1. Introduction
Optical coherence tomography (OCT) is a non-invasive imaging technique that uses low-coherence interferometry to produce high-resolution, cross-sectional images [
1]. In ophthalmology, OCT allows for detailed visualization of the retina, enabling early detection and continuous monitoring of various retinal diseases such as age-related macular degeneration (AMD) [
2] and diabetic macular edema (DME) [
3]. Deep learning (DL) has made significant breakthroughs in medical imaging, particularly for image classification and segmentation. Studies have demonstrated that using DL for interpreting OCT is efficient and accurate and performs well for discriminating eyes affected by a disease from normal [
4]. This suggests that incorporating DL technology in OCT addresses the gaps in the current practice and clinical workflow [
4]. Recent advances in DL algorithms offer accurate detection of complex image patterns, achieving accuracy levels in image classification and segmentation tasks. Segmentation analysis of the SD-OCT image may also eliminate the need for manual refinement of conventionally segmented retinal layers and biomarkers [
5].
Modern artificial intelligence (AI) methods have recently increased exponentially due to the advent of affordable computational capabilities. Many innovative segmentation methods are generalized on widespread datasets. For example, a novel Segment Anything Model (SAM) was mainly trained on datasets of ordinary objects [
6]. Although it contains microscopy [
7] and X-ray [
8] images, according to the original paper, it does not contain OCT or fundus images. Therefore, using such models in ophthalmology could be enhanced by fine-tuning the foundational models. Applying such innovative models in OCT can improve the quality of ophthalmologic service and assist physicians in decision-making [
9].
1.1. Deep Learning for Segmentation of OCT Biomarkers
In recent years, the application of DL in OCT has developed enormously, and many methods for biomarkers segmentation on OCT B-scans have been developed, addressing specific challenges like segmentation of macular holes (MH), intraretinal cysts (IRC), intraretinal fluid (IRF), and pigment epithelial detachment (PED).
Table 1 summarizes novel studies in the domain.
Ganjee et al. [
10] proposed an unsupervised segmentation approach in a three-level hierarchical framework for segmenting IRC regions in the SD-OCT images. In the first level, the ROI mask is built, keeping the exact retina area. The prune mask is built in the second level, reducing the search area. In the third level, the cyst mask is extracted by applying the Markov Random Field (MRF) model and employing intensity and contextual information. The authors tested their approach on the public datasets where B-scans with IRC are available: OPTIMA [
19], UMN [
20], and Kermany [
21].
Wang et al. [
14] proposed a new CNN-based architecture D3T-FCN by integrating the principle of Auto-Encoder and the U-Net model for joint segmentation of MH and cystoid macular edema (CME) in retinal OCT images. The authors evaluated the method with the AROI dataset.
U-Net-based models are still widespread. In contemporary works, authors still use U-Net and its modifications. Rahil et al. [
11] proposed an ensemble approach utilizing U-Net models with a combination of the relative layer distance, independently trained for IRF, SFR, and PED classes, available in the RETOUCH dataset. Ganjee et al. [
12] proposed a two-staged framework for IRC segmentation. In the first step, prior information is embedded, and the input data is adjusted. In the second step, an extended structure of the U-Net with an implemented connection module between the encoder and decoder parts was applied to the scans to predict the masks. Melinščak et al. [
13] used Attention U-Net and applied it to the AROI dataset, focusing on AMD biomarkers, particularly subretinal fluid (SRF), IRF, and PED. Attention added to U-Net architecture is a way to highlight relevant activations during the training, which potentially leads to better generalization. Daanouni et al. [
15] proposed a new architecture inspired by U-Net++ and a spatially adaptive denormalization unit with a class-guided module. The segmentation was performed in two stages to segment the layers and fluids in the AROI dataset. George et al. [
16] used the U-Net model trained on DME scans from Kermany dataset and evaluated the model on the Lu et al. [
22] dataset.
New works pioneered using SAM-based models for ophthalmology have recently begun to appear. Qiu et al. [
17] explored the Segment Anything Model (SAM) application in ophthalmology, including in their training process fundus, OCT, and OCT Angiography (OCTA) datasets. The authors proposed a fine-tuning method, training only the prompt layer and task head based on a one-shot mechanism. The model’s performance raised about 25% of the Dice score compared with the pure SAM. Fazekas et al. [
18] adapted SAM specifically for the OCT domain using a fine-tuning strategy on the RETOUCH dataset and compared IRF, SRF, and PED segmentation results for pure SAM, SAM with fine-tuned decoder and SAMed. Fine-tuning the decoder led to a significant performance improvement, with an increase in Dice score of up to 50% compared to zero-short segmentation using SAM.
These studies collectively showcase the ongoing advancements in OCT image segmentation. Models like U-Net and its variants continue to dominate, while newer approaches like SAM are beginning to emerge, promising enhanced segmentation performance across various retinal biomarkers.
1.2. OCT Biomarkers Segmentation
Identifying specific biomarkers in OCT images can help classify retinal diseases such as Age-related Macular Degeneration (AMD), Diabetic Retinopathy (DR), Diabetic Macular Edema (DME), Central Serous Chorioretinopathy (CSH), Retinal Vein Occlusion (RVO), Retinal Artery Occlusion (RAO), and Vitreoretinal Interface Disease (VID).
Modern literature uses various biomarker names to describe typical changes in the macula seen in OCT: disorganization of retinal inner layers (DRIL) [
23,
24], different stages of outer retina atrophy (ORA) [
25], soft drusen [
26], double-layer sign [
27], intraretinal fluid (IRF), subretinal fluid (SRF), pigment epithelial detachment (PED) [
28], macular hole (MH), intraretinal cyst (IRC) [
29], etc.
With the rapid development of OCT technology, new biomarkers regularly appear, and older classifications are revised. For example, IRC and IRF are very similar. Morphologically, they both refer to cavities in the retina’s neuroepithelium of different origins. Cavities found around a macular hole in VID are often called IRC, while cysts formed by fluid leaking from retinal vessels in DR, DME, or AMD are more often called IRF. The names of biomarkers often vary depending on the size and contents of the cavities caused by the disease’s pathogenesis.
In public datasets, we found four biomarkers that can describe volumetric changes in the macula. IRF and PED often appeared together when describing vascular-related pathologies (AMD, DME, RVO), while MH and IRC were paired to describe VID and DR,
Table 1.
