1. Introduction
Dengue fever, caused by dengue viruses (
Orthoflavivirus denguei, DENV), is a mosquito-borne viral disease that poses a significant public health concern worldwide, especially in tropical and subtropical regions [
1]. DENV, a single-stranded RNA virus, comprises four distinct serotypes (DENV1 to DENV4), exhibiting unique genetic and antigenic properties that contribute to a wide spectrum of clinical manifestations [
2,
3]. Since its introduction in Brazil in the 1980s, dengue has presented a major epidemiological challenge. In 2024, Brazil reported 6,486,906 probable cases of dengue, with over 1,696,426 cases documented in the state of Minas Gerais alone as of August 2024, making it the second highest incidence of the disease [
4]. This year, Minas Gerais, located in southeastern Brazil, has experienced the co-circulation of DENV1, DENV2, and DENV3 serotypes, further complicating the local epidemiological landscape [
5].
Recent advances in dengue prevention include the introduction of the Qdenga vaccine (TAK-003 by Takeda Pharmaceuticals) into the Brazilian public healthcare system. Qdenga is a live attenuated tetravalent vaccine derived from a DENV2 backbone incorporated with recombinant strains expressing surface proteins for DENV1, DENV3, and DENV4. Clinical trials have demonstrated its efficacy in reducing both the incidence and severity of dengue fever [
6], offering promising prospects for mitigating dengue-related morbidity and mortality in endemic regions. However, the vaccine leaflet warns of side effects observed during studies in children, adolescents, and adults [
7]. Commonly reported reactions include pain at the injection site, headache, muscle pain, general discomfort, weakness, respiratory infections, fever, irritability, and drowsiness. Less frequent adverse effects, occurring in 0.1% to 1% of patients, include gastrointestinal symptoms such as diarrhea, nausea, and vomiting, as well as dizziness, skin itching, hives, and fatigue. Rarely, up to 0.01% of patients experience rapid swelling under the skin, particularly on the face, throat, arms, and legs [
8,
9]. Additionally, transient viremia following Qdenga vaccination was observed in 49% of study participants who had no prior dengue infection and in 16% of those with a history of dengue infection [
10]. This scenario is particularly concerning for surveillance purposes, as dengue diagnostic tests may yield positive results during vaccine-induced viremia, potentially leading to misinterpretation as the emergence of serotypes or genotypes not currently circulating in the region.
Understanding dengue vaccine-associated infections is essential for preventing misdiagnoses and ensuring the effectiveness of surveillance programs globally. In this study, we provide a detailed characterization of the first Qdenga vaccine-associated case in the state of Minas Gerais, southeast Brazil, underscoring the urgent need for accurate monitoring to protect public health.
2. Materials and Methods
2.1. Sample Collection and Molecular Diagnostic Assay
A serum sample from a 7-year-old male suspected of DENV4 infection was submitted to the Central Laboratory of Public Health of Minas Gerais (Lacen-MG) at the Fundação Ezequiel Dias (Funed) for diagnostic confirmation. Viral RNA extraction was performed using an automated protocol on the Extracta 96 system (Loccus, Brazil) with the Extracta Kit DNA and RNA Pathogen MDx (Loccus), following the manufacturer's instructions. Molecular diagnosis was conducted on the CFX-96 Real Time System (Bio-Rad) using the Molecular ZDC-IBMP Kit, developed by the Instituto de Biologia Molecular do Paraná (Paraná, Brazil) and provided by the Brazilian Ministry of Health (BrMoH) to the public laboratory network. This kit enables detection and differential diagnosis of arboviruses through RT-qPCR multiplex reactions for DENV serotyping (DENV1-4), Zika virus (Orthoflavivirus zikaense), chikungunya virus (Alphavirus chikungunya), and the human RNAseP gene, serving as an endogenous control.
