and demonstrated superior capacity to modulate key metabolic pathways, including glycolysis inhibition, TCA cycle enhancement, PPP modulation, and antioxidant capacity improvement. These results suggest that while both species possess valuable anti-inflammatory properties, yellow-flowered Meconopsis may be a more promising candidate for anti-inflammatory therapeutic development.
Our findings have important implications for both traditional medicine and modern drug discovery. They provide scientific validation for the traditional use of Meconopsis in treating inflammatory conditions and offer new insights into potential clinical substitutes for blue-flowered Meconopsis. Furthermore, this comparative approach underscores the importance of species-specific evaluations within a genus, as closely related plants may exhibit varying degrees of bioactivity.
Collectively, the results establish yellow- and blue-flowered Meconopsis as credible alternatives with comparable anti-inflammatory properties, validating their interchangeable use based on activity and pharmacological criteria. The metabolic profiling provides initial clues regarding their multi-targeted modes of action, with M. integrifolia showing a greater capacity to modulate inflammatory metabolism. Further work characterizing specific bioactive constituents is warranted to dissect their precise mechanisms of action.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1: The 277 compounds identified through LC-MS analysis in the yellow-flowered and blue-flowered Meconopsis species.
Author Contributions
Conceptualization, P.C., R.G. and C.W.; methodology, K.N.; software, Q.X. and F.Z.; validation, S.K., Z.J, and D.J.; formal analysis, P.C. and Q.X.; writing—original draft preparation, R.G.; writing—review and editing, C.W; project administration, X.F.; funding acquisition, J.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Key Research and Development Plan of Tibet Autonomous Region, China (Grant No. XZ202101ZY0013G). The project, titled "Research on Key Technologies for Artificial Propagation of Endangered Tibetan Medicinal Plant Green Velvet Saussurea," was supervised by the State-owned Assets Supervision and Administration Commission of the People's Government of Tibet Autonomous Region.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Guo, Q.; Bai, R.; Zhao, B.; Feng, X.; Zhao, Y.; Tu, P.; Chai, X. An ethnopharmacological, phytochemical and pharmacological review of the genus Meconopsis. Am J Chin Med 2016, 44, 439–462. [Google Scholar] [CrossRef] [PubMed]
- Shi, N.; Wang, C.; Wang, J.; Wu, N.; Naudiyal, N.; Zhang, L.; Wang, L.; Sun, J.; Du, W.; Wei, Y.; et al. Biogeographic patterns and richness of the Meconopsis species and their influence factors across the Pan-Himalaya and adjacent regions. 2022, 14, 661. [Google Scholar] [CrossRef]
- Wang, W.; Guo, W.; Jarvie, S.; Svenning, J. The fate of Meconopsis species in the Tibeto-Himalayan region under future climate change. 2021, 11, 887–899. [Google Scholar] [CrossRef]