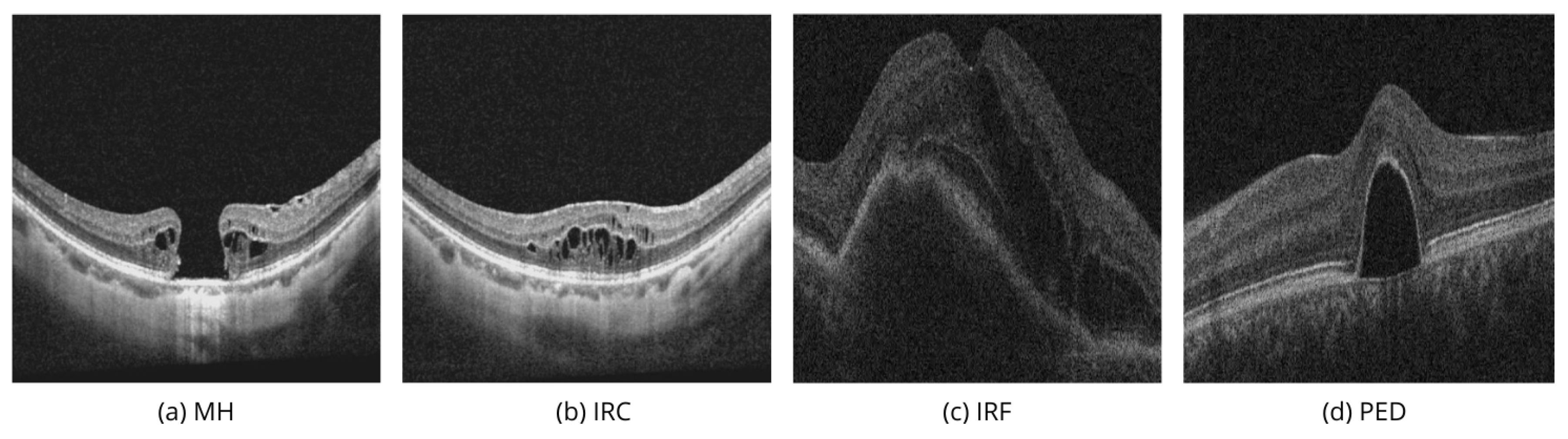
Figure 1a,b show two linear images taken from the same patient in different areas of the macula.
Figure 1a passes through the center of the macula, while
Figure 1b passes through an area next to the main defect.
Figure 1b shows multiple IRCs that form in the areas surrounding the macular hole in response to damage.
Figure 1a shows the MH biomarker, which provides important diagnostic information about a full-thickness defect in the macula, including the photoreceptor layer, which can severely affect vision. In modern vitreous surgery, the maximum width of this biomarker influences the choice of surgical treatment and vision prognosis. Automated analysis of the volumetric characteristics of this defect could improve this aspect of ophthalmology.
If it weren’t known that the images show adjacent areas of the same patient, the IRC biomarker in
Figure 1b could suggest vascular-related pathologies (AMD, DME, RVO), where fluid accumulation in the neuroepithelium is more commonly called IRF. Therefore,
Figure 1a,b highlight the importance of evaluating macular tissue conditions using multiple images. In similar cases, especially with smaller macular holes, there could be a diagnostic error and incorrect treatment.
Figure 1c illustrates IRF, or fluid accumulation within the retina, which may signal inflammatory changes or the result of neovascularization [
30].
Figure 1d demonstrates PED, which involves the detachment of the retinal pigment epithelium from the underlying choroid, commonly associated with AMD, DR, or CSH [
31].
It is important to accurately understand the volumetric characteristics of IRF and PED to assess disease stages, monitoring frequency, when to start invasive procedures, and choosing the right medications and their dosing schedule. Their detection and accurate segmentation in OCT images are essential for effective disease management and reducing the risk of blindness.
In this paper, we adapt the SAM 2 using MedSAM 2 for the OCT domain and examine the performance measuring IRC, IRF, PED, and MH biomarkers - the most important biomarkers for managing patients with macular, diabetic, and vascular retinal diseases. We evaluate different SAM 2 modalities on public datasets [
32,
33]. We apply modalities like point and box selections with fine-tuned MedSAM 2 for volumetric segmentation of 3D OCT and compare the metrics with pure SAM 2 and the classical U-Net model.
2. Methods
2.1. Segment Anything Models
SAM is a cutting-edge segmentation model that handles various segmentation tasks [
6]. SAM can perform zero-shot segmentation, which means it can segment objects in images without requiring task-specific training or fine-tuning. SAM architecture combines a robust image encoder, prompt encoder, and mask decoder. In the original work, the image encoder was an MAE [
34] pre-trained Vision Transformer (ViT) [
35]. A prompt encoder has two sets of prompts: sparse (points, boxes, text) and dense (masks). Points and boxes are represented by positional encodings summed with learned embeddings for each prompt type and free-form text with an off-the-shelf text encoder from CLIP [
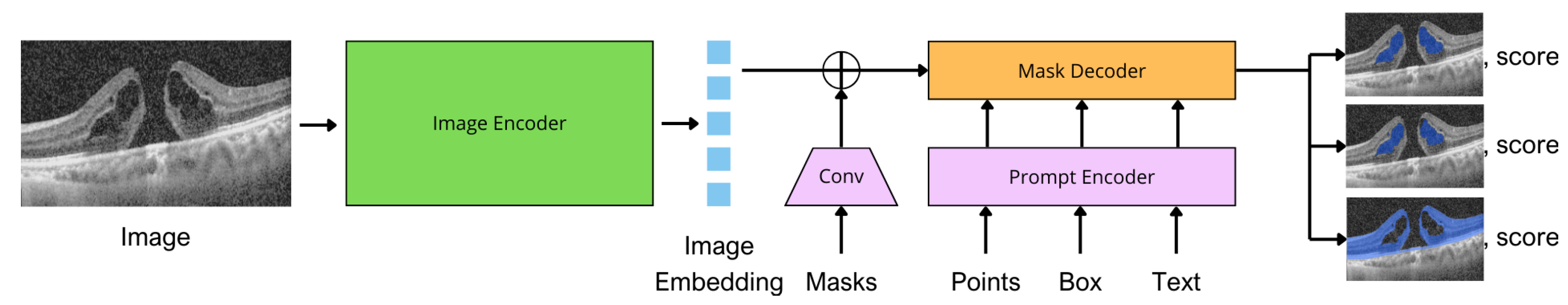
36]. Dense prompts are embedded using convolutions and summed element-wise with the image embedding. The image encoder outputs an image embedding that can be efficiently queried by various input prompts to produce the object masks. SAM can output multiple valid masks with corresponding confidence scores.