2.2. cDNA Synthesis and Whole-Genome Sequencing Using Nanopore Technology
Viral RNA was subjected to cDNA synthesis using the ProtoScript II First Strand cDNA Synthesis Kit (NEB), following the manufacturer's instructions. The resulting cDNA was then submitted to sequencing multiplex PCR (35 cycles) using Q5 High Fidelity Hot-Start DNA Polymerase (NEB) and a set of specific primers designed by the CADDE project (
https://www.caddecentre.org/) for sequencing the complete genomes of DENV1 to DENV4 serotypes. Amplicons were purified using 1x AMPure XP Beads (Beckman Coulter, USA) and quantified with a Qubit 3.0 fluorimeter (Thermofisher Scientific, USA) using the Qubit™ dsDNA HS Assay Kit (Thermofisher Scientific, USA). DNA library preparation was performed with the Ligation Sequencing Kit (SQK-LSK109) and the Native Barcoding Kit (EXP-NBD104) from Oxford Nanopore Technologies (ONT) [
11], followed by sequencing on an R9.4 flow cell using the MinION platform (ONT).
2.3. Generation of Consensus Sequence
Raw files were basecalled and demultiplexed using Guppy v.6.0 (Oxford Nanopore Technologies). Consensus sequences were generated by a hybrid approach using the Genome Detective online tool (
https://www.genomedetective.com/) [
12]. DENV genotyping was assessed using the Dengue Virus Typing Tool (
https://www.genomedetective.com/app/typingtool/dengue/) [
13], integrated within the Genome Detective platform. The newly generated DENV2 and DENV4 sequences has been deposited in GISAID under accession numbers EPI_ISL_19365344 and EPI_ISL_19365345, respectively.
2.4. Phylogenetic Analysis
To determine the origin of the DENV2 and DENV4 genomes from this case, we performed maximum likelihood (ML) phylogenetic analyses using two distinct datasets. The DENV2 dataset was constructed with our new DENV2 sequence, along with 292 reference DENV2 complete genomes representing various genotypes deposited in public database (GenBank and GISAID) (Genotype I n=20, Genotype II n=54, Genotype III n=89, Genotype IV n=35, Genotype V n=35, and Genotype VI n=3). This dataset also included the DENV2 genomes isolated from three Qdenga vaccine-associated cases in the Brazilian state of Mato Grosso do Sul (EPI_ISL_18877651, EPI_ISL_19115812, and EPI_ISL_19115813). Additionally, our dataset includes the sequence KU725663, which is the vaccine strain used by Takeda. Similarly, the DENV4 dataset comprised our new DENV4 sequence and 476 sequences from the prM/E region, representing various genotypes (Genotype I n=191, Genotype II n=277, and Genotype III n=8). This dataset also included the DENV4 sequence isolated from a Qdenga vaccine-associated case in Mato Grosso do Sul (EPI_ISL_18877652). Notably, our dataset also includes the sequence MW793460, which is the vaccine strain used by Takeda.
Sequence alignment for each dataset was performed using MAFFT v7.3.10 [
14] and manually curated to remove artifacts using AliView v1.28 [
15]. ML phylogenetic trees were estimated using IQTREE v2.3.6 [
16] applying the best-fit model inferred by the ModelFinder application implemented within IQTREE. Branch support was assessed using the approximate likelihood-ratio test incorporating bootstrap and the Shimodaira-Hasegawa-like procedure with 1,000 replicates.
3. Case Description
On January 29, 2024, a 7-year-old male child received a dose of the Qdenga vaccine. Six days later, on February 4, 2024, he developed symptoms including fever, myalgia, rash, and sore throat. On February 6, 2024, the patient was admitted to a public health unit in the city of Teófilo Otoni, Minas Gerais, Southeast Brazil. There, he underwent a dengue NS1 rapid test, yielding a negative result. On the same day, a serum sample was collected and sent to the Universidade Federal dos Vales do Jequitinhonha e Mucuri (UFVJM) campus in Teófilo Otoni, for molecular diagnosis of arboviruses. This institution is part of the accredited laboratory network of the Secretaria de Estado de Saúde de Minas Gerais (SES-MG). The RT-qPCR assay using the Molecular ZDC-IBMP Kit detected the DENV4 serotype, with a cycle threshold (CT) value of 29. Due to the epidemiological significance of this case, particularly because this serotype is not circulating in Brazil, the sample was forwarded to Lacen-MG, located at Funed, for confirmation and genetic characterization through next-generation sequencing.
At Funed, underwent further molecular diagnosis using the Molecular ZDC-IBMP Kit, which confirmed the detection of the DENV4 serotype, this time with a CT value of 32. To further investigate the possibility of a Qdenga vaccine-associated case, next-generation sequencing was performed using the MinION platform (ONT) and specific primers for each DENV serotype, as outlined in previous protocols (
https://www.caddecentre.org/protocols/).
