1. Introduction
Hepatoblastoma is the most common primary liver cancer in children. Before eighties, complete resection of the tumor was the only curative treatment for children with primary liver tumors. Nowadays, some treatment with chemotherapy were introduced which improved management of these patients but still poor outcome is observed for patients with distant metastasis [
1,
2].
Clinical evaluation of hepatoblastoma consists of several known prognostic factors, such as the metastatic disease at diagnosis or pre-treatment extent of tumor (PRETEXT) stage, histological subtypes, and serum alpha-fetoprotein (AFP) levels [
3,
4,
5]. Some prognosis staging systems such as the PRETEXT stages (for pre-treatment extent of tumor) were developed to limit the toxic side effects of chemotherapy during patient treatment. PRETEXT score is determined by the number of liver segments affected, the degree of local invasion, the involvement of regional lymph nodes, and the presence of distant metastasis [
1,
6,
7]. Additionally, the CHIC risk stratification system was created by the Children's Hepatic Tumor International Collaborative (CHIC)[
3,
8]. The CHIC classification defines Standard-risk disease as PRETEXT I/II tumors or PRETEXT III tumors without any high-risk PRETEXT annotation factors. Intermediate-risk disease includes PRETEXT IV tumors or PRETEXT I-III tumors that have positive PRETEXT annotation factors. High-risk disease is defined as metastatic hepatoblastoma. [
9].
The Warburg effect plays a significant role in the development of hepatoblastoma tumors. These tumors are often linked to various somatic mutations in the
CTNNB1 gene, which encodes β-catenin, a transcriptional co-factor that responds to Wnt signaling. In mouse models of hepatoblastoma, the tumor’s metabolic profile is strongly influenced by the specific type of β-catenin mutations present [
10]. This metabolic variation, based on different CTNNB1 mutations, was also observed in human hepatoblastoma cell lines, where it was shown that β-catenin can regulate the
GLUT3-SLC2A3 glucose transporter [
11]. Additionally, Brain-expressed X-linked protein 1 (BEX1) helps maintain the stem cell-like properties of hepatoblastoma cells by promoting the Warburg effect through a PPARγ/PDK1-dependent pathway [
12].
In parallel to these crucial steps, various cellular metabolites provide the chemical moieties for DNA and histone modifications, resulting in a complex interplay between metabolism and epigenetics [
10]. DNA methylation regulates several biological processes, including gene transcription, X chromosome inactivation, genomic imprinting, [
11]. During tumorigenesis, DNA methylation disturbances could induce repression of tumor suppressor genes by promoting hypermethylation, and hypomethylation at repetitive sequences, leading to genomic instability [
12]. The evaluation of hepatoblastoma methylation genomic profiles revealed a genome-wide methylation dysfunction, characterized by hypermethylation at specific CpG islands, in addition to a mild hypomethylation pattern in non-repetitive intergenic sequences [
13].
In this work, based on metabolic enzyme expression program of hepatoblastoma tumors, we characterized a dual overexpression of PFKFB4 and DNMT3B in patient samples at metastatic risk (High risk CHIC stratification). With combined tumor expression of DNMT3B and PFKFB4, a meta.score was computed and this parameter was confirmed as an independent adverse score to predict metastatic status upon hepatoblastoma. We suggest that the meta.score might be useful to improve the surveillance of patients at risk of tumor recurrence (i.e. >3 years, PRETEXT IV) (Fan Li, PLOS One, 2021, 16(11): e0259503).
2. Materials and Methods
2.1. Public Hepatoblastoma Tumor Transcriptome Dataset
GSE131329 [
9] transcriptome dataset was downloaded on Gene Expression Omnibus (GEO) with GEOquery R-package version 2.70.0 [
14,
15] in R software environment version 4.3.3. This microarray dataset is available at the following address:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131329 (accessed on 2024, September 3
rd). It comprised experiments performed with [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array microarray technology corresponding to annotation platform GPL6244 available at the following address:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL6244 (accessed on 2024, September 3
rd). This transcriptome dataset comprised experiments performed on 53 hepatoblastoma tissues and 14 noncancerous liver tissue samples (
Table 1, column “Total” for description of tumor samples
).
2.2. Expression of Metabolic Program
In our previous work [
16], starting from Mammalian Metabolic Enzyme Database [
17], it was identified a metabolic program of 41 enzymes overexpressed in hepatoblastoma tumors (HB metabolic-41) (
Table S1). With HB metabolic-41 expression program, an unsupervised principal component analysis was performed on the total transcriptome of the 53 hepatoblastoma tissues and 14 noncancerous liver tissue samples from GSE131329 dataset but also on the 53 tumors samples with FactoMineR R-package version 2.11 [
18]. Expression of predictive enzymes for metastasis status was used to performed unsupervised clustering (Euclidean distances and Ward.D2 method) with pheatmap R-package version 1.0.12. ROC curves and area under curve to predict metastasis status with expression data were determined with pROC R-package version 1.18.5 [
19].
2.3. Machine Learning Elasticnet Model on Metabolic Markers
For tumor samples, expression of the HB metabolic-41 program was extracted from dataset GSE131329 and combined to the metastasis status metadata as outcome. After data splitting in training and validation sets (0.7/0.3 ratio), Elasticnet model (tumor cell status binary outcome) was tuned on alpha and lambda parameters with caret R-package version 6.0-94 [
20]. Final Elasticnet was fitted with best alpha parameter (alpha=0.2) by using glmnet R-package version 4.1-8 [
21].
Loop of univariate binomial regression was performed on each enzyme of HB metabolic-41 program according to metastatic binomial outcome. These analyses were performed with logitloop R-package version 1.0.0 available at the following web address:
https://github.com/cdesterke/logitloop (accessed on 2024, September 3
rd). With most significant metabolic gene a meta.score was computed summing their expression and binomial beta score according the equation (1):
Comparisons between qualitative parameters were performed with chi.square test R function and corresponding mosaicplot were drawn with vcd R-package version 1.4-12[
22] . Optimal threshold on meta.score was determined with cutpointr R-package version 1.1.2. Multivariate binomial model with metastasis status as outcome (negative or positive) was built with generalized linear “glm” model R function with binomial logit family. This model integrated metabolic expression score (combined expression of DNMT3B and PFKFB4 in tumors) (meta.score) but also some epidemiological parameters: age at diagnosis, gender, clinical parameter: PRETEXT stages, and histological parameter: histological type with state of differentiation. Corresponding nomogram of multivariate binomial model was built with regplot R-package version 1.1.
2.4. C1/C2 Classifier
According to the previous publication [
23], hepatoblastoma C1/C2 16-gene classifier of was applied on GSE131329 transcriptome cohort. This signature included expression of 16 genes: GHR, APCS, C1S, AQP9, CYP2E1, APOC4, HPD, NLE, RPL10A, E2F5, BUB1, DLG7, IGSF1, AFP, DUSP9, and ALDH2. K-means classification in two groups was performed on selected matrix of expression. With group results confusion matrix and accuracy according metastasis prediction were evaluated caret R package version 6.0-94 [
20].