Figure 2 shows the model architecture adapted for OCT domain.
SAM achieved sufficient segmentation results primarily on the objects characterized by distinct boundaries. However, SAM has significant limitations in segmenting typical medical images with weak boundaries or low contrast.
MedSAM [
37] was proposed to overcome these limitations in the medical domain: this is a refined foundation model that improves the segmentation performance of SAM on medical images. MedSAM accomplishes this by fine-tuning SAM on a specific medical dataset with more than one million medical image-mask pairs. The MedSAM was initialized with the pre-trained SAM ViT-Base. The prompt encoder was frozen during the training since it could already encode the bounding box prompt. All the trainable parameters in the image encoder and mask decoder were updated. MedSAM was trained with a large-scale medical image dataset containing over 1.5 million image-mask pairs covering ten imaging modalities and over 30 cancer types.
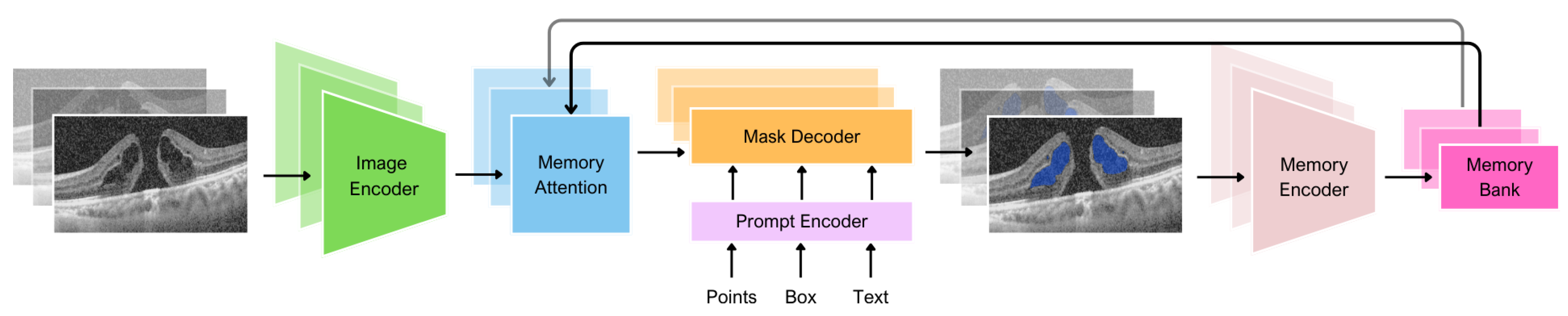
The first generation of SAM was designed to work with 2D images. Of course, using 3D images is possible with frame-by-frame analysis, but there is no connection between the images. Updated SAM 2 [
38] can produce segmentation masks of the object of interest across video frames. The model has a memory that stores information about the object from previous frames, which allows it to generate masks throughout the video and correct them based on the object’s stored memory context. This architecture generalizes SAM to the video domain and could also potentially be applied to 3D data.
MedSAM 2 [
39] adopted the philosophy of taking medical images as videos, adopting SAM 2 for the 3D medical data. Based on SAM 2, MedSAM 2 introduced an additional confidence memory bank and weighted pick-up strategy. In SAM 2, we merge all information equally when combining a new input image embedding with data from the memory bank. Alternatively, MedSAM 2 uses a weighted pick-up strategy to assign higher weights to images that are more similar to the input image. During training, MedSAM 2 employs a calibration head to ensure that the model associates higher confidence with more accurate segmentations and lower confidence with less accurate ones. This calibration aligns the model’s confidence with the accuracy of its predictions, improving the effectiveness of the confidence memory bank.
Thus, in this work, we applied MedSAM 2 to adapt SAM 2 for the segmentation of biomarkers in OCT volumes.
2.2. U-Net Model
The U-Net architecture [
40], initially proposed for biomedical segmentation problems, is a symmetric network with an encoder and a decoder connected by skip connections that efficiently combine low-level and high-level features to reconstruct spatial information accurately. In the original version of U-Net, the encoder successively reduces the image size with increasing feature depth, and the decoder restores image resolution using transposed convolutions. In this work, a modified version doubled the number of channels in the bottleneck layer, increasing the feature depth and helping to represent complex objects better. Here, we used the classical U-Net model as a baseline because, according to the number of publications, it is one of the most popular models for OCT biomarker segmentation.
2.3. Datasets
To evaluate the quality of model segmentation, we used datasets including labeled OCT volumes, in which there is a series of frame-by-frame OCT images per patient’s eye. AROI dataset [
32] consists of macular SD-OCT volumes recorded with the Zeiss Cirrus HD OCT 4000 device. Each OCT volume consists of 128 B-scans with a
pixels resolution and a pixel size of
. The dataset includes OCT volumes of 24 patients diagnosed with AMD undergoing active anti-VEGF therapy. The dataset has masks with eight classes: above internal limiting membrane (ILM); ILM - inner plexiform layer (IPL) / inner nuclear layer (INL); IPL / INL - retinal pigment epithelium (RPE); RPE - Bruch membrane (BM); under BM; PED; SRF; IRF. In total, the authors annotated 1136 B-scans of 24 patients.
The OIMHS dataset [
33] comprises 125 OCT volumes from 125 eyes, having 3859 OCT B-scans from 119 patients with macular holes (MH). OCT volumes were performed in all patients using the SD-OCT system (Spectralis HRA OCT, Heidelberg Engineering, Heidelberg, Germany). All the B-scans have
pixels resolution. The authors annotated each B-scan and provided the masks with four classes: MH, IRC, retina, and choroid.
2.4. Training
Given an input OCT volume , where H and W are the height and width of the frame, respectively, and N is the number of frames in the volume. We aim to obtain a segmentation mask , where C is the number of classes.