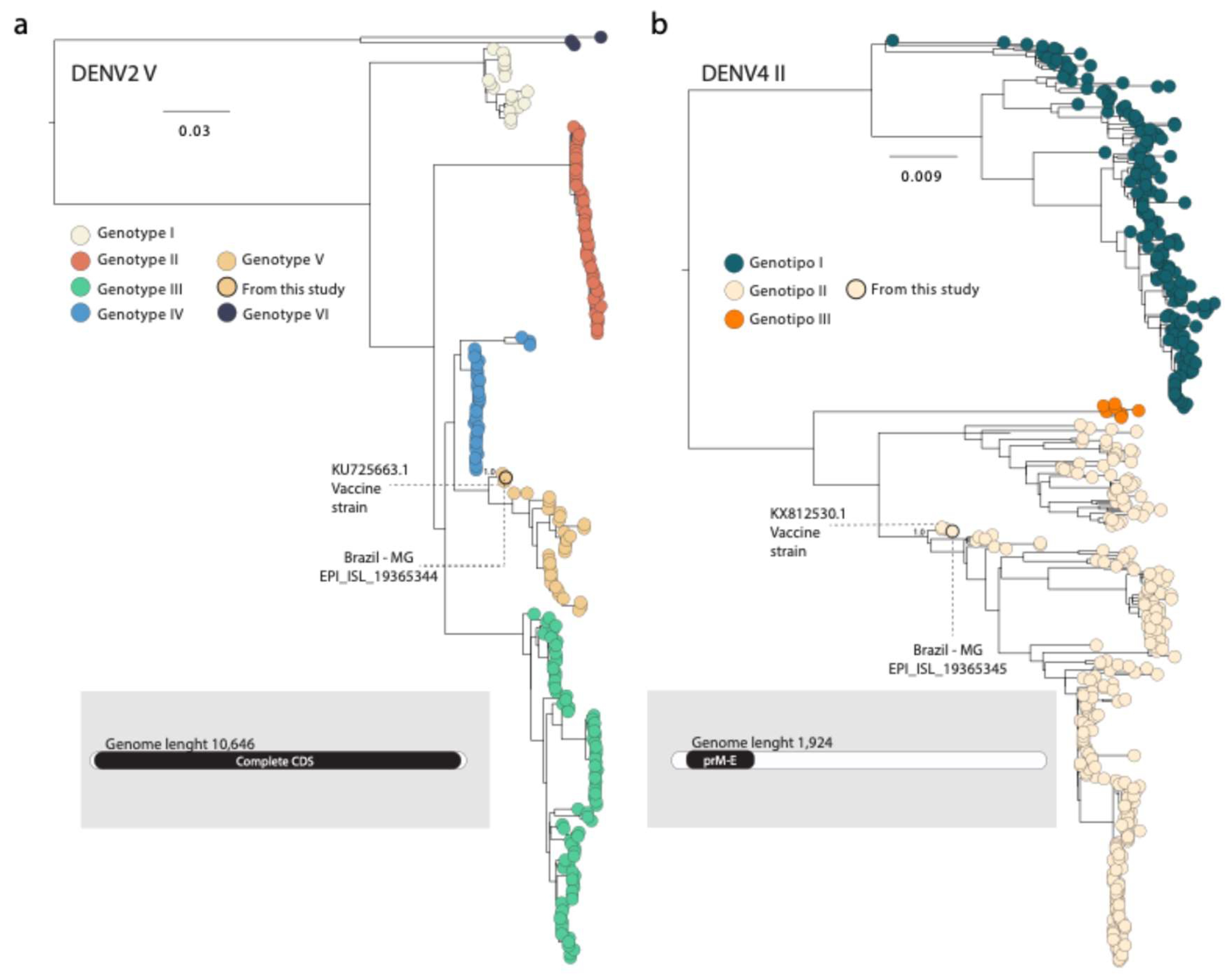
The sequencing procedures revealed the presence of both DENV2 and DENV4 serotypes. For the DENV2 a total of 484,441 reads were obtained, with an average depth of 49,563× and a coverage of 59.7%. In contrast, the DENV4 genome yielded 256,966 reads, with an average depth of 169.9× and a coverage of 9.3%, corresponding to a fragment of the prM/E region. Using the online Dengue Typing Tool, the DENV2 and DENV4 sequences were classified as DENV2 genotype V (DENV2-V), which is not recorded as circulanting in Brazil, and DENV4 genotype II (DENV4-II) respectively. Phylogenetic analysis subsequently confirmed the genotypes identified previously by the online tool (
Figure 1).