- Wangchuk, P.; Samten; Jamtsho, T. Phytopharmaceutical properties and quality assessment of two Himalayan medicinal plants, Meconopsis horridula and Meconopsis simplicifolia. Journal of Herbal Medicine 2023, 38, 100628. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Q.; Wang, R.; Yang, Y.; Wang, Y.; Liu, X.; Zhang, X.; Qiao, X.; Zhong, G.; Wei, J.; et al. Preliminary study on the effective site and mechanism of action of Meconopsis quintuplinervia regel in alleviating acute alcoholic liver injury in mice. Journal of Ethnopharmacology 2023, 308, 116230. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Burgess, K.; Yang, X.; Ahrends, A.; Gao, L.; Li, D. Upward elevation and northwest range shifts for alpine Meconopsis species in the Himalaya–Hengduan Mountains region. 2019, 9, 4055–4064. [Google Scholar] [CrossRef]
- He, J.; Huang, B.; Ban, X.; Tian, J.; Zhu, L.; Wang, Y. In vitro and in vivo antioxidant activity of the ethanolic extract from Meconopsis quintuplinervia. Journal of Ethnopharmacology 2012, 141, 104–110. [Google Scholar] [CrossRef]
- Zhao, F.; Bai, R.; Li, J.; Feng, X.; Jiao, S.; Wuken, S.; Ge, F.; Zhang, Q.; Zhou, X.; Tu, P.; et al. Meconopsis horridula Hook. f. & Thomson extract and its alkaloid oleracein E exert cardioprotective effects against acute myocardial ischaemic injury in mice. Journal of Ethnopharmacology 2020, 258, 112893. [Google Scholar] [CrossRef]
- Yang, F.; Qin, A.; Li, Y.; Wang, X. Great genetic differentiation among populations of Meconopsis integrifolia and its implication for plant speciation in the Qinghai-Tibetan Plateau. PLOS ONE 2012, 7, e37196. [Google Scholar] [CrossRef]
- Fan, J.; Wang, Y.; Wang, X.; Wang, P.; Tang, W.; Yuan, W.; Kong, L.; Liu, Q. The antitumor activity of Meconopsis Horridula Hook, a traditional Tibetan medical plant, in murine leukemia L1210 cells. Cellular Physiology and Biochemistry 2015, 37, 1055–1065. [Google Scholar] [CrossRef]
- Sergeant, S.; Rahbar, E.; Chilton, F.H. Gamma-linolenic acid, Dihommo-gamma linolenic, eicosanoids and inflammatory processes. European Journal of Pharmacology 2016, 785, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Kikut, J.; Komorniak, N.; Ziętek, M.; Palma, J.; Szczuko, M. Inflammation with the participation of arachidonic (AA) and linoleic acid (LA) derivatives (HETEs and HODEs) is necessary in the course of a normal reproductive cycle and pregnancy. Journal of Reproductive Immunology 2020, 141, 103177. [Google Scholar] [CrossRef] [PubMed]
- Sul, O.-J.; Ra, S.W. Quercetin prevents LPS-induced oxidative stress and inflammation by modulating NOX2/ROS/NF-kB in lung epithelial cells. Molecules 2021, 26(22), 6949. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Xiao, Y.; Hu, H.; Zou, Q.; Li, Y.; Gao, Y.; Ge, W.; Cheng, X.; Sun, S.C. Proinflammatory TLR signalling is regulated by a TRAF2-dependent proteolysis mechanism in macrophages. Nat Commun 2015, 6, 5930. [Google Scholar] [CrossRef]
- Mohammadi, A.; Sharifi, A.; Pourpaknia, R.; Mohammadian, S.; Sahebkar, A. Manipulating macrophage polarization and function using classical HDAC inhibitors: implications for autoimmunity and inflammation. Crit Rev Oncol Hematol 2018, 128, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Mass spectrometry-based metabolomics: applications to biomarker and metabolic pathway research. Biomed. Chromatogr. 2016, 30, 7–12. [Google Scholar] [CrossRef]
- Hanna, V.S.; Hafez, E.A.A. Synopsis of arachidonic acid metabolism: a review. J Adv Res 2018, 11, 23–32. [Google Scholar] [CrossRef]
- Herschman, H.R. , Xie, W., Reddy, S. (1999). Function and regulation of prostaglandin synthase 2. in: Honn, K.V., Marnett, L.J., Nigam, S., Dennis, E.A. (eds) Eicosanoids and other bioactive lipids in cancer, inflammation, and radiation injury, 4. Advances in Experimental Medicine and Biology, vol 469. Springer, Boston, MA. [CrossRef]