We split the volumes from the AROI and OIMHS datasets into training and test subsets at the patient level with a training-test ratio of 80:20 for both datasets, such that all dataset biomarkers were performed in both subsets.
For the training, we used the pretrained SAM 2 weights "sam2_hiera_small.pt" available at SAM 2 official repository [
41]. The input image_size was set to 1024. The training was conducted on NVIDIA A100 (40GB) GPU using a conda virtual environment with Python 3.12, CUDA 12.1, and PyTorch 2.4.0. Training on 100 epochs on one dataset takes up to 3 hours. The training loss on AROI dataset was 0.0298, and 0.0653 on OIMHS dataset.
For the training of the U-Net model, we applied the Adam optimizer, categorical cross-entropy loss function, and ReLU activation function. The batch size was set to 8, contributing to the efficient use of computing resources and speeding up the training process. The learning rate was . To work with GPUs, the scaler GradScaler was chosen. The model training was carried out on 2x Nvidia Tesla T4 GPUs. For the AROI dataset, the average loss was 0.0219, and for the OIMHS dataset, the average loss was 0.0495.
2.5. Metrics
For the model evaluation, we applied two of the most popular metrics for segmentation tasks: intersection-over-union (IoU), also known as the Jaccard index, and Dice score, also known as the F1 score. IoU is calculated by dividing the intersection between the predicted (
) and ground truth (
) regions by the area of their union:
where
,
, and
. The Dice score equals twice the size of the intersection of predicted (A) and ground truth (B) regions divided by their sum:
Dice score and IoU are the most commonly used metrics for semantic segmentation as they penalize the FP, a common factor in highly class-imbalanced datasets [
41].
3. Results
For the evaluation, we compared different SAM 2 prompt modes with the results of the segmentation using U-Net for four types of biomarkers separately. The selected biomarkers were MH, IRC, IRF, and PED.
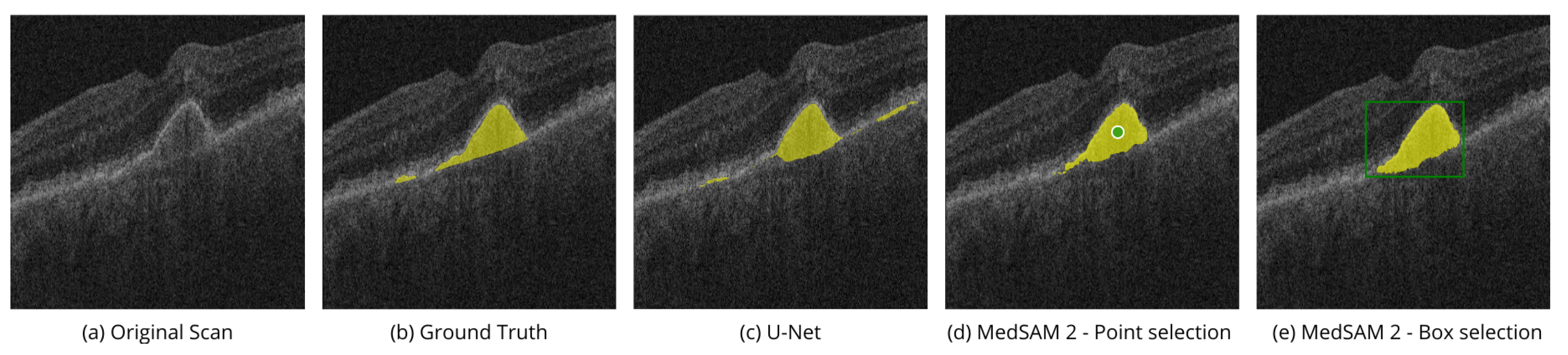
Table 2 shows the IoU and Dice scores for all of the experiments. Fine-tuned MedSAM 2 models are better than U-Net in most cases. Also, the results show that using box selection is more efficient than point selection, although the opposite was true for IRF.
Table 2 shows the superiority of MedSAM 2 in the box selection mode compared to U-Net by 8.2% - 9.2% in IoU and 9.5% in Dice score. MedSAM 2 with box selection mode is better than U-Net, with a 5.7% improvement in the IoU score and a 3.3% improvement in the Dice score. In the case of the IRF biomarker, MedSAM 2 with point selection mode achieves the best result, outperforming U-Net by 5% and 2.8% in the IoU and Dice scores, respectively.
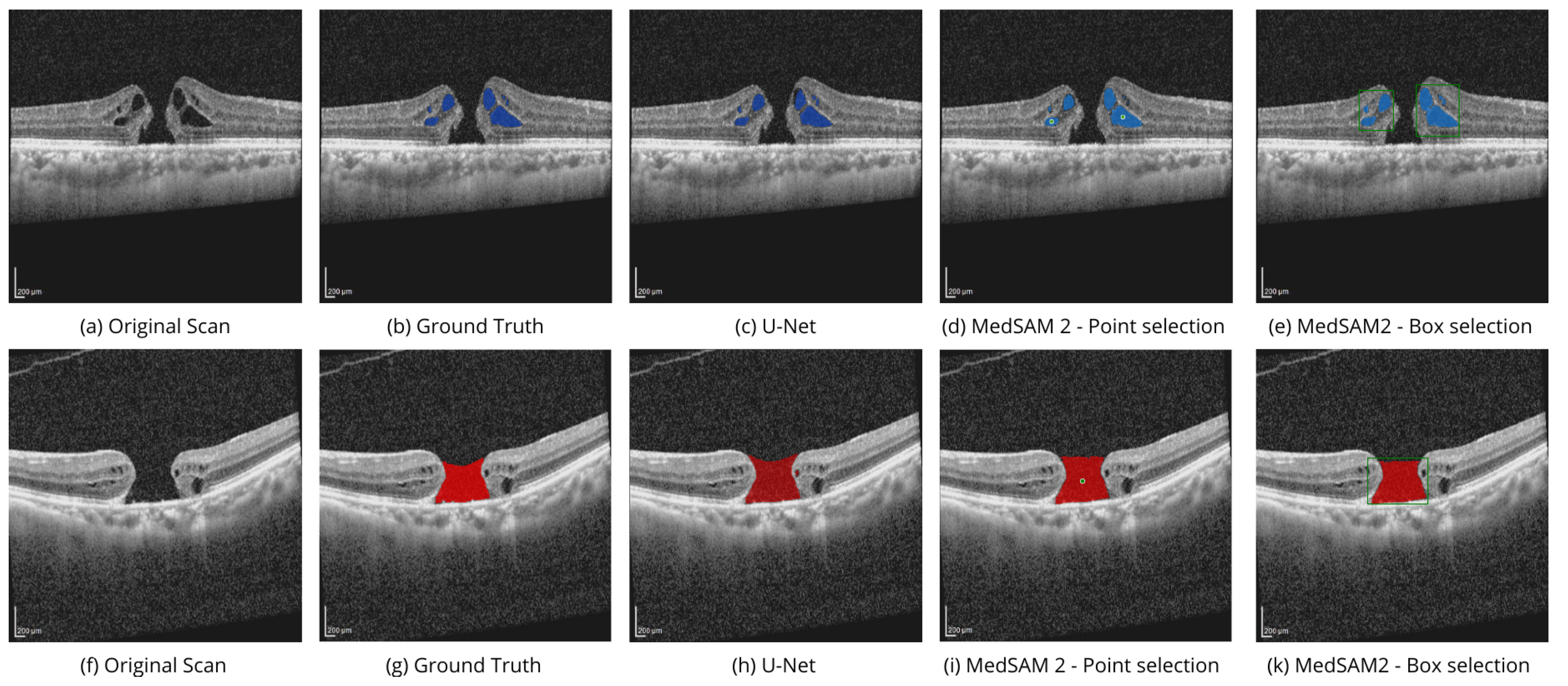
Figure 4 shows the performance of MedSAM 2 segmentation modes on the OIMHS dataset, displaying IRC and MH segmentation examples, which are shown in blue and red, respectively.
