Submitted:
11 September 2024
Posted:
11 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection and RNA Extraction
2.2. Reverse-Transcriptase and Double Conventional PCR Amplification
2.3. Gel Electrophoresis
2.4. Pre-Sequencing Samples Purification and Sequencing
2.5. Bioinformatics Analysis and Genetic Characterization
3. Results
3.1. Double Conventional PCR Amplification
3.2. Sequencing and Genetic Characterization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- International Committee on Taxonomy of Viruses. Virus taxonomy: 2022, [released, 2022; modified, March 2023; cited, Sept. 2023]. Available from:https://talk.ictvonline.org/taxonomy/.
- Cook, J.K.A.; Cavanagh, D. Detection and differentiation of avian pneumoviruses (metapneumoviruses). Avian Pathol. 2002, 31, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.A.; Allée, C.; Courtillon, C.; Szerman, N.; Lemaitre, E.; Toquin, D.; Mangart, J.M.; Amelot, M.; Eterradossi, N. Host specificity of avian metapneumoviruses. Avian Pathol. 2019, 48, 311–318. [Google Scholar] [CrossRef]
- Canuti, M.; Kroyer, A.N.K.; Ojkic, D.; Whitney, H.G.; Robertson, G.J.; Lang, A.S. Discovery and characterization of novel rna viruses in aquatic North American wild birds. Viruses 2019, 11, 1–14. [Google Scholar] [CrossRef]
- Retallack, H.; Clubb, S.; DeRisi, J.L. Genome Sequence of a Divergent Avian Metapneumovirus from a Monk Parakeet (Myiopsitta monachus). Microbiol. Resour. Announc. 2019, 8, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Bexter, F.; Rüger, N.; Sid, H.; Herbst, A.; Gabriel, G.; Osterhaus, A.; Rautenschlein, S. In Vitro Investigation of the Interaction of Avian Metapneumovirus and Newcastle Disease Virus with Turkey Respiratory and Reproductive Tissue. Viruses 2023, 15(4) 907, 1–19. [Google Scholar] [CrossRef]
- Hartmann, S.; Sid, H.; Rautenschlein, S. Avian metapneumovirus infection of chicken and turkey tracheal organ cultures: comparison of virus–host interactions. Avian Pathol. 2015, 44, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Kaboudi, K.; Lachheb, J. Avian metapneumovirus infection in turkeys: a review on turkey rhinotracheitis. J Appl Poult Res 2021, 100211, 1–17. [Google Scholar] [CrossRef]
- Lupini, C.; Legnardi, M.; Graziosi, G.; Cecchinato, M.; Listorti, V.; Terregino, C.; Catelli, E. Vaccine Interaction and Protection against Virulent Avian Metapneumovirus (aMPV) Challenge after Combined Administration of Newcastle Disease and aMPV Live Vaccines to Day-Old Turkeys. Vaccines 2023, 708, 1–10. [Google Scholar] [CrossRef]
- Tucciarone, C.M.; Franzo, G.; Lupini, C.; Alejo, C.T.; Listorti, V.; Mescolini, G.; Brandão, P.E.; Martini, M.; Catelli, E.; Cecchinato, M. Avian Metapneumovirus circulation in Italian broiler farms. Poult. Sci. 2018, 97, 503–509. [Google Scholar] [CrossRef]
- Suarez, D.L.; Miller, P.J.; Koch, G.; Mundt, E.; Rautenschlein, S. Newcastle disease, other avian paramyxoviruses, and avian metapneumovirus infections. In Diseases of Poultry, 14th ed.; Swayne, D.E., Boulianne, M., Logue, C.M., McDougald, L.R., Nair, V., Suarez, D.L., de Wit, S., Grimes, T., Johnson, D., Kromm, M., Prajitno, T.Y., Rubinoff, I., Zavala, G. Eds.; Wiley-Blackwell. Hoboken New Jersey, USA, 2020; pp. 111–166. [CrossRef]
- Jbenyeni, A.; Croville, G.; Cazaban, C.; Guérin, J.L. Predominance of low pathogenic avian influenza virus H9N2 in the respiratory co-infections in broilers in Tunisia: a longitudinal field study, 2018 – 2020. Vet. Res. 2023, 1–14. [Google Scholar] [CrossRef]