In the phylogenetic tree, the DENV2 genome obtained from Minas Gerais clustered with other Brazilian strains sampled in 2024 (EPI_ISL_19115812, EPI_ISL_19115813, and EPI_ISL_18877651). These sequences were isolated from suspected cases of Adverse Event Following Immunization (AEFI) in the Brazilian state of Mato Grosso do Sul, where patients had recently received the Qdenga vaccine. Additionally, this clade included the DENV2 genome strain 16681 (Mahidol strain, KU725663), which is the original strain used as backbone for Takeda’s Qdenga vaccine and, after attenuation, became known as DENV2 PDK-53 [
17,
18] (
Figure 1).
Regarding the DENV4 serotype, the newly generated sequence from Minas Gerais clustered with another Brazilian strain sampled in 2024 (EPI_ISL_18877652), isolated from a suspected AEFI case in Mato Grosso do Sul state. This clade also included the sequences MW793460 and KX812530, corresponding to DENV4 isolate 1036 (
Figure 1), which was used in the production of the Qdenga vaccine [
17,
19].
4. Discussion
In this study, we describe the first Qdenga vaccine-associated case in the state of Minas Gerais, southeastern Brazil, involving a male child from Teófilo Otoni, as indicated by sequencing results and phylogenetic analyses. Therefore, the purpose of this case report is to alert public and private laboratories about the importance of accurately interpreting diagnoses of DENV serotypes that are not currently circulating in a country. The introduction of a new DENV serotype could result to an increase in dengue cases and severity of clinical symptoms [
20,
21,
22]. This highlights the critical need for accurate monitoring to protect public health and ensure the effectiveness of public policies.
Phylogenetic analyses revealed that the newly sequenced DENV2 genome belongs to genotype V. Since its initial detection in Brazil in the early 1990s [
23], DENV2 have been responsible for widespread epidemics [
22]. Genotype III, also known as the Southeast Asian-American, was the predominant genotype in Brazil until 2021, when genotype II, also known as Cosmopolitan, was first detected in the country [
24]. In contrast, genotype V is mostly confined to Asia [
25], with no evidence of autochthonous circulation in Brazil to date. Our new DENV2-V strain from Minas Gerais clustered with other Brazilian isolates from suspected cases of AEFI and with the DENV2 genome strain 16681, which, after attenuation, was used as the backbone for the Qdenga vaccine [
17,
18]. The sequencing results also revealed the presence of the DENV4 serotype, specifically corresponding to the prM/E region, similar to the insert used in the Qdenga vaccine [
8]. Taken together, these findings suggest that this case is result of transient viremia following the administration of the Qdenga vaccine.
Vaccine-induced viremia, as observed in this case report, is a phenomenon seen with other vaccines, such as those for Rubella and Yellow Fever [
26,
27]. This occurs because these vaccines contain attenuated or inactivated forms of the virus that can temporarily replicate in the body to stimulate an immune response. Temporary viremia is usually short-lived and does not cause illness, but it is sufficient to induce the production of antibodies and provide immunity against the actual infection [
28]. The primary mechanism of action of Qdenga vaccine is to replicate locally and elicit neutralizing antibodies to confer protection against dengue disease caused by any of the four DENV serotypes. The vaccine activates multiple arms of the immune system, including binding antibodies, complement-fixing antibodies, functional antibodies to dengue NS1, and cell-mediated immune responses [
9]. During clinical trials of the Qdenga vaccine, efficacy varied by individual serotypes (DENV 1, 69.8%; DENV 2, 95.1%; DENV 3, 48.9%; DENV 4, 51%). Cumulative rates of serious adverse events were similar in experimental group (4%) and placebo (4.8%) recipients and were consistent with expected medical disorders in the study population. Among those who presented with a febrile illness within 30 days of vaccination, vaccine viraemia was detected in 7% of participants after the first dose. The Qdenga vaccine was well tolerated and efficacious against symptomatic dengue regardless of serostatus before immunization. Thus, it was deemed safe for administration by regulatory agencies [
28].