- Granström, E. The arachidonic acid cascade. Inflammation 1984, 8, S15–S25. [Google Scholar] [CrossRef]
- Brenna, J.T.; Salem, N., Jr.; Sinclair, A.J.; Cunnane, S.C. α-Linolenic acid supplementation and conversion to n-3 long-chain polyunsaturated fatty acids in humans. Prostaglandins Leukot Essent Fatty Acids 2009, 80, 85–91. [Google Scholar] [CrossRef]
- Averill-Bates, D.A. The antioxidant glutathione. Vitamins and Hormones 2023, 121, 109–141. [Google Scholar] [CrossRef]
- Lever, M.; McEntyre, C.J.; George, P.M.; Chambers, S.T. Is N,N-dimethylglycine N-oxide a choline and betaine metabolite? Biol. Chem. 2017, 398, 775–784. [Google Scholar] [CrossRef]
- Magnusson, M.; Wang, T.J.; Clish, C.; Engström, G.; Nilsson, P.; Gerszten, R.E.; Melander, O. Dimethylglycine deficiency and the development of diabetes. Diabetes 2015, 64, 3010–3016. [Google Scholar] [CrossRef] [PubMed]
- Sutter, B.M.; Wu, X.; Laxman, S.; Tu, B.P. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 2013, 154, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Gresser, M.J.; Ford-Hutchinson, A.W. Kinetic mechanism of glutathione conjugation to leukotriene A4 by leukotriene C4 synthase. Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism 1998, 1391, 157–168. [Google Scholar] [CrossRef]
- Wijnands, K.A.P.; Castermans, T.M.R.; Hommen, M.P.J.; Meesters, D.M.; Poeze, M. Arginine and citrulline and the immune response in sepsis. Nutrients 2015, 7(3), 1426–1463. [Google Scholar] [CrossRef]
- Moncada, S.; Higgs, A. The L-Arginine-Nitric Oxide Pathway. N Engl J Med 1993, 329, 2002–2012. [Google Scholar] [CrossRef]
- Martí i Líndez, A.-A.; Reith, W. Arginine-dependent immune responses. Cellular and Molecular Life Sciences 2021, 78, 5303–5324. [Google Scholar] [CrossRef]
- Tsai, WB. , Long, Y., Savaraj, N., Feun, L.G., Kuo, M.T. (2017). Mechanisms of l-Arginine-Auxotrophic response and their cancer therapeutic implications. In: Patel, V., Preedy, V., Rajendram, R. (eds) L-Arginine in clinical nutrition. Nutrition and Health. Humana Press, Cham. [CrossRef]
- Wang, X.; Liu, Y.; Wang, S.; Pi, D.; Leng, W.; Zhu, H.; Zhang, J.; Shi, H.; Li, S.; Lin, X.; et al. Asparagine reduces the mRNA expression of muscle atrophy markers via regulating protein kinase B (Akt), AMP-activated protein kinase α, toll-like receptor 4 and nucleotide-binding oligomerisation domain protein signalling in weaning piglets after lipopolysaccharide challenge. British Journal of Nutrition 2016, 116, 1188–1198. [Google Scholar] [CrossRef]
- Chen, S.; Liu, Y.; Wang, X.; Wang, H.; Li, S.; Shi, H.; Zhu, H.; Zhang, J.; Pi, D.; Hu, C.-A.A.; et al. Asparagine improves intestinal integrity, inhibits TLR4 and NOD signaling, and differently regulates p38 and ERK1/2 signaling in weanling piglets after LPS challenge. Innate Immun. 2016, 22, 577–587. [Google Scholar] [CrossRef]
- Kieler, M.; Hofmann, M.; Schabbauer, G. More than just protein building blocks: how amino acids and related metabolic pathways fuel macrophage polarization. FEBS J. 2021, 288, 3694–3714. [Google Scholar] [CrossRef]
- Vettore, L.A.; Westbrook, R.L.; Tennant, D.A. Proline metabolism and redox; maintaining a balance in health and disease. Amino Acids 2021, 53, 1779–1788. [Google Scholar] [CrossRef]
- Ji, D.; Yin, J.; Li, D.; Zhu, C.; Ye, J.; Pan, Y. Effects of inflammatory and anti-inflammatory environments on the macrophage mitochondrial function. Scientific Reports 2020, 10, 20324. [Google Scholar] [CrossRef]