- Lachheb, J.; Bouslama, Z.; Nsiri, J.; Badr, C.; Al Gallas, N.; Souissi, N.; Khazri, I.; Larbi, I.; Kaboudi, K.; Ghram, A. Phylogenetic and phylodynamic analyses of subtype-B metapneumovirus from chickens in Tunisia. Poult. Sci. 2022, 102, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sid, H.; Benachour, K.; Rautenschlein, S. Co-infection with Multiple Respiratory Pathogens Contributes to Increased Mortality Rates in Algerian Poultry Flocks. Avian Dis. 2015, 59, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Mernizi, A.; Bouziane, S.; Fathi, H.; Criado, J.; Bouslikhane, M.; Ghram, A.; Catelli, E.; Mouahid, M.; Nassik, S. First seroepidemiological and risk factor survey of avian metapneumovirus circulation in Moroccan broiler farms. Vet. Glas. 2022, 77, 1–16. [Google Scholar] [CrossRef]
- Mernizi, A.; Kadiri, O.; Criado, J.L.; Bouslikhane, M.; Ghram, A.; Mouahid, M.; Catelli, E.; Nassik, S. Detection of Avian Metapneumovirus Subtypes A and B in Moroccan Broiler. Iran J Vet Med 2023, 1–28. [Google Scholar] [CrossRef]
- Juhasz, K.; Easton, A.J. Extensive sequence variation in the attachment (G) protein gene of avian pneumovirus: Evidence for two distinct subgroups. J. Gen. Virol. 1994, 75, 2873–2880. [Google Scholar] [CrossRef] [PubMed]
- Cecchinato, M.; Catelli, E.; Lupini, C.; Ricchizzi, E.; Clubbe, J.; Battilani, M.; Naylor, C.J. Avian metapneumovirus (AMPV) attachment protein involvement in probable virus evolution concurrent with mass live vaccine introduction. Vet. Microbiol. 2010, 146, 24–34. [Google Scholar] [CrossRef]
- Mescolini, G.; Lupini, C.; Franzo, G.; Quaglia, G.; Legnardi, M.; Cecchinato, M.; Tucciarone, C.M.; Blanco, A.; Turblin, V.; Biarnés, M.; Tatone, F.; Falchieri, M.; Catelli, E. What is new on molecular characteristics of Avian metapneumovirus strains circulating in Europe? Transbound. Emerg. Dis. 2021, 68, 1314–1322. [Google Scholar] [CrossRef]
- Bayraktar, E.; Umar, S.; Yilmaz, A.; Turan, N.; Franzo, G.; Tucciarone, C.M.; Cecchinato, M.; Cakan, B.; Iqbal, M.; Yilmaz, H. First molecular characterisation of avian metapneumovirus (aMPV) in Turkish broiler flocks. Avian Dis. 2018, 62, 425–430. [Google Scholar] [CrossRef]
- Owoade, A.A.A.; Ducatez, M.F.; Hübschen, J.M.; Sausy, A.; Chen, H.; Guan, Y.; Muller, C.P. Avian Metapneumovirus Subtype A in China and Subtypes A and B in Nigeria. Avian Dis. 2008, 52, 502–506. [Google Scholar] [CrossRef]
- Kariithi, H.M.; Volkening, J.D.; Veiga Alves, V.; Reis-cunha, J.L.; Cury Rocha Veloso Arantes, L.; Santos Fernando, F.; Fernandes Filho, T.; da Silva Martins, N.R.; Lemière, S.; de Freitas Neto, O.C.; Decanini, E.L.; Afonso, C.L.; Suarez, D.L. Complete Genome Sequences of Avian Metapneumovirus. Microbiol. Resour. Announc. 2023, 7, 8–11. [Google Scholar] [CrossRef]
- Luqman, M.; Duhan, N.; Temeeyasen, G.; Selim, M.; Jangra, S.; Mo, S.K. Geographical Expansion of Avian Metapneumovirus Subtype B: First Detection and Molecular Characterization of Avian Metapneumovirus Subtype B in US Poultry. Viruses 2024, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- El Houafdi, M.; Hamam, J.; Vanmarche, J.; Cook, J.K.A. Swollen head syndrome in broiler chicken in Morocco. In Proceedings of 40th Western Poultry Disease Conference, Acapulco, Mexico, Mar 10-13; Jensen, M.; Sarfati, D., Eds; Asociación Nacional de Especialistas en Ciencias Avícolas, Cancun, Mexico, 1991, p. 126–127.