3. Results
3.1. Hepatoblastoma Tumor Metabolic Program Predicted Distant Metastasis Status
Previous studies, using data from the Mammalian Metabolic Enzyme Database [
20,
26], identified a metabolic program consisting of 41 enzymes that are overexpressed in hepatoblastoma tumor cells. In an independent cohort of transcriptome (dataset GSE131329) [
9] (
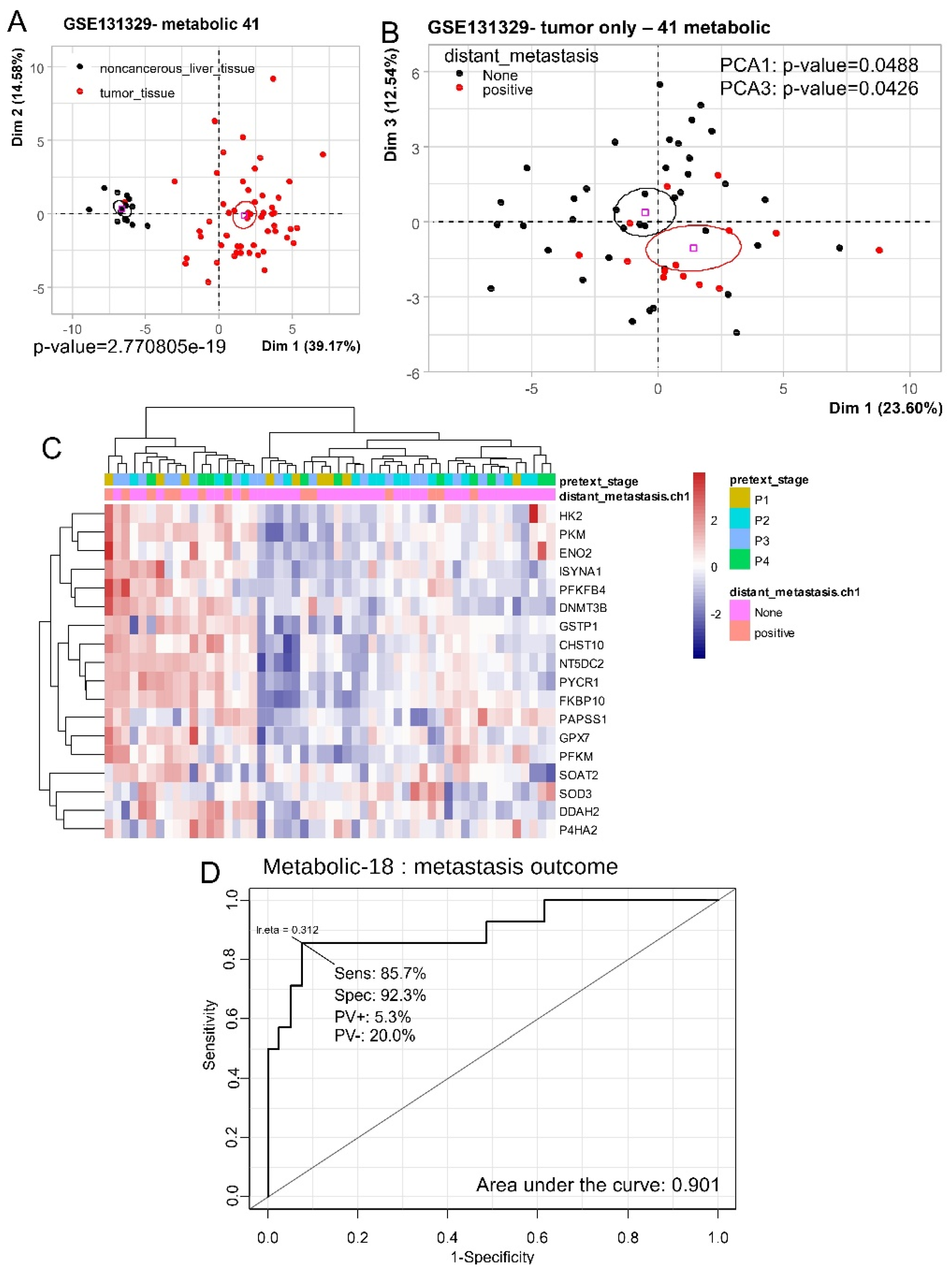
Table 1), by unsupervised principal component analysis it was confirmed that HB metabolic-41 program well stratified noncancerous liver tissue samples from HB tumor samples (p-value=2.77E-19,
Figure 1A).
Unsupervised principal component analysis performed only on the 53 tumor samples with metabolic-41 expression program allowed us to stratify metastatic outcomes of patients (PCA1 p-value=0.048 and PCA3 p-value=0.042,
Figure 1B). Analyses of loading for respective axes PCA1 and PCA3 allowed to identify 18 enzymes overexpressed in positive status of metastasis for these samples. By unsupervised clustering (Euclidean distances), expression of these 18 enzymes allowed to stratify the majority of the tumor samples according to their metastatic status (P=0.0492, OR=3.9 95%CI=1.1-12.1) (
Figure 1C). These predictive enzymes comprised: HK2 (hexokinase 2), PKM (pyruvate kinase M1/2), ENO2 (enolase 2), ISYNA1 (inositol-3-phosphate synthase 1), PFKFB4 (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4), DNMT3B (DNA methyltransferase 3 beta), GSTP1 (glutathione S-transferase pi 1), CHST10 (carbohydrate sulfotransferase 10), NT5DC2 ( 5'-nucleotidase domain containing 2), PYCR1 (pyrroline-5-carboxylate reductase 1), FKBP10 (FKBP prolyl isomerase 10), PAPSS1 (3'-phosphoadenosine 5'-phosphosulfate synthase 1), GPX7 (glutathione peroxidase 7), PFKM (phosphofructokinase, muscle), SOAT2 (sterol O-acyltransferase 2), SOD3 (superoxide dismutase 3), DDAH2 (dimethylarginine dimethylaminohydrolase 2), and P4HA2 (prolyl 4-hydroxylase subunit alpha 2). ROC curve analysis of the combined expression of these eighteen enzymes allowed to predict positive status of metastasis with an area under curve (AUC) of 0.901 with a sensibility of 85.7% and a specificity of 92.3% (
Figure 1D). These results suggest that metabolic expression program is affected in hepatoblastoma tumors in context of metastasis.