Figure 5 shows the performance of MedSAM 2 segmentation modes on the AROI dataset, displaying PED segmentation examples, shown in yellow.
Point selection modality is performed by selection of the related to the region points of interest on a single OCT image slice, as shown in
Figure 4d,i, and
Figure 5d. Box selection modality is done by selecting the related square area where the biomarker is located, as shown in
Figure 4e,k, and
Figure 5e. Segmentation of the corresponding biomarkers is then carried out on all OCT slices.
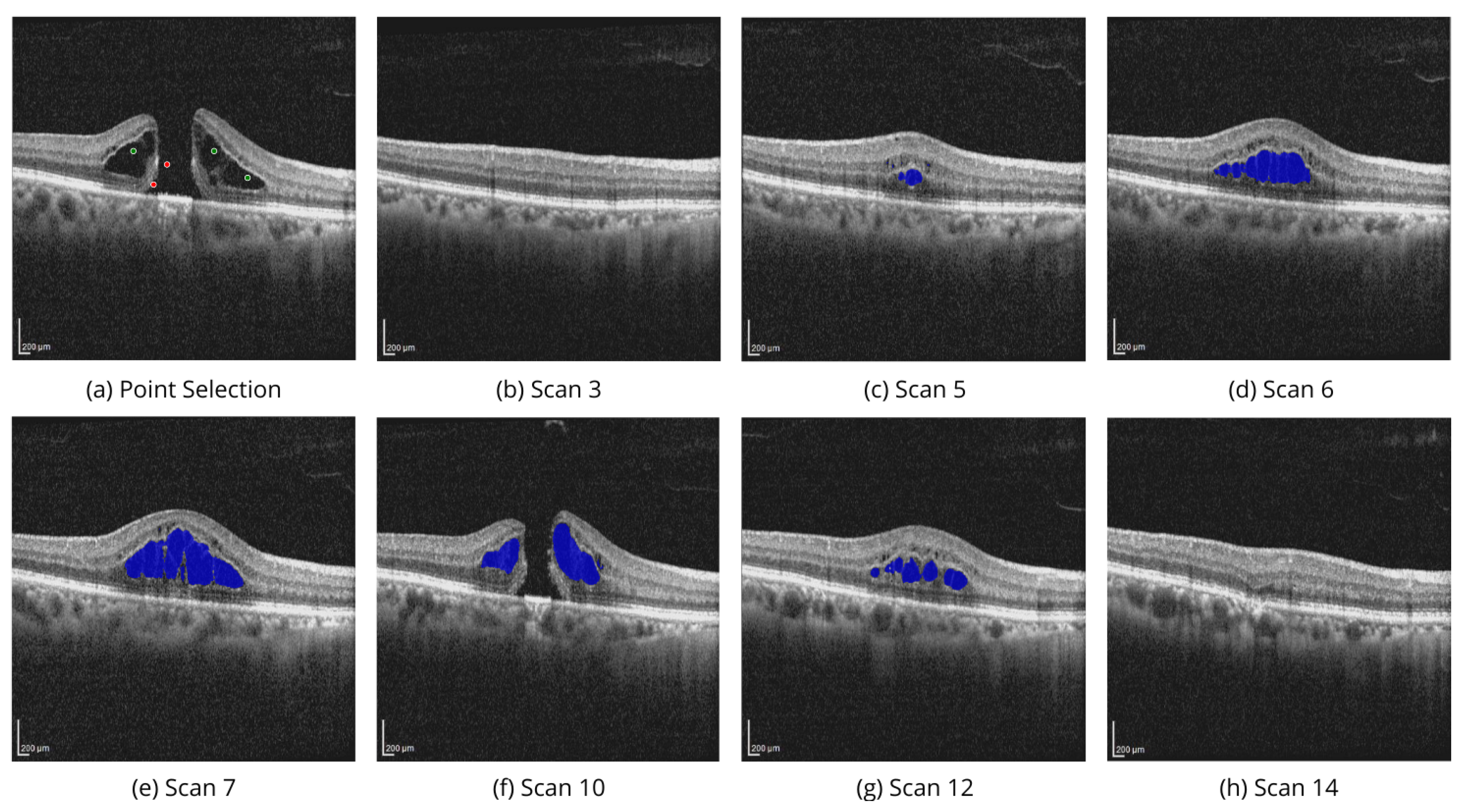
Figure 6 demonstrates the volumetric segmentation of IRC using the point selection modality on the scans from OIMHS dataset. In the example, point selection was done with three positive prompts shown in green and two negative prompts shown in red. IRC mask predictions are shown in blue.