The side effects observed in this report were not different from those seen during the clinical testing phase of the vaccine, and the patient described did not experience any severe effects. However, caution must be taken when interpreting positive diagnostic tests for non-circulating strains within the country, especially in post-vaccination patients. A thorough patient history should be conducted, and public health laboratories that perform viral sequencing should be involved to better determine the clinical case before notifying epidemiological authorities.
In summary, our results suggest that the identification of both DENV2 and DENV4 serotypes in the analyzed sample is not related to the recent emergence of wild-type strains, but rather to the presence of a genetically modified vaccine clone of the Qdenga vaccine. This clone consists of a DENV2 backbone and expresses the prM/E region of the DENV4 serotype, which has the highest titer in the vaccine (≥ 4.5 log10 PFU/dose) [
7]. In this context, understanding dengue vaccine-associated infections informs public health strategies, emphasizing the importance of surveillance, rapid diagnosis, and genomic characterization. Such insights are crucial for developing more effective preventive measures and enhancing vaccine safety, thereby mitigating the impact of dengue.
Author Contributions
Conceptualization, T.E.R.A.; M.G., and F.C.M.I; methodology, T.E.R.A., M.L., L.M.R.T.; N.R.G., P.E.S.S.; K.M.F.M.; A.C.A.S.; and N.R.P.; formal analysis, T.E.R.A., L.M.T., V.F., and M.G.; investigation, T.E.R.A., S.H.S.P.P., M.L., L.M.R.T., N.R.G., C.S.A.S., L.C.J.A., M.G., and F.C.M.I.; resources, L.C.J.A., and M.G.; writing—original draft preparation, T.E.R.A., S.H.S.P.P., M.G., and F.C.M.I.; writing—review and editing, T.E.R.A., S.H.S.P.P., M.L., L.M.R.T., N.R.G.; V.F., P.E.S.S., C.S.A.S., L.C.J.A., M.G., and F.C.M.I. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Pan American Health Organization (PAHO/WHO), by the Brazilian Ministry of Health grant SCON2021-00180 (Coordenação Geral de Laboratório de Saúde Pública—CGLAB and Coordenação Geral de Vigilância de Arboviroses—CGARB), by the Programa Inova Fiocruz/Oswaldo Cruz Foundation, by the National Institutes of Health USA grant U01 AI151698 for the United World Arbovirus Research Network (UWARN), by the Rede Unificada de Análises Integradas de Arbovírus de Minas Gerais (REDE UAI-ARBO-MG, financed by Fundação de Amparo à Pesquisa do Estado de Minas Gerais/FAPEMIG, grant number RED-00234-23), and in part by the CRP-ICGEB RESEARCH GRANT 2020 Project CRP/BRA20-03, Contract CRP/20/03. T.E.R.A., S.H.S.P.P., and M.L. are supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) under the process numbers 153597/2024, 172892/2023-6, and 173680/2023-2, respectively. M.G. was funded by PON “Ricerca e Innovazione” 2014–2020. F.C.M.I. is supported by FAPEMIG under process number BIP-00123-23. L.M.R.T., N.R.G., and V.F. receive BDCTI-I schorlarships from FAPEMIG.
Institutional Review Board Statement
This project was reviewed and approved by the Pan American Health Organization Ethics Review Committee (reference no. PAHO-2016-08-0029) and Research Ethics Committee (Comitê de Ética em Pesquisa, CEP) of Federal University of Minas Gerais, Belo Horizonte, Brazil (CEP no. 32912820.6.1001.5149), as part of the arboviral genomic surveillance efforts, in accordance with the terms of the National Health Council Resolution no. 510/2016 and National Research Ethics Commission, Ministry of Health, Brazil (CONEP, Comissão Nacional de Ética em Pesquisa, Ministério da Saúde). This resolution authorizes the use of clinical samples collected in the Brazilian Central Public Health Laboratory without informed consent to accelerate the acquisition of knowledge and contribute to surveillance and outbreak responses. The sample processed in this study was obtained anonymously from material that exceeded the routine diagnosis of arboviruses at the Central Laboratory of Public Health of Minas Gerais (Lacen-MG), which is part of BrMoH's public network.
Informed Consent Statement
Not applicable.
Data Availability Statement
Newly generated DENV2 and DENV4 sequences have been deposited in GISAID under accession numbers EPI_ISL_19365344 and EPI_ISL_19365345.