3.2. Ranking of Metabolic Enzymes to Predict Metastasis status in Hepatoblastoma Tumors
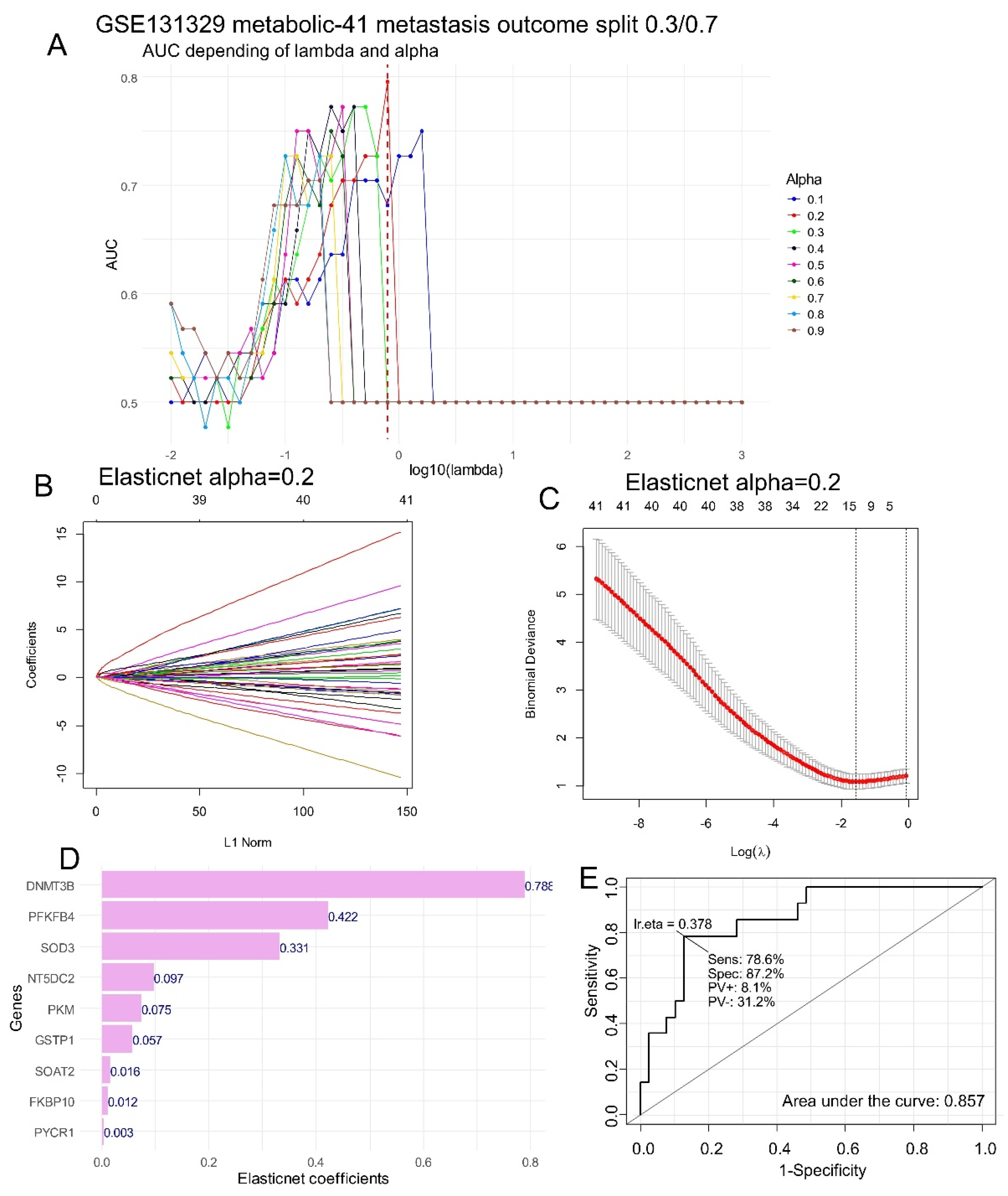
To evaluate the importance of each enzyme which predict metastasis status of hepatoblastoma, machine learning tuning was performed by Elasticnet on the expression of HB metabolic-41 expression program. After splitting cohort (
Table 1) in two datasets: training dataset (70 percent of samples) and validation dataset (30% of samples) a sequence of Elasticnet models were turned on a training set with variation of lambda and alpha parameters (
Figure 2A). Best prediction on validation set was obtained for alpha parameter fixed to 0.2 with an area under curve of 0.78 for optimal lambda (
Figure 2A). Elasticnet model was fit with optimal alpha parameter fixed to 0.2 (
Figure 2B) and the coefficient of variation of these models (
Figure 2C) allowed us to identify 9 metabolic enzymes: DNMT3B, PFKFB4, SOD3, NT5DC2, PKM, GSTP1, SOAT2, FKBP10, and PYCR1 with positive individual coefficients (
Figure 2D). DNMT3B followed by PFKFB4 and SOD3 was identified having the highest importance to predict the positive status of metastasis in HB (
Figure 2D). Combined expression of these 9 enzymes allowed to still predict positive metastasis status with an area under curve of 0.86, a sensibility of 78.6% and a specificity of 87.2% (
Figure 2E).
3.3. Combined Tumor Expression of DNMT3B and PFKFB4 Allowed to Predict Metastasis and CHIC Risk Stratification during Hepatoblastoma
A univariate logistic regression analysis was performed for each enzyme in the HB metabolic-41 expression program, based on the binary outcome of metastasis in hepatoblastoma (
Figure 3A). These analyses confirmed major importance of DNMT3B and PFKFB4 expression in HB tumor to predict metastasis (
Figure 3A). During univariate analyses against metastasis outcome, DNMT3B expression harbored an odds-ratios of 29.64 and PFKFB4 expression and odds ratio of 10.07 (
Table 2).
Indeed, difference in DNMT3B expression was confirmed between HB tumors with negative and positive status for metastasis (2 sided ttest p-value=0.007,
Figure 3B), and it was the same case for PFKFB4 expression (2 sided ttest p-value=0.020,
Figure 3C). Based on the expression levels of the two markers, DNMT3B and PFKFB4, a metabolic score (meta.score) was calculated, and this score was found to be significantly different between hepatoblastoma tumors with positive and negative metastasis status (two-sided t-test, p-value = 0.0054,
Figure 3D). As reported in cohort metastatic status, optimal threshold on meta.score parameter was determined with ROC analysis: a threshold of 38.3 allowed to obtain an area under curve of 0.78 to predict metastasis (
Figure 3E). In relation to this meta.score threshold, we performed a cohort stratification in two groups: low (30 patients) and HIGH (23 patients) (
Table 1). Patients belonging to these two groups did not shown difference for sex (p-value=1,
Table 1) or age of diagnosis (p-value=0,27,
Table 1). Based on meta.score stratification a significant difference was found for clinical course (p-value=0.01408,