- Soto-Heredero, G.; Gómez de las Heras, M.M.; Gabandé-Rodríguez, E.; Oller, J.; Mittelbrunn, M. Glycolysis – a key player in the inflammatory response. FEBS J. 2020, 287, 3350–3369. [Google Scholar] [CrossRef]
- Chiba, S.; Hisamatsu, T.; Suzuki, H.; Mori, K.; Kitazume, M.T.; Shimamura, K.; Mizuno, S.; Nakamoto, N.; Matsuoka, K.; Naganuma, M.; et al. Glycolysis regulates LPS-induced cytokine production in M2 polarized human macrophages. Immunology Letters 2017, 183, 17–23. [Google Scholar] [CrossRef]
- Boscá, L.; González-Ramos, S.; Prieto, P.; Fernández-Velasco, M.; Mojena, M.; Martín-Sanz, P.; Alemany, S. Metabolic signatures linked to macrophage polarization: from glucose metabolism to oxidative phosphorylation. Biochemical Society Transactions 2015, 43, 740–744. [Google Scholar] [CrossRef]
- Palmieri, E.M.; Holewinski, R.; McGinity, C.L.; Pierri, C.L.; Maio, N.; Weiss, J.M.; Tragni, V.; Miranda, K.M.; Rouault, T.A.; Andresson, T.; et al. Pyruvate dehydrogenase operates as an intramolecular nitroxyl generator during macrophage metabolic reprogramming. Nature Communications 2023, 14, 5114. [Google Scholar] [CrossRef]
- Bonvini, A.; Rogero, M.M.; Coqueiro, A.Y.; Raizel, R.; Bella, L.M.; Fock, R.A.; Borelli, P.; Tirapegui, J. Effects of different branched-chain amino acids supplementation protocols on the inflammatory response of LPS-stimulated RAW 264.7 macrophages. Amino Acids 2021, 53, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Wang, S.; Zhang, C.; Zhao, Y. Coordinated Modulation of Energy Metabolism and Inflammation by Branched-Chain Amino Acids and Fatty Acids. Front. Endocrinol. 2020, 11, 617. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, R.; Gu, H.; Zhang, E.; Qu, J.; Cao, W.; Huang, X.; Yan, H.; He, J.; Cai, Z. Metabolic reprogramming in macrophage responses. Biomarker Research 2021, 9, 1. [Google Scholar] [CrossRef]
- Ham, M.; Lee, J.-W.; Choi, A.H.; Jang, H.; Choi, G.; Park, J.; Kozuka, C.; Sears, D.D.; Masuzaki, H.; Kim, J.B. Macrophage glucose-6-phosphate dehydrogenase stimulates proinflammatory responses with oxidative stress. Molecular and Cellular Biology 2013, 33, 2425–2435. [Google Scholar] [CrossRef] [PubMed]
- Linden, J.; Koch-Nolte, F.; Dahl, G. Purine release, metabolism, and signaling in the inflammatory response. Annu Rev Immunol 2019, 37, 325–347. [Google Scholar] [CrossRef]
- John, S.V.; Seim, G.L.; Erazo-Flores, B.J.; Steill, J.; Freeman, J.; Votava, J.A.; Arp, N.L.; Qing, X.; Stewart, R.; Knoll, L.J.; et al. Macrophages undergo functionally significant reprograming of nucleotide metabolism upon classical activation. bioRxiv 2023, 12, 27–573447. [Google Scholar] [CrossRef]
- Haskó, G.; Pacher, P. Regulation of Macrophage Function by Adenosine. J. Am. Heart Assoc. 2012, 32, 865–869. [Google Scholar] [CrossRef]
- Yanaka, N.; Koyama, T.-A.; Komatsu, S.-I.; Nakamura, E.; Kanda, M.; Kato, N. Vitamin B6 suppresses NF-κB activation in LPS-stimulated mouse macrophages. Int J Mol Med 2005, 16, 1071–1075. [Google Scholar] [CrossRef]
- Shan, M.-R.; Zhou, S.-N.; Fu, C.-N.; Song, J.-W.; Wang, X.-Q.; Bai, W.-W.; Li, P.; Song, P.; Zhu, M.-L.; Ma, Z.-M.; et al. Vitamin B6 inhibits macrophage activation to prevent lipopolysaccharide-induced acute pneumonia in mice. J. Cell. Mol. Med. 2020, 24, 3139–3148. [Google Scholar] [CrossRef]
Figure 1.