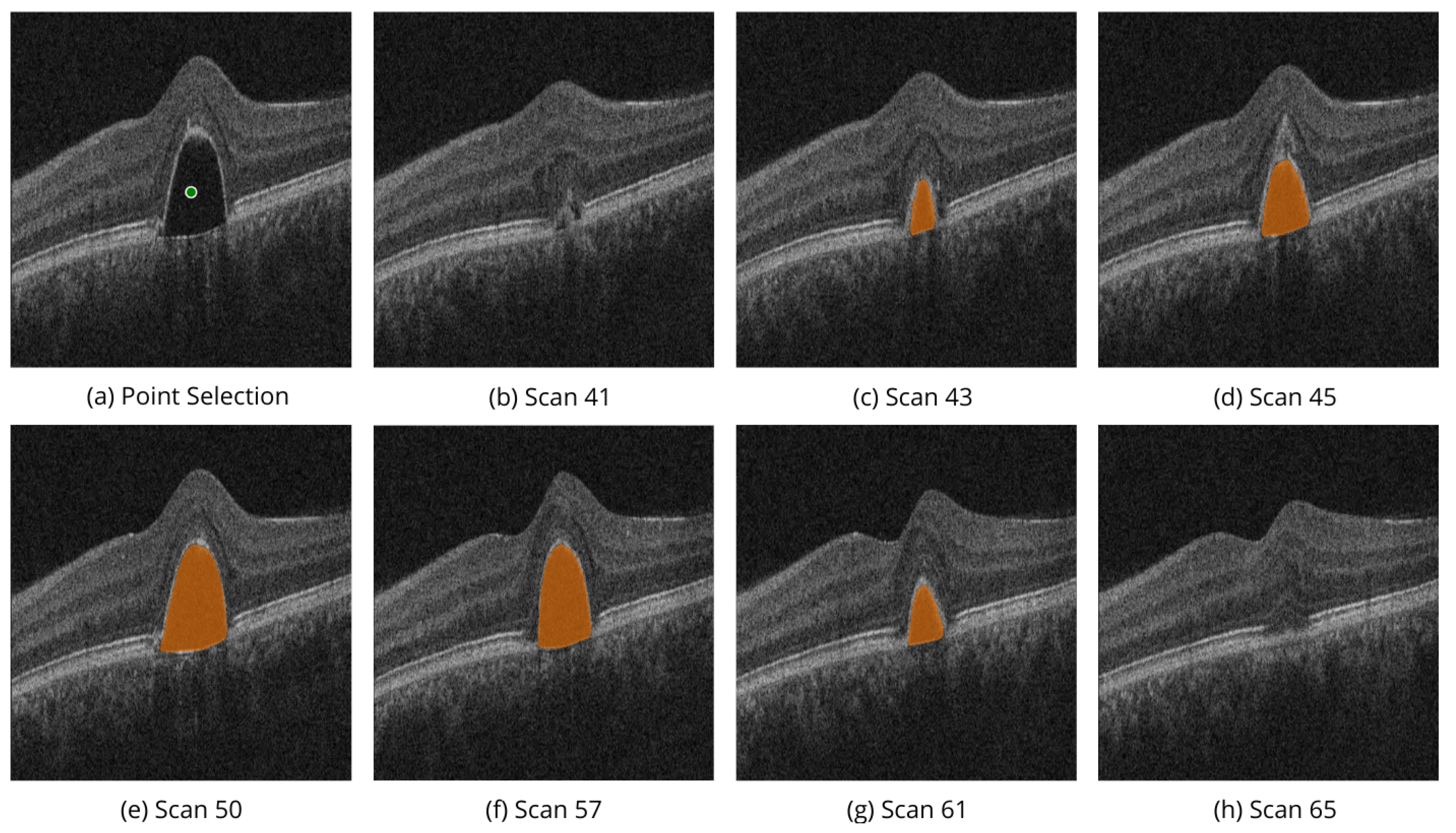
Similarly,
Figure 7 shows the volumetric segmentation of PED using the point selection modality on scans from AROI dataset. In the example, point selection was done with one positive prompt shown in green. PED mask predictions are shown in orange.
These examples illustrate the model’s ability to perform high-quality segmentation across various OCT image slices, ensuring accurate identification of regions of interest, specifically the PED and IRC biomarkers.
4. Discussion
The metrics obtained from MedSAM 2 are higher than those from U-Net for all classes of biomarkers used in the evaluation. The inclusion of spatial information improves the quality of segmentation. The model has a wide range of applications. Aside from automated segmentation, it can also be used in medical data labeling applications. This model will significantly reduce the time required for a medical worker to label specific biomarkers, unlike U-Net, which does not offer real-time supervision. This is especially useful for 3D segmentation, such as when a medical worker needs to label the same biomarker throughout an entire 3D scan volume.
Exploring the model’s potential on diverse datasets, such as the OCTDL dataset [
9] categorized by pathological conditions, could open up new research and application routes. In this scenario, images with similar biomarkers could be used instead of the frames of the volume scan, offering a new perspective of the memory bank and the model’s capabilities.
In modern ophthalmologic practice, many classic and contemporary biomarkers are highlighted to help the practitioner identify the correct macular disease, choose tactics, and predict treatment outcomes. Often, such marking is manual and time-consuming; some information may be missed due to the abundance of scans. From the parameters automated for analysis in modern tomography software, available in the broad world of ophthalmic practice, single biomarkers (central retinal thickness, avascular area) are used, obtained by summing up linear measurements on thousands of B-scan scans. Volumetric measures (MH, IRF, IRC, PED, etc.) are not automatically analyzed. However, this information could make it easier and more accurate for ophthalmologists to combat macular diseases, the leading causes of blindness worldwide.
5. Conclusions
The rapidly advancing field of OCT imaging has seen significant progress with DL methods. U-Net and its variants have been widely used due to their reliable performance in segmenting OCT biomarkers, such as MH, IRC, and PED. However, recent developments, like SAM and its medical adaptations, particularly MedSAM 2, are beginning to outperform U-Net models.
SAM2, specifically designed for the labeling of spatial data, introduces an advanced approach with memory-based learning to enhance segmentation accuracy that can be employed for the segmentations of medical volumetric scans. With the availability of several prompt modes, SAM-based architectures allow control of the segmentation process and are promising to be utilized in semi-automatic labeling tools for medical data.