Table 1) with higher proportion of patients alive in group of low meta.score (
Figure 3F), also a significant difference was observed concerning clinical events during follow up (p-value=0.013,
Table 1) with increased proportion of positive clinical events in group meta.score HIGH (
Figure 3G), and a significant difference of CHIC risk stratification.
3.4. Metabolic Expression Score (DNMT3B & PFKFB4) Better Predict METASTASIS as compared to C1-C2 Classifier
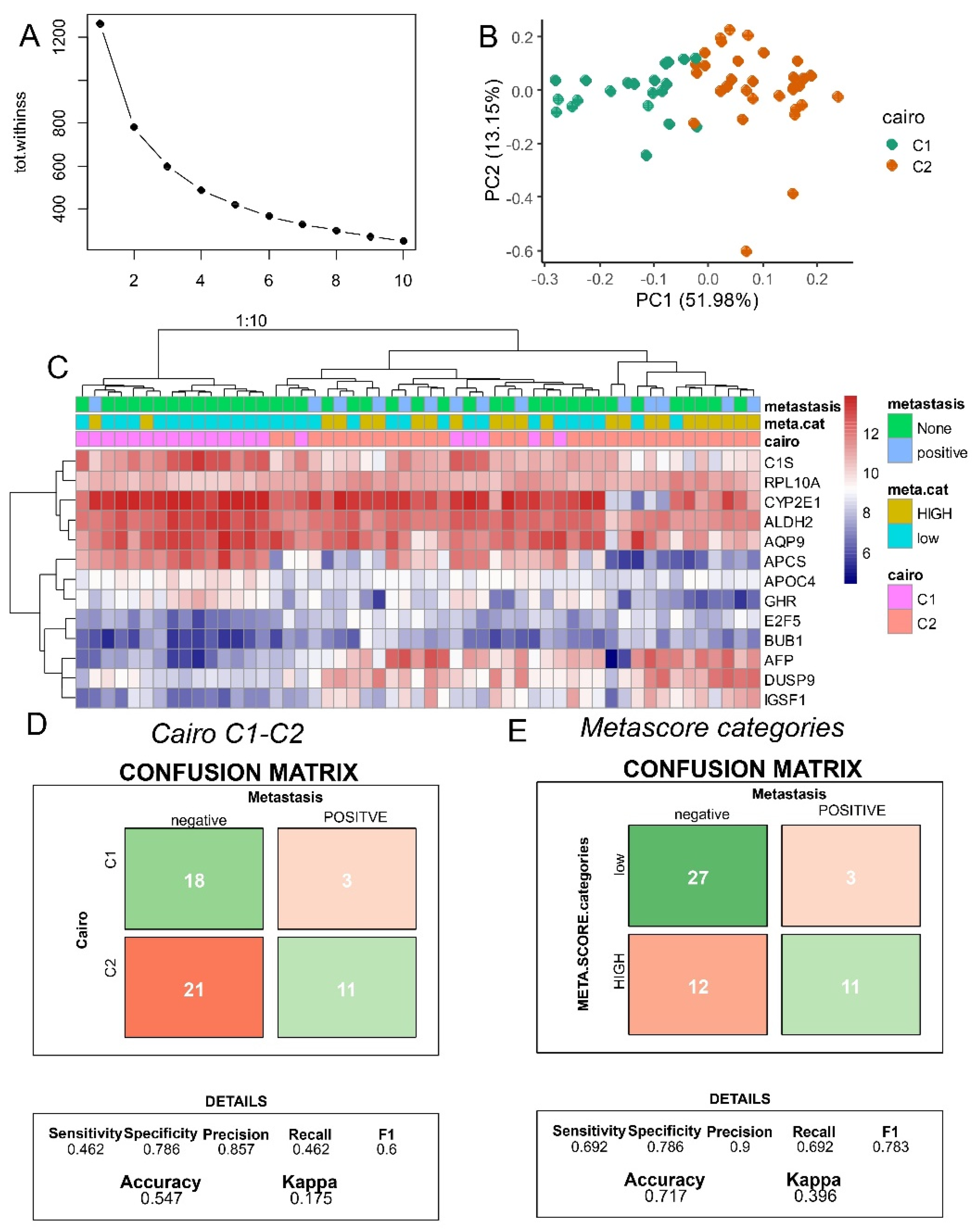
During hepatoblastoma C1/C2 16 gene classifier distinct two types of tumors with different expression of hepatic stem/progenitor markers in immature tumors in relation with activation state of beta-catenin[
23]. This referent classifier was applied to GSE131329 transcriptome dataset[
9]. In context of Affymetrix Human Gene 1.0 ST Array technology employed on this cohort, it was possible 13 of the 16 markers. Based on expression of these marker a kmeans classifier was stratified in two groups (
Figure 4A) with well stratified Cairo groups by principal component analysis (
Figure 4B). Unsupervised clustering by Euclidean distances aggregate majority of Cairo-C1 samples on left cluster and majority of Cairo-C2 samples on right cluster(
Figure 4C). Confusion matrix between Cairo C1-C2 prediction and distant metastasis allowed to evaluate a metastasis prediction accuracy of 0.55 with 46% of sensibility and 79% of specificity (
Figure 4D). Concerning meta.score allowed to evaluate a metastasis prediction accuracy of 0.72 with 69% of sensibility and 79% of specificity (
Figure 4E). In comparison, to C1/C2 classifier, meta.score better predict metastasis status in hepatoblastoma in terms of accuracy and sensibility. Specificity of the 2 parameters was found equivalent.
3.5. Metabolic Expression Score (DNMT3B & PFKFB4) is an Independent Adverse Parameter to Predict Metastasis in Tumor from Hepatoblastoma
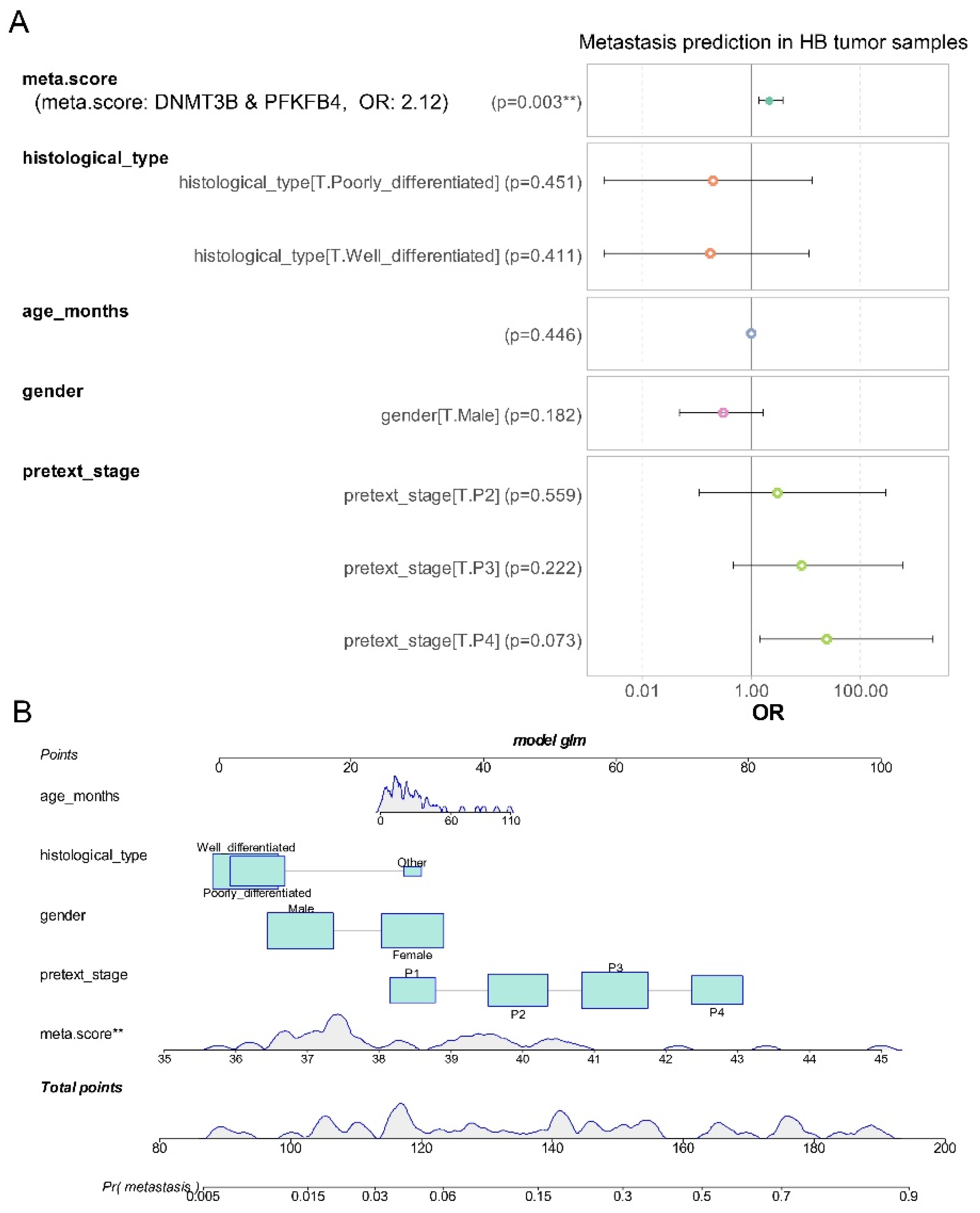
At the univariate level, we demonstrated that the meta.score, derived from the combined expression of DNMT3B and PFKFB4, can predict metastasis status in hepatoblastoma tumors. Using the generalized linear model R function, we constructed a logistic multivariate model to address the binomial outcome of metastasis. This model incorporated meta.score expression along with epidemiological parameters such as age at diagnosis and sex, as well as tissue differentiation and PRETEXT stages (see
Figure 5A). In this multivariate model, the meta.score continued to be a significant adverse factor (multivariate p-value = 0.003,
Figure 5A), with an odds ratio of 2.12 for predicting a positive metastatic status in hepatoblastoma. The nomogram for this model (
Figure 5B) showed that the distribution of meta.score is well-represented within the total points, especially between 0 and 60. These results suggest that, despite the integration of epidemiological, histological, and clinical parameters, the meta.score remains a significant independent predictor of metastasis in hepatoblastoma.