Effects of MIE (yellow) and MBE (blue) extracts on RAW264.7 cell viability and nitric oxide (NO) production under inflammatory conditions. A. Cytotoxicity profiles: cell survival rate. B. Dose-dependent inhibition of NO production: NO inhibition rate in LPS-stimulated cells.
Figure 1.
Effects of MIE (yellow) and MBE (blue) extracts on RAW264.7 cell viability and nitric oxide (NO) production under inflammatory conditions. A. Cytotoxicity profiles: cell survival rate. B. Dose-dependent inhibition of NO production: NO inhibition rate in LPS-stimulated cells.
Figure 2.
Dose-dependent antioxidant and anti-inflammatory effects of MIE (yellow) and MBE (blue). A. Impact on oxidative stress markers GSH, MDA, and SOD levels in cells. B. Effects on inflammatory gene expression of IL-1A, IL-1B, IL-6, and TNF in cells.
Figure 2.
Dose-dependent antioxidant and anti-inflammatory effects of MIE (yellow) and MBE (blue). A. Impact on oxidative stress markers GSH, MDA, and SOD levels in cells. B. Effects on inflammatory gene expression of IL-1A, IL-1B, IL-6, and TNF in cells.
Figure 3.
Comparative metabolomics analysis of extracts of yellow (MIE) and blue (MBE) Meconopsis varieties. The heatmap illustrates the relative abundance of compounds across different categories in the two Meconopsis species. Compounds are classified into 12 major categories and further grouped as yellow-specific, common to both species, or blue-specific. The color scale ranges from blue (lower abundance) to red (higher abundance).
Figure 3.
Comparative metabolomics analysis of extracts of yellow (MIE) and blue (MBE) Meconopsis varieties. The heatmap illustrates the relative abundance of compounds across different categories in the two Meconopsis species. Compounds are classified into 12 major categories and further grouped as yellow-specific, common to both species, or blue-specific. The color scale ranges from blue (lower abundance) to red (higher abundance).
Figure 4.
Gene Ontology (GO) enrichment analysis of common feature targets in Meconopsis species. GO enrichment analysis of 413 genes from two Meconopsis varieties. GO terms are categorized into Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). The color intensity represents the -log10(p-value) of enrichment, while the size of each square indicates the count of genes associated with each GO term.
Figure 4.
Gene Ontology (GO) enrichment analysis of common feature targets in Meconopsis species. GO enrichment analysis of 413 genes from two Meconopsis varieties. GO terms are categorized into Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). The color intensity represents the -log10(p-value) of enrichment, while the size of each square indicates the count of genes associated with each GO term.
Figure 5.
Sankey diagram illustrating the relationships between major compound categories (left) and their associated enriched GO terms (right) identified in the common feature targets of both yellow and blue Meconopsis species. The width of the flows represents the strength of the association between compound categories and GO terms.
Figure 5.
Sankey diagram illustrating the relationships between major compound categories (left) and their associated enriched GO terms (right) identified in the common feature targets of both yellow and blue Meconopsis species. The width of the flows represents the strength of the association between compound categories and GO terms.
Figure 6.
Gene-disease enrichment of common feature targets in Meconopsis species. A. Gene-disease network. B. Bubble plot of enriched diseases. Diseases were categorized by their corresponding therapeutic efficacy and represented by different colors. The size of the disease name and each bubble correlates with the adjusted p-value, where larger text sizes and bubbles indicate higher enrichment significance for the associated disease.