Acknowledgments
The authors acknowlegde the important contributions of the Comitê Técnico Científico Multidisciplinar from the Universidade Federal dos Vales do Jequitinhonha e Mucuri and the Minas Gerais State Health Department (SES-MG).
Conflicts of Interest
The authors declare no competing interests.
References
- Nanaware, N.; Banerjee, A.; Mullick Bagchi, S.; Bagchi, P.; Mukherjee, A. Dengue Virus Infection: A Tale of Viral Exploitations and Host Responses. Viruses 2021, 13, 1967. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol 2002, 10, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Rico-Hesse, R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 1990, 174, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Ministério da Saúde (MS/Brasil). Atualização de Casos de Arboviroses. 2024. Available online: https://www.gov.br/saude/pt-br/assuntos/saude-de-a-a-z/a/aedes-aegypti/monitoramento-das-arboviroses (accessed on 25 August 2024).
- Adelino, T.; Lima, M.; Guimarães, N.R.; Xavier, J.; Fonseca, V.; Tomé, L.M.R.; Pereira, M.A.; Machado, V.F.; Alcantara, L.C.J.; Iani, F.C.M.; et al. Resurgence of Dengue Virus Serotype 3 in Minas Gerais, Brazil: A Case Report. Pathogens 2024, 13, 202. [Google Scholar] [CrossRef]
- Rivera, L.; Biswal, S.; Sáez-Llorens, X.; Reynales, H.; López-Medina, E.; Borja-Tabora, C.; Bravo, L.; Sirivichayakul, C.; Kosalaraksa, P.; Martinez Vargas, L.; et al. Three-year Efficacy and Safety of Takeda's Dengue Vaccine Candidate (TAK-003). Clin Infect Dis 2022, 75, 107–117. [Google Scholar] [CrossRef]
- Takeda Pharmaceuticals. QDENGA® (Dengue Tetravalent Vaccine [Live, Attenuated]). Singen, Germany: Takeda Pharmaceuticals. 2022. Available from: https://assets-dam.takeda.com/raw/upload/v1674594191/legacy-dotcom/siteassets/system/what-we-do/areas-of-focus/vaccines/pdf/ACC_Qdenga_SmPC.pdf. Approval code: DISETUJUI OLEH BPOM: 08/08/2022ID EREG10023512100029-32 (accessed on 31August 2024).
- Takeda Pharmaceuticals. Dossiê de avaliação de tecnologias em saúde preparado para a CONITEC – Qdenga (Vacina Dengue 1, 2, 3 e 4 – atenuada) indicada para a prevenção de dengue em indivíduos de 4 aos 60 anos de idade. 2023. Available online: https://www.gov.br/conitec/pt-br/midias/consultas/dossie/2023/DossietakedaVacinadengue.pdf (accessed on 25 August 2024).
- European Medicines Agency. Assessment report: Qdenga (Common name: dengue tetravalent vaccine [live, attenuated]), Procedure No. EMEA/H/C/005155/0000. 2022. Available online: https://www.ema.europa.eu/en/documents/assessment-report/qdenga-epar-public-assessment-report_en.pdf (accessed on 25 August 2024).