4. Discussion
During this work starting from Mammalian Metabolic Enzyme Database [
17] we established an expression metabolic score based on combined expression of DNMT3B and PFKFB4 in hepatoblastoma. This metabolic score allowed to predict positive metastatic status of the tumor and high CHIC risk stratification during hepatoblastoma. Almost high-risk CHIC hepatoblastoma patients had distant metastasis and high expression of histone cluster genes and small nucleolar RNA, suggesting that distant metastasis of hepatoblastoma may be correlated with epigenetic regulation[
9].
Among the epigenetic regulation mechanisms, DNA methylation occurs through the addition of a methyl group to a cytosine (5mC) by DNA methyltransferase enzymes (DNMTs) [
24]. During hepatoblastoma, enrichment of 5 hydroxymethyl cytosine associated to expression disruption of UHRF1, TET1, and TET2 was already described [
25]. DNMT3B with DNMT3A in opposite to DNMT1 led to the establishment of de novo methylation[
26]. In HCT116 colorectal cancer cell line, disruption of DNMT1 and DMNT3B and pharmacologic inhibition with 5-Aza-2'-deoxycytidine (5-Aza-dC, decitabine) activated demethylation of the MEG3-DMR and expression of 14q32 miRNAs, which suppressed adhesion, invasion, and migration (AIM) properties of metastatic tumor cells [
27]. During human adult hepatocellular carcinoma, microRNA-26a inhibits proliferation and metastasis of cancer by regulating DNMT3B-MEG3 axis [
28].
Phosphofructokinase 2 (PFK2) is a bifunctional enzyme with both kinase and phosphatase activities encoded by four PFK2 isozymes in human: PFKFB1, PFKFB2, PFKFB3, and PFKFB4 [
29]. Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4) could impact on tumor development by regulating the flux through the glycolytic and pentose phosphate pathways and ATP synthesis in the cells during hypoxic response [
30,
31]. PFKFB4 was shown to be induced by hypoxia in multiple cancer cell lines and over-expressed in matched human lung, breast and colon tumor tissues relative to normal tissues from the same patients [
32,
33]. During adult hepatocellular carcinoma, PFKFB4 is known to be a metabolic driver of disease progression and chemoresistance through ROS mitigation [
34]. PFKFB4 has been already shown to be implicated in metastatic process during cancer progression. During breast cancer, hypoxia induced PFKFB4 in tumor microenvironment to shape metabolic and cellular plasticity with increase of metastatic competence [
35]. The Warburg pathway enzyme PFKFB4 acts as a molecular fulcrum that couples sugar metabolism to transcriptional activation by stimulating SRC-3 to promote aggressive metastatic tumors [
36]. During melanoma, PFKFB4 is known to activate RAS/AKT pathway to impact on cell migration [
37]. PFKFB4 expression is known to be increased by carbonic anhydrase IX to promote motility of human cervical cancer cells [
38].
To finish, we would like to mention that the identification of DNMT3B and PFKFB4 as predictive biomarkers for metastasis in hepatoblastoma opens promising avenues for targeted therapeutic interventions. Given their roles in epigenetic regulation and metabolic pathways, respectively, these proteins represent potential druggable targets. DNMT3B, an enzyme involved in DNA methylation, could be targeted with existing DNMT inhibitors like azacitidine or decitabine, which are currently used in other cancers but not in hepatoblastoma. Similarly, PFKFB4, a key regulator of glycolysis, could be targeted with glycolysis inhibitors such as 3PO or PFK15, which have shown efficacy in preclinical models of other cancers. By developing or repurposing drugs that specifically inhibit DNMT3B and PFKFB4, it may be possible to disrupt the metastatic process and improve treatment outcomes in hepatoblastoma. Furthermore, the use of combination therapies that address both epigenetic and metabolic dysregulations could enhance the effectiveness of these treatments and provide a more comprehensive approach to managing this aggressive cancer.
5. Conclusions
Based on hepatoblastoma metabolic enzyme expression program, we characterized a dual overexpression of PFKFB4 and DNMT3B in patients’ samples at metastatic risk (High risk CHIC stratification). With combined tumor expression of DNMT3B and PFKFB4, a meta.score was computed and this parameter was confirmed as an independent adverse score to predict metastatic status during hepatoblastoma.
Author Contributions
C.D. and J.M-G. designed the study. C.D. and J.M-G. analyzed and interpreted data and wrote the manuscript. C.D. performed most of the experiments, with contributions from R.F. and C.M. A.M. and P.P. contributed to manuscript correction and data analysis. All authors have read and agreed to the published version of the manuscript.
Funding
P.P. and J.M-G. provided the funding of this article by MEAE AMBASS FRANCE AU PEROU FSPI - S-AC23007, Filière Santé Maladie Rare du Foie de l’Adulte et de l’Enfant.
Data Availability Statement
Acknowledgments
Many thanks to Eiso Hiyama and collaborators of Japanese Study Group for Pediatric Liver Tumors-2 (JPLT-2) group to submit transcriptome dataset GSE131329 with associated phenotype data.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Perilongo, G.; Shafford, E.; Plaschkes, J. Liver Tumour Study Group of the International Society of Paediatric Oncology SIOPEL Trials Using Preoperative Chemotherapy in Hepatoblastoma. Lancet Oncol 2000, 1, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, F.; Matsunaga, T.; Iwafuchi, M.; Hayashi, Y.; Ohkawa, H.; Ohira, M.; Okamatsu, T.; Sugito, T.; Tsuchida, Y.; Toyosaka, A.; et al. Outcome of Hepatoblastoma Treated with the JPLT-1 (Japanese Study Group for Pediatric Liver Tumor) Protocol-1: A Report from the Japanese Study Group for Pediatric Liver Tumor. Journal of Pediatric Surgery 2002, 37, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.L.; Maibach, R.; Hiyama, E.; Häberle, B.; Krailo, M.; Rangaswami, A.; Aronson, D.C.; Malogolowkin, M.H.; Perilongo, G.; von Schweinitz, D.; et al. Risk-Stratified Staging in Paediatric Hepatoblastoma: A Unified Analysis from the Children’s Hepatic Tumors International Collaboration. Lancet Oncol 2017, 18, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Koh, K.N.; Park, M.; Kim, B.E.; Bae, K.W.; Kim, K.M.; Im, H.J.; Seo, J.J. Prognostic Implications of Serum Alpha-Fetoprotein Response during Treatment of Hepatoblastoma. Pediatr Blood Cancer 2011, 57, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.L.; Rowland, J.R.; Krailo, M.; Chen, Z.; Katzenstein, H.M.; Malogolowkin, M.H. Predictive Power of Pretreatment Prognostic Factors in Children with Hepatoblastoma: A Report from the Children’s Oncology Group. Pediatric Blood & Cancer 2009, 53, 1016–1022. [Google Scholar] [CrossRef]
- Hata, Y. The Clinical Features and Prognosis of Hepatoblastoma: Follow-up Studies Done on Pediatric Tumors Enrolled in the Japanese Pediatric Tumor Registry between 1971 and 1980. Part I. The Japanese Journal of Surgery 1990, 20, 498–502. [Google Scholar] [CrossRef]
- Hishiki, T.; Matsunaga, T.; Sasaki, F.; Yano, M.; Ida, K.; Horie, H.; Kondo, S.; Watanabe, K.-I.; Oue, T.; Tajiri, T.; et al. Outcome of Hepatoblastomas Treated Using the Japanese Study Group for Pediatric Liver Tumor (JPLT) Protocol-2: Report from the JPLT. Pediatr Surg Int 2011, 27, 1–8. [Google Scholar] [CrossRef]
- Czauderna, P.; Haeberle, B.; Hiyama, E.; Rangaswami, A.; Krailo, M.; Maibach, R.; Rinaldi, E.; Feng, Y.; Aronson, D.; Malogolowkin, M.; et al. The Children’s Hepatic Tumors International Collaboration (CHIC): Novel Global Rare Tumor Database Yields New Prognostic Factors in Hepatoblastoma and Becomes a Research Model. European Journal of Cancer 2016, 52, 92–101. [Google Scholar] [CrossRef]
- Hiyama, E. Gene Expression Profiling in Hepatoblastoma Cases of the Japanese Study Group for Pediatric Liver Tumors-2 (JPLT-2) Trial; Science Repository OU, 2019.