- Andreopoulou, M.; Franzo, G.; Tucciarone, C.M.; Prentza, Z.; Koutoulis, K.C.; Cecchinato, M.; Chaligianni, I. Molecular epidemiology of infectious bronchitis virus and avian metapneumovirus in Greece. Poult. Sci. 2019, 98, 5374–5384. [Google Scholar] [CrossRef]
- Banet-Noach, C.; Simanov, L.; Perk, S. Characterization of Israeli avian metapneumovirus strains in turkeys and chickens. Avian Pathol. 2005, 34, 220–226. [Google Scholar] [CrossRef]
- Franzo, G.; Tucciarone, C.M.; Enache, M.; Bejan, V.; Ramon, G.; Koutoulis, K.C.; Cecchinato, M. First Report of Avian Metapneumovirus Subtype B Field Strain in a Romanian Broiler Flock during an Outbreak of Respiratory Disease. Avian Dis. 2017, 61, 250–254. [Google Scholar] [CrossRef]
- Lupini, C.; Tucciarone, C.M.; Mescolini, G.; Quaglia, G.; Graziosi, G.; Turblin, V.; Brown, P.; Cecchinato, M.; Legnardi, M.; Delquigny, T.; Lemière, S.; Perreul, S.; Catelli, E. Longitudinal Survey on aMPV Circulation in French Broiler Flocks following Different Vaccination Strategies. Animals 2023, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tucciarone, C.M.; Andreopoulou, M.; Franzo, G.; Prentza, Z.; Chaligiannis, I.; Cecchinato, M. First Identification and Molecular Characterization of Avian metapneumovirus Subtype B from Chickens in Greece. Avian Dis. 2017, 61, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.A.; Allée, C.; Courtillon, C.; Szerman, N.; Lemaitre, E.; Toquin, D.; Mangart, J.M.; Amelot, M.; Eterradossi, N. Host specificity of avian metapneumoviruses. Avian Pathol. 2019, 48, 311–318. [Google Scholar] [CrossRef]
- Loor-Giler, A.; Muslin, C.; Santander-Parra, S.; Coello, D.; de la Torre, D.; Abad, H.; Nuñez, L. Simultaneous detection of infectious bronchitis virus and avian metapneumovirus genotypes A, B, and C by multiplex RT-qPCR assay in chicken tracheal samples in Ecuador. Front. vet. sci. 2024, 11, 1–10. [Google Scholar] [CrossRef]
- Graziosi, G.; Lupini, C.; Catelli, E. Disentangling the role of wild birds in avian metapneumovirus (aMPV) epidemiology: A systematic review and meta-analysis. Transbound. Emerg. Dis. 2022, 69, 3285–3299. [Google Scholar] [CrossRef]
- Tucciarone, C.M.; Franzo, G.; Legnardi, M.; Pasotto, D.; Lupini, C.; Catelli, E.; Quaglia, G.; Graziosi, G.; Dal Molin, E.; Gobbo, F.; Cecchinato, M. Molecular Survey on A, B, C and New Avian Metapneumovirus (aMPV) Subtypes in Wild Birds of Northern-Central Italy. Vet. Sci. 2022, 9, 1–11. [Google Scholar] [CrossRef]