Figure 6.
Gene-disease enrichment of common feature targets in Meconopsis species. A. Gene-disease network. B. Bubble plot of enriched diseases. Diseases were categorized by their corresponding therapeutic efficacy and represented by different colors. The size of the disease name and each bubble correlates with the adjusted p-value, where larger text sizes and bubbles indicate higher enrichment significance for the associated disease.
Figure 7.
Metabolomics analysis of anti-inflammatory effects of Meconopsis varieties in LPS-stimulated RAW264.7 cells. A. Positive ion mode PCA score plot. B. Negative ion mode PCA score plot. C. Positive ion mode PLS-DA score plot. D. Negative ion mode PLS-DA score plot.
Figure 7.
Metabolomics analysis of anti-inflammatory effects of Meconopsis varieties in LPS-stimulated RAW264.7 cells. A. Positive ion mode PCA score plot. B. Negative ion mode PCA score plot. C. Positive ion mode PLS-DA score plot. D. Negative ion mode PLS-DA score plot.
Figure 8.
Heatmap of differentially abundant metabolites across key metabolic pathways in response to LPS stimulation and Meconopsis treatments. This heatmap illustrates the relative abundance of metabolites involved in nine key pathways: Arachidonic Acid Metabolism, Glutathione and Methionine Metabolism, Arginine and Proline Metabolism, Energy Metabolism, Purine Metabolism, Pyrimidine Metabolism, Amino Sugar Metabolism, Tyrosine Metabolism, and Vitamin B Metabolism. The color scale ranges from blue (lower abundance) to red (higher abundance). Asterisks indicate the significance level of differential abundance compared to the model group (M).
Figure 8.
Heatmap of differentially abundant metabolites across key metabolic pathways in response to LPS stimulation and Meconopsis treatments. This heatmap illustrates the relative abundance of metabolites involved in nine key pathways: Arachidonic Acid Metabolism, Glutathione and Methionine Metabolism, Arginine and Proline Metabolism, Energy Metabolism, Purine Metabolism, Pyrimidine Metabolism, Amino Sugar Metabolism, Tyrosine Metabolism, and Vitamin B Metabolism. The color scale ranges from blue (lower abundance) to red (higher abundance). Asterisks indicate the significance level of differential abundance compared to the model group (M).
Figure 9.
Network visualization of metabolite-gene interactions in enriched pathways. This figure presents a comprehensive network analysis of the interactions between differentially abundant metabolites and their corresponding genes within the enriched pathways. Nodes representing differential metabolites and their associated genes, and edges connecting these nodes signify the biochemical reactions or regulatory interactions that occur between them.
Figure 9.
Network visualization of metabolite-gene interactions in enriched pathways. This figure presents a comprehensive network analysis of the interactions between differentially abundant metabolites and their corresponding genes within the enriched pathways. Nodes representing differential metabolites and their associated genes, and edges connecting these nodes signify the biochemical reactions or regulatory interactions that occur between them.
Table 1.
Primer Sequences for Real-Time Quantitative PCR in Mice.
Table 1.
Primer Sequences for Real-Time Quantitative PCR in Mice.
| Gene |
Forward primer (5'-3') |
Reverse primer (3'-5') |
| IL-1β |
GAAATGCCACCTTTTGACAGTG |
TGGATGCTCTCATCAGGACAG |
| IL-1α |
TCTCAGATTCACAACTGTTCGTG |
AGAAAATGAGGTCGGTCTCACTA |
| TNF-α |
GTAGCCCACGTCGTAGCAAA |
ACAAGGTACAACCCATCGGC |
| IL-6 |
TGGAGTACCATAGCTACCTGGA |
TGGAAATTGGGGTAGGAAGGAC |
| β-actin |
GATATCGCTGCGCTGGTCG |
CATTCCCACCATCACACCCT |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).