- Dai, Z.; Ramesh, V.; Locasale, J.W. The Evolving Metabolic Landscape of Chromatin Biology and Epigenetics. Nat Rev Genet 2020, 21, 737–753. [Google Scholar] [CrossRef]
- Geiman, T.M.; Robertson, K.D. Chromatin Remodeling, Histone Modifications, and DNA Methylation—How Does It All Fit Together? J of Cellular Biochemistry 2002, 87, 117–125. [Google Scholar] [CrossRef]
- Rodríguez-Paredes, M.; Esteller, M. Cancer Epigenetics Reaches Mainstream Oncology. Nat Med 2011, 17, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Liu, B.; Zheng, S.; Dong, K.; Dong, R. Genome-Wide Analysis of DNA Methylation in Hepatoblastoma Tissues. Oncology Letters 2016, 12, 1529–1534. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A Bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets--Update. Nucleic Acids Res. 2013, 41, D991–995. [Google Scholar] [CrossRef]
- Monge, C.; Francés, R.; Marchio, A.; Pineau, P.; Desterke, C.; Matta-Garrido, J. Activated Metabolic Transcriptional Program in Tumor Cells from Hepatoblastoma. International Journal of Molecular Sciences 2024, preprints–117670. [Google Scholar] [CrossRef]
- Corcoran, C.C.; Grady, C.R.; Pisitkun, T.; Parulekar, J.; Knepper, M.A. From 20th Century Metabolic Wall Charts to 21st Century Systems Biology: Database of Mammalian Metabolic Enzymes. American Journal of Physiology-Renal Physiology 2017, 312, F533–F542. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. Journal of Statistical Software 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinformatics 2011, 12, 77. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Soft. 2008, 28. [Google Scholar] [CrossRef]
- Tay, J.K.; Narasimhan, B.; Hastie, T. Elastic Net Regularization Paths for All Generalized Linear Models. J. Stat. Soft. 2023, 106. [Google Scholar] [CrossRef]
- Zeileis, A.; Meyer, D.; Hornik, K. Residual-Based Shadings for Visualizing (Conditional) Independence. Journal of Computational and Graphical Statistics 2007, 16, 507–525. [Google Scholar] [CrossRef]
- Cairo, S.; Armengol, C.; De Reyniès, A.; Wei, Y.; Thomas, E.; Renard, C.-A.; Goga, A.; Balakrishnan, A.; Semeraro, M.; Gresh, L.; et al. Hepatic Stem-like Phenotype and Interplay of Wnt/Beta-Catenin and Myc Signaling in Aggressive Childhood Liver Cancer. Cancer Cell 2008, 14, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. The DNA Methylation Paradox. Trends in Genetics 1999, 15, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.P.; Aguiar, T.F.M.; Fernandes, G.R.; Caires-Júnior, L.C.; Goulart, E.; Telles-Silva, K.A.; Cypriano, M.; De Toledo, S.R.C.; Rosenberg, C.; Carraro, D.M.; et al. TET Upregulation Leads to 5-Hydroxymethylation Enrichment in Hepatoblastoma. Front. Genet. 2019, 10, 553. [Google Scholar] [CrossRef]
- Qureshi, M.Z.; Sabitaliyevich, U.Y.; Rabandiyarov, M.; Arystanbekuly, A.T. Role of DNA Methyltransferases (DNMTs) in Metastasis. Cell Mol Biol (Noisy-le-grand) 2022, 68, 226–236. [Google Scholar] [CrossRef]
- Oshima, G.; Poli, E.C.; Bolt, M.J.; Chlenski, A.; Forde, M.; Jutzy, J.M.S.; Biyani, N.; Posner, M.C.; Pitroda, S.P.; Weichselbaum, R.R.; et al. DNA Methylation Controls Metastasis-Suppressive 14q32-Encoded miRNAs. Cancer Research 2019, 79, 650–662. [Google Scholar] [CrossRef]
- Li, Y.; Ren, M.; Zhao, Y.; Lu, X.; Wang, M.; Hu, J.; Lu, G.; He, S. MicroRNA-26a Inhibits Proliferation and Metastasis of Human Hepatocellular Carcinoma by Regulating DNMT3B-MEG3 Axis. Oncology Reports 2017, 37, 3527–3535. [Google Scholar] [CrossRef]
- Rider, M.H.; Bertrand, L.; Vertommen, D.; Michels, P.A.; Rousseau, G.G.; Hue, L. 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase: Head-to-Head with a Bifunctional Enzyme That Controls Glycolysis. Biochem J 2004, 381, 561–579. [Google Scholar] [CrossRef]
- Chesney, J.; Clark, J.; Klarer, A.C.; Imbert-Fernandez, Y.; Lane, A.N.; Telang, S. Fructose-2,6-Bisphosphate Synthesis by 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase 4 (PFKFB4) Is Required for the Glycolytic Response to Hypoxia and Tumor Growth. Oncotarget 2014, 5, 6670–6686. [Google Scholar] [CrossRef]
- Gao, R.; Li, D.; Xun, J.; Zhou, W.; Li, J.; Wang, J.; Liu, C.; Li, X.; Shen, W.; Qiao, H.; et al. CD44ICD Promotes Breast Cancer Stemness via PFKFB4-Mediated Glucose Metabolism. Theranostics 2018, 8, 6248–6262. [Google Scholar] [CrossRef]
- Minchenko, O.H.; Ochiai, A.; Opentanova, I.L.; Ogura, T.; Minchenko, D.O.; Caro, J.; Komisarenko, S.V.; Esumi, H. Overexpression of 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase-4 in the Human Breast and Colon Malignant Tumors. Biochimie 2005, 87, 1005–1010. [Google Scholar] [CrossRef]
- Minchenko, O.H.; Ogura, T.; Opentanova, I.L.; Minchenko, D.O.; Ochiai, A.; Caro, J.; Komisarenko, S.V.; Esumi, H. 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase Gene Family Overexpression in Human Lung Tumor. Ukr Biokhim Zh (1999) 2005, 77, 46–50. [Google Scholar] [PubMed]
- Olaizola, P.; Banales, J.M. PFKFB4 Is a Metabolic Driver of HCC Progression and Chemoresistance Through ROS Mitigation. Cellular and Molecular Gastroenterology and Hepatology 2023, 15, 1527–1529. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Rosario, S.R.; Katsuta, E.; Sawant Dessai, A.; Paterson, E.J.; Novickis, A.T.; Cortes Gomez, E.; Zhu, B.; Liu, S.; Wang, H.; et al. Hypoxic Activation of PFKFB4 in Breast Tumor Microenvironment Shapes Metabolic and Cellular Plasticity to Accentuate Metastatic Competence. Cell Reports 2022, 41, 111756. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, S.; Rajapakshe, K.; Zhu, B.; Nikolai, B.C.; Yi, P.; Putluri, N.; Choi, J.M.; Jung, S.Y.; Coarfa, C.; Westbrook, T.F.; et al. Metabolic Enzyme PFKFB4 Activates Transcriptional Coactivator SRC-3 to Drive Breast Cancer. Nature 2018, 556, 249–254. [Google Scholar] [CrossRef]
- Sittewelle, M.; Kappès, V.; Zhou, C.; Lécuyer, D.; Monsoro-Burq, A.H. PFKFB4 Interacts with ICMT and Activates RAS/AKT Signaling-Dependent Cell Migration in Melanoma. Life Sci. Alliance 2022, 5, e202201377. [Google Scholar] [CrossRef]
- Hsin, M.-C.; Hsieh, Y.-H.; Hsiao, Y.-H.; Chen, P.-N.; Wang, P.-H.; Yang, S.-F. Carbonic Anhydrase IX Promotes Human Cervical Cancer Cell Motility by Regulating PFKFB4 Expression. Cancers (Basel) 2021, 13, 1174. [Google Scholar] [CrossRef]
Figure 1.