- Nguyen, V.G.; Chung, H.C.; Do, H.Q.; Nguyen, T.T.; Cao, T.B.P.; Truong, H.T.; Mai, T.N.; Le, T.T.; Nguyen, T.H.; Le, T.L.; Huynh, T.M.L. Serological and molecular characterization of avian metapneumovirus in chickens in Northern Vietnam. Vet. Sci. 2021, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Salles, G.B.C.; Pilati, G.V.T.; Savi, B.P.; Muniz, E.C.; Dahmer, M.; Vogt, J.R.; de Lima Neto, A.J.; Fongaro, G. Surveillance of Avian Metapneumovirus in Non-Vaccinated Chickens and Co-Infection with Avian Pathogenic Escherichia coli. Microorganisms 2024, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lupini, C.; Cecchinato, M.; Ricchizzi, E.; Naylor, C.J.; Catelli, E. A turkey rhinotracheitis outbreak caused by the environmental spread of a vaccine-derived avian metapneumovirus. Avian Pathol. 2011, 40, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Salles, G.B.C.; Pilati, G.V.T.; Muniz, E.C.; de Lima Neto, A.J.; Vogt, J.R.; Dahmer, M.; Savi, B.P.; Padilha, D.A.; Fongaro, G. Trends and Challenges in the Surveillance and Control of Avian Metapneumovirus. Viruses 2023, 15, 1–11. [Google Scholar] [CrossRef]
- Catelli, E.; Lupini, C.; Cecchinato, M.; Ricchizzi, E.; Brown, P.; Naylor, C.J. Field avian Metapneumovirus evolution avoiding vaccine induced immunity. Vaccine 2010, 28, 916–921. [Google Scholar] [CrossRef]
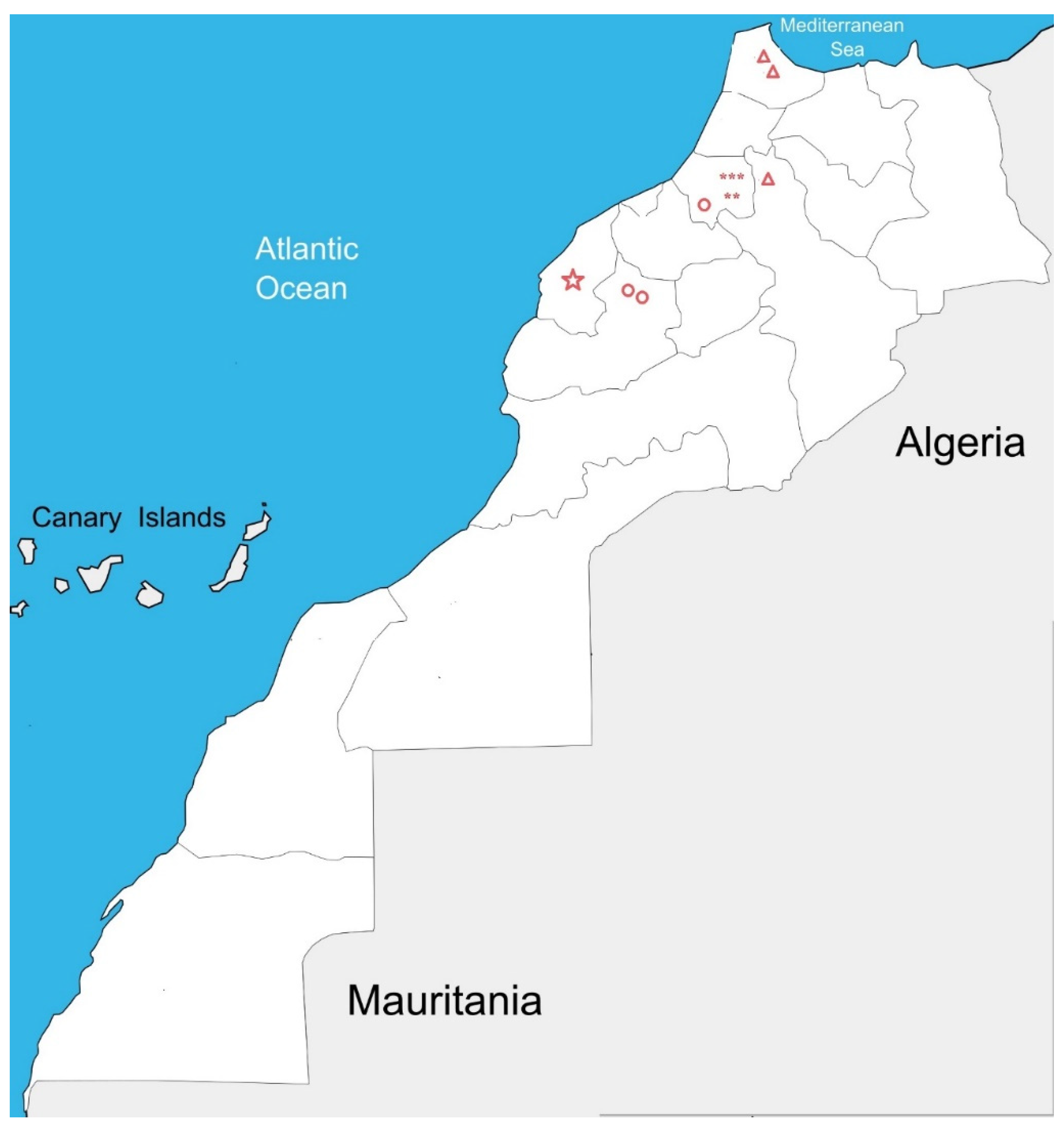

| Sample identification | Number of samples | Origin | Nature of RNA samples |
|---|---|---|---|
| P1 to P22 | 22 | Broilers | Extracted from aMPV-positive swabs [16] |
| 727 to 731 | 05 | Broilers | Extracted from birds with SHS |
| 764 to 766 | 03 | Turkeys | Extracted from birds with TRT |
| Primers | Sequence (5’-3’) | References |
|---|---|---|
| G Start+ | CAAGTATCCAGATGGGGTC | [17] |
| G5- | CAAAGAA/G CCAATAAGCCCA | [18] |
| G6- | CTGACAAATTGGTCCTGATT | [18] |
| G8+A | CACTCACTGTTAGCGTCATA | [18] |
| G9+B | TAGTCCTCAAGCAAGTCCTC | [18] |
| 1st amplification | 2nd amplification | |||
|---|---|---|---|---|
| Steps | Composition | Volume (µl) | Composition | Volume (µl) |
| Preparation of the PCR mix | Mastermix DreamTaq® Green PCR | 25 | Mastermix DreamTaq® Green PCR | 25 |
| Forward G6- | 2 | Forward G5- | 2 | |
| Reverse G START+ | 2 | Reverse G8+A Reverse G9+B |
2 2 |
|
| H2O | 11 | H2O | 9 | |
| cDNA | 10 | cDNA | 10 | |
| Total | 40 | Total | 40 | |
| Hybridization | 57°C | 58°C | ||
| Code | Sequence name on the submission | Accession Number |
|---|---|---|
| 727G | aMPV/B/Morocco/Ck/MA-1/2023 | PQ202992 |
| 727P | aMPV/B/Morocco/Ck/MA-2/2023 | PQ202991 |
| 731G | aMPV/B/Morocco/Ck/MA-3/2023 | PQ202994 |
| 764P | aMPV/B/Morocco/Ty/MA-4/2023 | PQ202998 |