Evaluations across public datasets reveal that fine-tuned MedSAM 2, specifically designed for medical data, generally outperform U-Net, especially in scenarios requiring detailed segmentation of biomarkers. Thus, MedSAM 2 is a promising tool for segmenting OCT biomarkers, especially when segmenting 3D data is necessary.
Author Contributions
Conceptualization M.K.; methodology, M.K. and A.Z.; software, M.K., A.P., and G.S.; validation, M.K., A.P., and A.N.; formal analysis, M.K.; investigation, M.K. and A.N.; writing—original draft preparation, M.K., A.Z., and A.N.; writing—review and editing, V.B. and M.R.; visualization, M.K.; supervision, A.M. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The data used in this study are available in the public domain [
32,
42].
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Huang, D.; Swanson, E.A.; Lin, C.P.; Schuman, J.S.; Stinson, W.G.; Chang, W.; Hee, M.R.; Flotte, T.; Gregory, K.; Puliafito, C.A.; others. Optical coherence tomography. science 1991, 254, 1178–1181. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Baughman, D.M.; Lee, A.Y. Deep learning is effective for classifying normal versus age-related macular degeneration OCT images. Ophthalmology Retina 2017, 1, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Lemaître, G.; Rastgoo, M.; Massich, J.; Cheung, C.Y.; Wong, T.Y.; Lamoureux, E.; Milea, D.; Mériaudeau, F.; Sidibé, D. Classification of SD-OCT Volumes Using Local Binary Patterns: Experimental Validation for DME Detection. Journal of ophthalmology 2016, 2016, 3298606. [Google Scholar] [CrossRef] [PubMed]
- Ran, A.R.; Tham, C.C.; Chan, P.P.; Cheng, C.Y.; Tham, Y.C.; Rim, T.H.; Cheung, C.Y. Deep learning in glaucoma with optical coherence tomography: a review. Eye 2021, 35, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.C.; Jammal, A.A.; Berchuck, S.I.; Mariottoni, E.B.; Medeiros, F.A. Assessment of a segmentation-free deep learning algorithm for diagnosing glaucoma from optical coherence tomography scans. JAMA ophthalmology 2020, 138, 333–339. [Google Scholar] [CrossRef]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; others. Segment anything. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2023; 4015–4026. [Google Scholar]
- Caicedo, J.C.; Goodman, A.; Karhohs, K.W.; Cimini, B.A.; Ackerman, J.; Haghighi, M.; Heng, C.; Becker, T.; Doan, M.; McQuin, C.; others. Nucleus segmentation across imaging experiments: the 2018 Data Science Bowl. Nature methods 2019, 16, 1247–1253. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, L.; Wen, L.; Liu, X.; Wu, Y. Towards real-world prohibited item detection: A large-scale x-ray benchmark. Proceedings of the IEEE/CVF international conference on computer vision. 2021; 5412–5421. [Google Scholar]
- Kulyabin, M.; Zhdanov, A.; Nikiforova, A.; Stepichev, A.; Kuznetsova, A.; Ronkin, M.; Borisov, V.; Bogachev, A.; Korotkich, S.; Constable, P.A.; others. Octdl: Optical coherence tomography dataset for image-based deep learning methods. Scientific Data 2024, 11, 365. [Google Scholar] [CrossRef]
- Ganjee, R.; Ebrahimi Moghaddam, M.; Nourinia, R. An unsupervised hierarchical approach for automatic intra-retinal cyst segmentation in spectral-domain optical coherence tomography images. Medical Physics 2020, 47, 4872–4884. [Google Scholar] [CrossRef]
- Rahil, M.; Anoop, B.N.; Girish, G.N.; Kothari, A.R.; Koolagudi, S.G.; Rajan, J. A deep ensemble learning-based CNN architecture for multiclass retinal fluid segmentation in OCT images. IEEE Access 2023, 11, 17241–17251. [Google Scholar] [CrossRef]
- Ganjee, R.; Ebrahimi Moghaddam, M.; Nourinia, R. A generalizable approach based on the U-Net model for automatic intraretinal cyst segmentation in SD-OCT images. International Journal of Imaging Systems and Technology 2023, 33, 1647–1660. [Google Scholar] [CrossRef]
- Melinščak, M. Attention-based U-net: Joint Segmentation of Layers and Fluids from Retinal OCT Images. 2023 46th MIPRO ICT and Electronics Convention (MIPRO). IEEE. 2023; 391–396. [Google Scholar]
- Wang, M.; Lin, T.; Peng, Y.; Zhu, W.; Zhou, Y.; Shi, F.; Chen, X. Self-guided optimization semi-supervised method for joint segmentation of macular hole and cystoid macular edema in retinal OCT images. IEEE Transactions on Biomedical Engineering 2023, 70, 2013–2024. [Google Scholar] [CrossRef]
- Daanouni, O.; Cherradi, B.; Tmiri, A. Automated end-to-end Architecture for Retinal Layers and Fluids Segmentation on OCT B-scans. Multimedia Tools and Applications, 2024; 1–24. [Google Scholar]
- George, N.; Shine, L.; Ambily, N.; Abraham, B.; Ramachandran, S. A two-stage CNN model for the classification and severity analysis of retinal and choroidal diseases in OCT images. International Journal of Intelligent Networks 2024, 5, 10–18. [Google Scholar] [CrossRef]
- Qiu, Z.; Hu, Y.; Li, H.; Liu, J. Learnable ophthalmology SAM. arXiv preprint arXiv:2304.13425, arXiv:2304.13425 2023.
- Fazekas, B.; Morano, J.; Lachinov, D.; Aresta, G.; Bogunović, H. Adapting Segment Anything Model (SAM) for Retinal OCT. Proceedings of the International Workshop on Ophthalmic Medical Image Analysis. Springer Nature Switzerland, 2023, pp. 92–101.