Hepatoblastoma tumor metabolic program predicted distant metastasis status: dataset GSE131329, A/ Principal component analysis performed on expression of hepatoblastoma (HB) metabolic-41 program with tissue type stratification (non-cancerous liver tissue versus tumor), p-value was obtained on first principal axis; B/ Principal component analysis performed on expression of HB metabolic-41 program with stratification for metastasis status, p-values were respectively obtained on first and third principal axes; C/ Unsupervised clustering (Euclidean distances) and expression heatmap for 18 best metabolic markers in HB tumors (metastasis prediction selected on PCA axes); ROC and area and under curve for expression of 18 best metabolic markers to predict metastasis status in hepatoblastoma tumors (Sens: sensibility, Spe: specificity, PV+: positive prevalence, PV-: negative prevalence).
Figure 1.
Hepatoblastoma tumor metabolic program predicted distant metastasis status: dataset GSE131329, A/ Principal component analysis performed on expression of hepatoblastoma (HB) metabolic-41 program with tissue type stratification (non-cancerous liver tissue versus tumor), p-value was obtained on first principal axis; B/ Principal component analysis performed on expression of HB metabolic-41 program with stratification for metastasis status, p-values were respectively obtained on first and third principal axes; C/ Unsupervised clustering (Euclidean distances) and expression heatmap for 18 best metabolic markers in HB tumors (metastasis prediction selected on PCA axes); ROC and area and under curve for expression of 18 best metabolic markers to predict metastasis status in hepatoblastoma tumors (Sens: sensibility, Spe: specificity, PV+: positive prevalence, PV-: negative prevalence).
Figure 2.
DNMT3B expression is the best metabolic marker to predict metastasis in hepatoblastoma tumors: A/ Elasticnet tuning (lambda and alpha parameters) performed on HB metabolic-41 expression program to predict metastasis status in tumors (AUC: area under curve evaluated on validation cohort after training-validation split: 0.7/0.3), B/ Elasticnet fit with best alpha parameter fixed to 0.2; C/ Elasticnet coefficient of variation with best alpha parameter fixed to 0.2; D/ Barplot of best positive Elasticnet coefficients to predict metastasis concerning metabolic markers. E/ ROC curve and area under curve to predict metastasis with combination of the nine elasticnet metabolic markers: DNMT3B, PFKFB4, SOD3, NT5DC2, PKM, GSTP1, SOAT2, FKBP10, and PYCR1.
Figure 2.
DNMT3B expression is the best metabolic marker to predict metastasis in hepatoblastoma tumors: A/ Elasticnet tuning (lambda and alpha parameters) performed on HB metabolic-41 expression program to predict metastasis status in tumors (AUC: area under curve evaluated on validation cohort after training-validation split: 0.7/0.3), B/ Elasticnet fit with best alpha parameter fixed to 0.2; C/ Elasticnet coefficient of variation with best alpha parameter fixed to 0.2; D/ Barplot of best positive Elasticnet coefficients to predict metastasis concerning metabolic markers. E/ ROC curve and area under curve to predict metastasis with combination of the nine elasticnet metabolic markers: DNMT3B, PFKFB4, SOD3, NT5DC2, PKM, GSTP1, SOAT2, FKBP10, and PYCR1.
Figure 3.
DNMT3B and PFKFB4 combined expression predicts metastasis and CHIC risk stratification in hepatoblastoma tumors: A/ Univariate binomial analyses for best metabolic markers in hepatoblastoma tumor according metastasis status as outcome; B/ Boxplot of DNMT3B expression stratified on metastatic status and colored according CHIC risk stratification, p-value obtained by two-tailed ttest; C/ Boxplot of PFKFB4 expression stratified on metastatic status and colored according CHIC risk stratification, p-value obtained by two-sided t test; D/ Boxplot of meta.score (metabolic/metastatic score: DNMT3B & PFKFB4 combined expression) stratified on metastasis status and colored according CHIC risk stratification, p-value obtained by twosided ttest; E/ Optimal cutpoint determination on meta.score to predict metastasis status; F/ mosaicplot crossing meta.score categories with clinical course status (p-value of chi-square test); G/ mosaicplot crossing meta.score categories with clinical event (p-value of chi-square test); H/ mosaicplot crossing meta.score categories with CHIC risk stratification (p-value of chi-square test).
Figure 3.
DNMT3B and PFKFB4 combined expression predicts metastasis and CHIC risk stratification in hepatoblastoma tumors: A/ Univariate binomial analyses for best metabolic markers in hepatoblastoma tumor according metastasis status as outcome; B/ Boxplot of DNMT3B expression stratified on metastatic status and colored according CHIC risk stratification, p-value obtained by two-tailed ttest; C/ Boxplot of PFKFB4 expression stratified on metastatic status and colored according CHIC risk stratification, p-value obtained by two-sided t test; D/ Boxplot of meta.score (metabolic/metastatic score: DNMT3B & PFKFB4 combined expression) stratified on metastasis status and colored according CHIC risk stratification, p-value obtained by twosided ttest; E/ Optimal cutpoint determination on meta.score to predict metastasis status; F/ mosaicplot crossing meta.score categories with clinical course status (p-value of chi-square test); G/ mosaicplot crossing meta.score categories with clinical event (p-value of chi-square test); H/ mosaicplot crossing meta.score categories with CHIC risk stratification (p-value of chi-square test).