| 766G | aMPV/B/Morocco/Ty/MA-5/2023 | PQ202995 |
| 766P | aMPV/B/Morocco/Ty/MA-6/2023 | PQ202996 |
| P5 | aMPV/B/Morocco/Ck/MA-7/2023 | PQ202993 |
| SF1 | aMPV/B/Morocco/Ck/FS-1/2023 | PQ202997 |
| Code | Documented GenBank strains | Similarity (%) |
Accession Number | Ref. |
|---|---|---|---|---|
|
727G |
Avian Metapneumovirus strain aMPV/B/Romania/Ty/67/17 glycoprotein (G) gene, partial cds Avian Metapneumovirus isolate 2018_0404_Chicken_Turkey_2018 attachment protein (G) gene, partial cds |
90.76 |
MT432878.1 MH352465.1 |
[19] [20] |
|
727P |
Avian Metapneumovirus isolate Algeria/26/aMPVB/turkey attachment protein gene, partial cds Avian Metapneumovirus partial mRNA for attachment protein (G gene), strain aMPV/chicken/Nigeria/NIR89/2006 |
96.79 96.09 |
KP892758.1 AM490057.1 |
[14] [21] |
|
731G |
Avian Metapneumovirus isolate aMPV-B/BR/1890/E1/19, complete genome Avian Metapneumovirus strain aMPV/B/Italy/Ty/742-01/17 glycoprotein (G) gene, partial cds Avian Metapneumovirus isolate 101/2011 attachment protein (G) gene, partial cds |
99.05 |
OP572408.1 MT436229.1 KC954647.1 |
[22] [19] |
|
764P |
Avian Metapneumovirus strain aMPV/B/France/GuineaFowl/1060/18 glycoprotein (G) gene, partial cds Avian Metapneumovirus strain aMPV/B/Romania/Ty/85/17 glycoprotein (G) gene, partial cds |
98.96 98.63 |
MT432904.1 MT432883.1 |
[19] [19] |
|
766G |
Avian Metapneumovirus isolate aMPV-B/BR/1890/E1/19, complete genome Avian Metapneumovirus strain aMPV/B/Italy/Ty/742-01/17 glycoprotein (G) gene, partial cds Avian Metapneumovirus isolate 101/2011 attachment protein (G) gene, partial cds |
99.70 |
OP572408.1 MT436229.1 KC954647.1 |
[22] [19] |
|
766P |
Avian Metapneumovirus isolate aMPV-B/BR/1890/E1/19, complete genome Avian Metapneumovirus strain aMPV/B/Italy/Ty/742-01/17 glycoprotein (G) gene, partial cds Avian Metapneumovirus isolate 101/2011 attachment protein (G) gene, partial cds |
98.70 |
OP572408.1 MT436229.1 KC954647.1 |
[22] [19] |
|
P5 |
Avian Metapneumovirus isolate aMPV/Chicken/PCRLAB/HG/2010 attachment protein (G) gene, partial cds Avian Metapneumovirus isolate 101/2011 attachment protein (G) gene, partial cds |
96.76 |
MN108496.1 KC954647.1 |
|
|
SF1 |
Avian Metapneumovirus strain aMPV/B/Spain/Ty/4954-1/15 glycoprotein (G) gene, partial cds Avian Metapneumovirus strain aMPV/B/France/Ck/785/17 glycoprotein (G) gene, partial cds Avian Metapneumovirus isolate aMPV-B/turkey/VA/USA/ADRDL-5, complete genome |
99.10 |
MT432835.1 MT432891.1 PP273460.1 |
[19] [19] [23] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