- Tricou, V.; Low, J.G.; Oh, H.M.; Leo, Y.S.; Kalimuddin, S.; Wijaya, L.; Pang, J.; Ling, L.M.; Lee, T.H.; Brose, M.; et al. Safety and immunogenicity of a single dose of a tetravalent dengue vaccine with two different serotype-2 potencies in adults in Singapore: A phase 2, double-blind, randomised, controlled trial. Vaccine 2020, 38, 1513–1519. [Google Scholar] [CrossRef]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 2017, 12, 1261–1276. [Google Scholar] [CrossRef]
- Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; Pauwels, R.; Alcantara, L.C.; Vanden Eynden, E.; Vandamme, A.M.; et al. Genome Detective: an automated system for virus identification from high-throughput sequencing data. Bioinformatics 2019, 35, 871–873, Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; Pauwels, R.; Alcantara, L.C.; Vanden Eynden, E.; Vandamme, A.M.; et al. Genome Detective: an automated system for virus identification from high-throughput sequencing data. Bioinformatics 2019, 35,
871–873. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, V.; Libin, P.J.K.; Theys, K.; Faria, N.R.; Nunes, M.R.T.; Restovic, M.I.; Freire, M.; Giovanetti, M.; Cuypers, L.; Nowé, A.; et al. A computational method for the identification of Dengue, Zika and Chikungunya virus species and genotypes. PLoS Negl Trop Dis 2019, 13, e0007231. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Larsson, A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Butrapet, S.; Tsuchiya, K.R.; Bhamarapravati, N.; Gubler, D.J.; Kinney, R.M. Dengue 2 PDK-53 virus as a chimeric carrier for tetravalent dengue vaccine development. J Virol 2003, 77, 11436–11447. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.S.; Winkle, P.; Faccin, A.; Nordio, F.; LeFevre, I.; Tsoukas, C.G. An open-label, Phase 3 trial of TAK-003, a live attenuated dengue tetravalent vaccine, in healthy US adults: immunogenicity and safety when administered during the second half of a 24-month shelf-life. Hum Vaccin Immunother 2023, 19, 22549–22564. [Google Scholar] [CrossRef]
- Gowri Sankar, S.; Mowna Sundari, T.; Alwin Prem Anand, A. Emergence of Dengue 4 as Dominant Serotype During 2017 Outbreak in South India and Associated Cytokine Expression Profile. Front Cell Infect Microbiol 2021, 11, 681937. [Google Scholar] [CrossRef]
- Goncalvez, A.P.; Escalante, A.A.; Pujol, F.H.; Ludert, J.E.; Tovar, D.; Salas, R.A.; Liprandi, F. Diversity and evolution of the envelope gene of dengue virus type 1. Virology 2002, 303, 110–9, Goncalvez AP, Escalante AA, Pujol FH, Ludert JE, Tovar D, Salas RA, Liprandi F. 2002. Diversity and evolution of the envelope gene of dengue virus type 1. Virology 303(1): 110–119. [Google Scholar] [CrossRef] [PubMed]
- Klungthong, C.; Putnak, R.; Mammen, M.P.; Li, T.; Zhang, C. Molecular genotyping of dengue viruses by phylogenetic analysis of the sequences of individual genes. J Virol Methods 2008, 154, 175–181. [Google Scholar] [CrossRef]
- Adelino, T.E.R.; Giovanetti, M.; Fonseca, V.; Xavier, J.; de Abreu, A.S.; do Nascimento, V.A.; Demarchi, L.H.F.; Oliveira, M.A.A.; da Silva, V.L.; de Mello, A.L.E.S.; et al. Field and classroom initiatives for portable sequence-based monitoring of dengue virus in Brazil. Nat Commun. 2021, 12, 2296. [Google Scholar] [CrossRef]
- Nogueira, R.M.; Miagostovich, M.P.; Lampe, E.; Schatzmayr, H.G. Isolation of dengue virus type 2 in Rio de Janeiro. Mem Inst Oswaldo Cruz 1990, 85, 253. [Google Scholar] [CrossRef]
- Giovanetti, M.; Pereira, L.A.; Santiago, G.A.; Fonseca, V.; Mendoza, M.P.G.; de Oliveira, C.; de Moraes, L.; Xavier, J.; Tosta, S.; Fristch, H.; et al. Emergence of Dengue Virus Serotype 2 Cosmopolitan Genotype, Brazil. Emerg Infect Dis 2022, 28, 1725–1727. [Google Scholar] [CrossRef]
- Harapan, H.; Michie, A.; Sasmono, R.T.; Imrie, A. Dengue: A Minireview. Viruses 2020, 12, 829. [Google Scholar] [CrossRef] [PubMed]
- Balfour, H.H.Jr.; Groth, K.E.; Edelman, C.K.; Amren, D.P.; Best, J.M.; Banatvala, J.E. Rubella viraemia and antibody responses after rubella vaccination and reimmunization. Lancet 1981, 1, 1078–1080. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P. Yellow fever vaccine. Expert Rev Vaccines 2005, 4, 553–574. [Google Scholar] [CrossRef] [PubMed]
- Biswal, S.; Borja-Tabora, C.; Martinez Vargas, L.; Velásquez, H.; Theresa Alera, M.; Sierra, V.; Johana Rodriguez-Arenales, E.; Yu, D.; Wickramasinghe, V.P.; Duarte Moreira, E.Jr.; et al. Efficacy of a tetravalent dengue vaccine in healthy children aged 4-16 years: a randomised, placebo-controlled, phase 3 trial. Lancet 2020, 395, 1423–1433. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).