- Wu, J.; Philip, A.M.; Podkowinski, D.; Gerendas, B.S.; Langs, G.; Simader, C.; Waldstein, S.M.; Schmidt-Erfurth, U.M. Multivendor spectral-domain optical coherence tomography dataset, observer annotation performance evaluation, and standardized evaluation framework for intraretinal cystoid fluid segmentation. Journal of Ophthalmology 2016, 2016, 3898750. [Google Scholar] [CrossRef] [PubMed]
- Rashno, A.; Koozekanani, D.D.; Drayna, P.M.; Nazari, B.; Sadri, S.; Rabbani, H.; Parhi, K.K. Fully automated segmentation of fluid/cyst regions in optical coherence tomography images with diabetic macular edema using neutrosophic sets and graph algorithms. IEEE Transactions on Biomedical Engineering 2017, 65, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; others. Identifying medical diagnoses and treatable diseases by image-based deep learning. cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Heisler, M.; Lee, S.; Ding, G.W.; Navajas, E.; Sarunic, M.V.; Beg, M.F. Deep-learning based multiclass retinal fluid segmentation and detection in optical coherence tomography images using a fully convolutional neural network. Medical image analysis 2019, 54, 100–110. [Google Scholar] [CrossRef]
- Zur, D.; Iglicki, M.; Feldinger, L.; Schwartz, S.; Goldstein, M.; Loewenstein, A.; Barak, A. Disorganization of retinal inner layers as a biomarker for idiopathic epiretinal membrane after macular surgery—the DREAM study. American journal of ophthalmology 2018, 196, 129–135. [Google Scholar] [CrossRef]
- Joltikov, K.A.; Sesi, C.A.; de Castro, V.M.; Davila, J.R.; Anand, R.; Khan, S.M.; Farbman, N.; Jackson, G.R.; Johnson, C.A.; Gardner, T.W. Disorganization of retinal inner layers (DRIL) and neuroretinal dysfunction in early diabetic retinopathy. Investigative ophthalmology & visual science 2018, 59, 5481–5486. [Google Scholar]
- de Sá Quirino-Makarczyk, L.; Ugarte, M.d.F.S. OCT Biomarkers for AMD. In Recent Advances and New Perspectives in Managing Macular Degeneration; IntechOpen, 2021.
- Thomson, R.J.; Chazaro, J.; Otero-Marquez, O.; Ledesma-Gil, G.; Tong, Y.; Coughlin, A.C.; Teibel, Z.R.; Alauddin, S.; Tai, K.; Lloyd, H.; others. Subretinal drusenoid deposits and soft drusen: are they markers for distinct retinal diseases? Retina 2022, 42, 1311–1318. [Google Scholar] [CrossRef]
- Konana, V.K.; Bhagya, M.; Babu, K. Double-Layer sign: a new OCT finding in active tubercular Serpiginous-like choroiditis to monitor activity. Ophthalmology Retina 2020, 4, 336–342. [Google Scholar] [CrossRef]
- Metrangolo, C.; Donati, S.; Mazzola, M.; Fontanel, L.; Messina, W.; D’alterio, G.; Rubino, M.; Radice, P.; Premi, E.; Azzolini, C. OCT Biomarkers in Neovascular Age-Related Macular Degeneration: A Narrative Review. Journal of ophthalmology 2021, 2021, 9994098. [Google Scholar] [CrossRef] [PubMed]
- Ritter, M.; Simader, C.; Bolz, M.; Deák, G.G.; Mayr-Sponer, U.; Sayegh, R.; Kundi, M.; Schmidt-Erfurth, U.M. Intraretinal cysts are the most relevant prognostic biomarker in neovascular age-related macular degeneration independent of the therapeutic strategy. British Journal of Ophthalmology 2014, 98, 1629–1635. [Google Scholar] [CrossRef] [PubMed]
- Tomkins-Netzer, O.; Niederer, R.; Greenwood, J.; Fabian, I.D.; Serlin, Y.; Friedman, A.; Lightman, S. Mechanisms of blood-retinal barrier disruption related to intraocular inflammation and malignancy. Progress in Retinal and Eye Research 2024, 101245. [Google Scholar] [CrossRef] [PubMed]
- Veritti, D.; Sarao, V.; Gonfiantini, M.; Rubinato, L.; Lanzetta, P. Faricimab in Neovascular AMD Complicated by Pigment Epithelium Detachment: An AI-Assisted Evaluation of Early Morphological Changes. Ophthalmology and Therapy.
- Melinščak, M.; Radmilović, M.; Vatavuk, Z.; Lončarić, S. Annotated retinal optical coherence tomography images (AROI) database for joint retinal layer and fluid segmentation. Automatika: časopis za automatiku, mjerenje, elektroniku, računarstvo i komunikacije 2021, 62, 375–385. [Google Scholar] [CrossRef]
- Ye, X.; He, S.; Zhong, X.; Yu, J.; Yang, S.; Shen, Y.; Chen, Y.; Wang, Y.; Huang, X.; Shen, L. OIMHS: An Optical Coherence Tomography Image Dataset Based on Macular Hole Manual Segmentation. Scientific Data 2023, 10, 769. [Google Scholar] [CrossRef]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Dollár, P.; Girshick, R. Masked autoencoders are scalable vision learners. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2022, pp. 16000–16009.
- Alexey, D. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv: 2010.11929, arXiv:2010.11929 2020.
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J. ; others. Learning transferable visual models from natural language supervision. International conference on machine learning. PMLR, 2021, pp. 8748–8763.
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nature Communications 2024, 15, 654. [Google Scholar] [CrossRef]
- Ravi, N.; Gabeur, V.; Hu, Y.T.; Hu, R.; Ryali, C.; Ma, T.; Khedr, H.; Rädle, R.; Rolland, C.; Gustafson, L. ; others. Sam 2: Segment anything in images and videos. arXiv preprint arXiv:2408.00714, arXiv:2408.00714 2024.
- Zhu, J.; Qi, Y.; Wu, J. Medical sam 2: Segment medical images as video via segment anything model 2. arXiv preprint arXiv:2408.00874, arXiv:2408.00874 2024.
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, -9, 2015, proceedings, part III 18. Springer, 2015, pp. 234–241. 5 October.
- Müller, D.; Soto-Rey, I.; Kramer, F. Towards a guideline for evaluation metrics in medical image segmentation. BMC Research Notes 2022, 15, 210. [Google Scholar] [CrossRef]
- Shen, L.; Ye, X.; He, S.; Zhong, X.; Shen, Y.; Yang, S.; Chen, Y.; Huang, X. OIMHS dataset 2023. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).