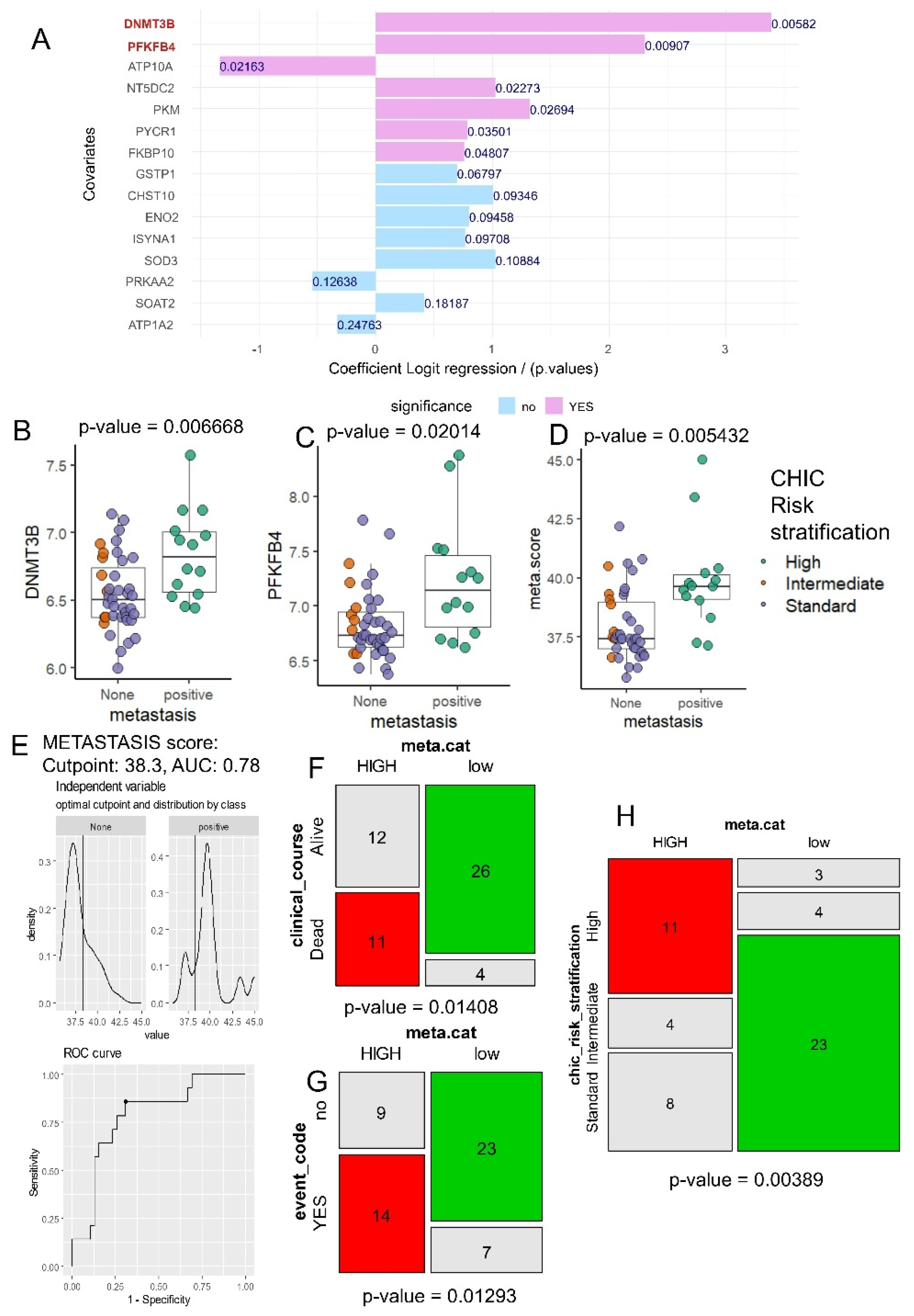
Figure 4.
meta.score better predict metastasis than C1-C2 classifier: dataset GSE131329: A/ Efficiency yield of cluster numbers during kmeans clustering based on C1-C2 expression signature; B/ Principal component analysis with stratification C1/C2 group based on Cairo signature; C/ Unsupervised clustering on C1/C2 signature with Cairo prediction, metastasis, and meta.score (metabolism) stratification; D/ Confusion matrix testing accuracy of Cairo C1-C2 classifier to predict metastasis status; E/ Confusion matrix testing accuracy of meta.score to predict metastasis status.
Figure 4.
meta.score better predict metastasis than C1-C2 classifier: dataset GSE131329: A/ Efficiency yield of cluster numbers during kmeans clustering based on C1-C2 expression signature; B/ Principal component analysis with stratification C1/C2 group based on Cairo signature; C/ Unsupervised clustering on C1/C2 signature with Cairo prediction, metastasis, and meta.score (metabolism) stratification; D/ Confusion matrix testing accuracy of Cairo C1-C2 classifier to predict metastasis status; E/ Confusion matrix testing accuracy of meta.score to predict metastasis status.
Figure 5.
meta.score is an independent adverse parameter to predict metastasis status in hepatoblastoma tumors: A/ Forestplot of the multivariate binomial clinical-biological model with metastasis status as outcome incorporing distinc parameters: age of diagnosis (age_months), meta.score (DNMT3B & PFKFB4 combined expression), gender of patients (T: female as reference), histological type of tumors (T: other as reference), PRETEXT stage (T: pretext stage 1 (P1) as reference, P2: stage 2, P3: stage 3, P4: stage 4), OR: odds ratios); B/ Nomogram of the multivariate metastasis model.
Figure 5.
meta.score is an independent adverse parameter to predict metastasis status in hepatoblastoma tumors: A/ Forestplot of the multivariate binomial clinical-biological model with metastasis status as outcome incorporing distinc parameters: age of diagnosis (age_months), meta.score (DNMT3B & PFKFB4 combined expression), gender of patients (T: female as reference), histological type of tumors (T: other as reference), PRETEXT stage (T: pretext stage 1 (P1) as reference, P2: stage 2, P3: stage 3, P4: stage 4), OR: odds ratios); B/ Nomogram of the multivariate metastasis model.
Table 1.
Clinical stratification (low and HIGH metabolic categories) of hepatoblastoma patients according to their metabolic score on metastasis (GSE131329).
Table 1.
Clinical stratification (low and HIGH metabolic categories) of hepatoblastoma patients according to their metabolic score on metastasis (GSE131329).
| Variable |
Level |
low (n=30) |
HIGH (n=23) |
Total (n=53) |
p-value |
| age_months |
mean (sd) |
24 (22.8) |
31.4 (25.7) |
27.2 (24.1) |
0.26766 |
CHIC_risk_stratification |
Standard |
23 (76.7) |
8 (34.8) |
31 (58.5) |
|
| |
High |
3 (10.0) |
11 (47.8) |
14 (26.4) |
|
| |
Intermediate |
4 (13.3) |
4 (17.4) |
8 (15.1) |
0.00389 |
clinical_course |
Alive |
26 (86.7) |
12 (52.2) |
38 (71.7) |
|
| |
Dead |
4 (13.3) |
11 (47.8) |
15 (28.3) |
0.01408 |
Clinical event during follow up |
no |
23 (76.7) |
9 (39.1) |
32 (60.4) |
|
| |
YES |
7 (23.3) |
14 (60.9) |
21 (39.6) |
0.01293 |
| histological_type |
Well diff. |
17 (56.7) |
13 (56.5) |
30 (56.6) |
|
| |
Other |
1 (3.3) |
1 (4.3) |
2 (3.8) |
|
| |
Poorly diff. |
12 (40.0) |
9 (39.1) |
21 (39.6) |
0.98116 |
sex |
Female |
14 (46.7) |
11 (47.8) |
25 (47.2) |
|
| |
Male |
16 (53.3) |
12 (52.2) |
28 (52.8) |
1.00000 |
PRETEXT stage |
P3 |
10 (33.3) |
8 (34.8) |
18 (34.0) |
|
| |
P2 |
10 (33.3) |
5 (21.7) |
15 (28.3) |
|
| |
P4 |
4 (13.3) |
7 (30.4) |
11 (20.8) |
|
| |
P1 |
6 (20.0) |
3 (13.0) |
9 (17.0) |
0.41827 |
Table 2.
Univariate binomial analyses for expression of the best metabolic markers to predict metastasis status in hepatoblastoma tumors.
Table 2.
Univariate binomial analyses for expression of the best metabolic markers to predict metastasis status in hepatoblastoma tumors.
| predictors |
beta coefficients |
Odds-ratios |
P-values |
| DNMT3B |
3.389 |
29.638 |
5.82E-03 |
| PFKFB4 |
2.310 |
10.071 |
9.07E-03 |
| NT5DC2 |
1.030 |
2.801 |
2.27E-02 |
| PKM |
1.321 |
3.745 |
2.69E-02 |
| PYCR1 |
0.792 |
2.208 |
3.50E-02 |
| FKBP10 |
0.764 |
2.146 |
4.81E-02 |
| GSTP1 |
0.702 |
2.017 |
6.80E-02 |
| CHST10 |
1.009 |
2.742 |
9.35E-02 |
| ENO2 |
0.802 |
2.230 |
9.46E-02 |
| ISYNA1 |
0.772 |
2.163 |
9.71E-02 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).