Submitted:
15 September 2024
Posted:
16 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
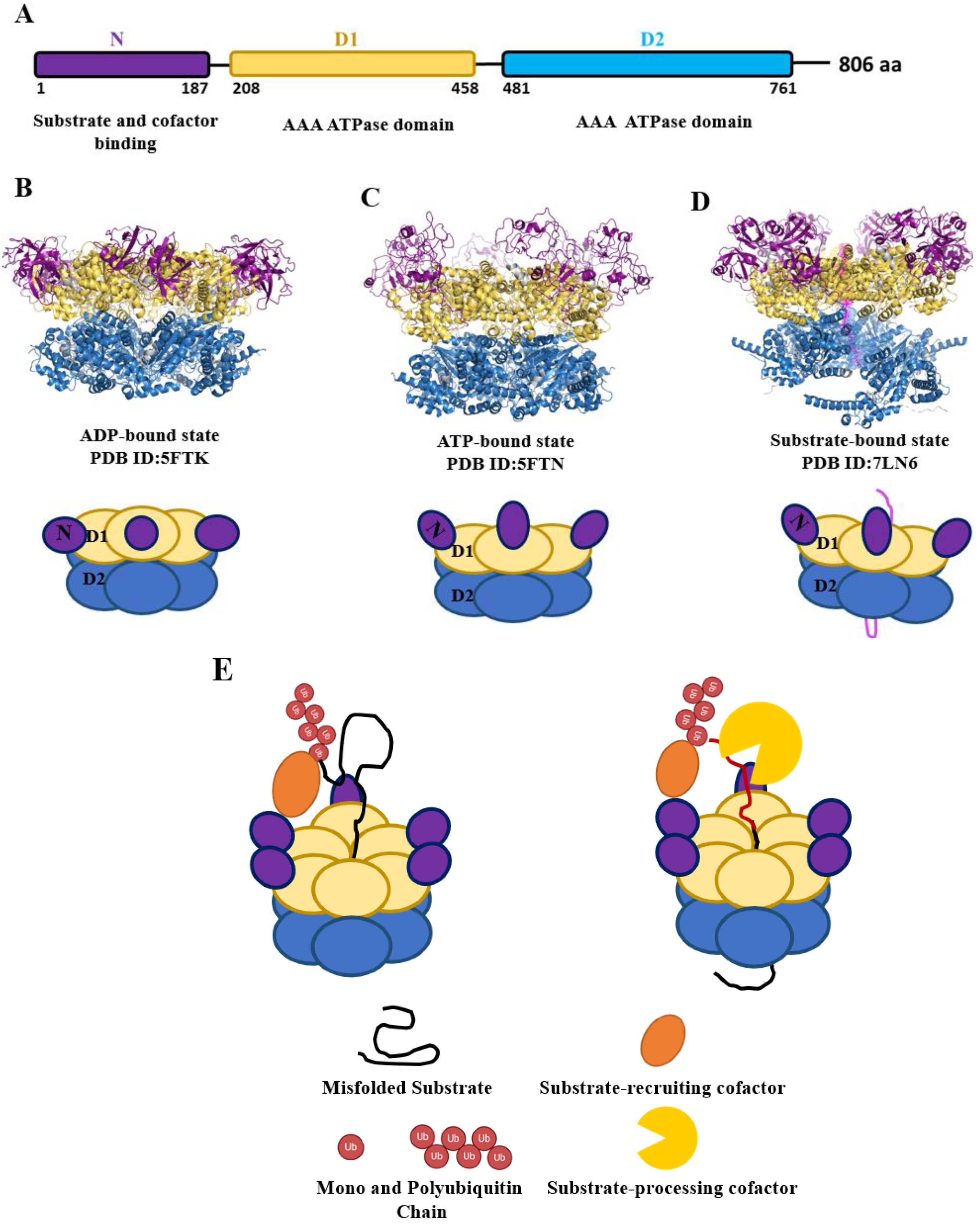
2. p97 Oligomerization
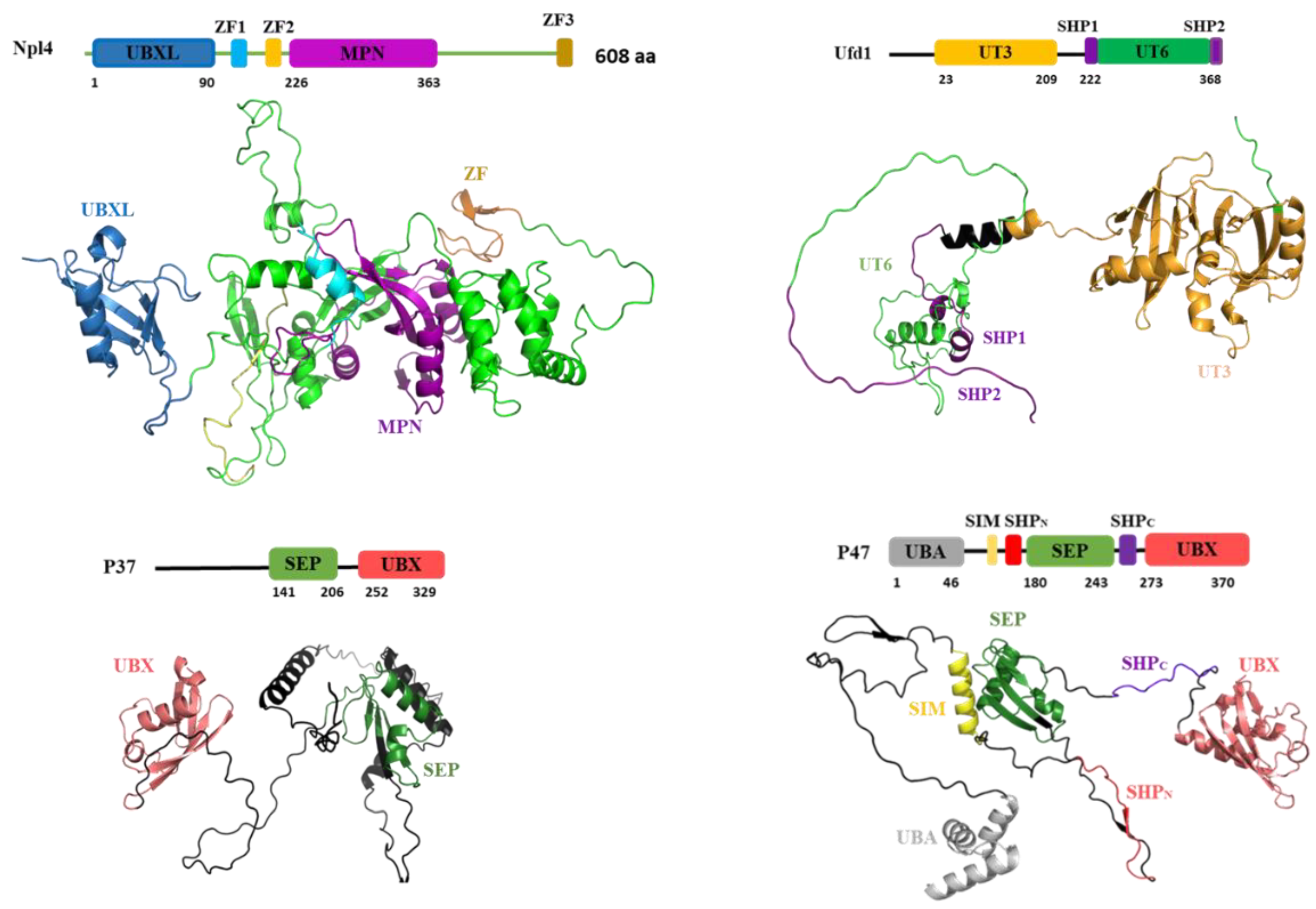
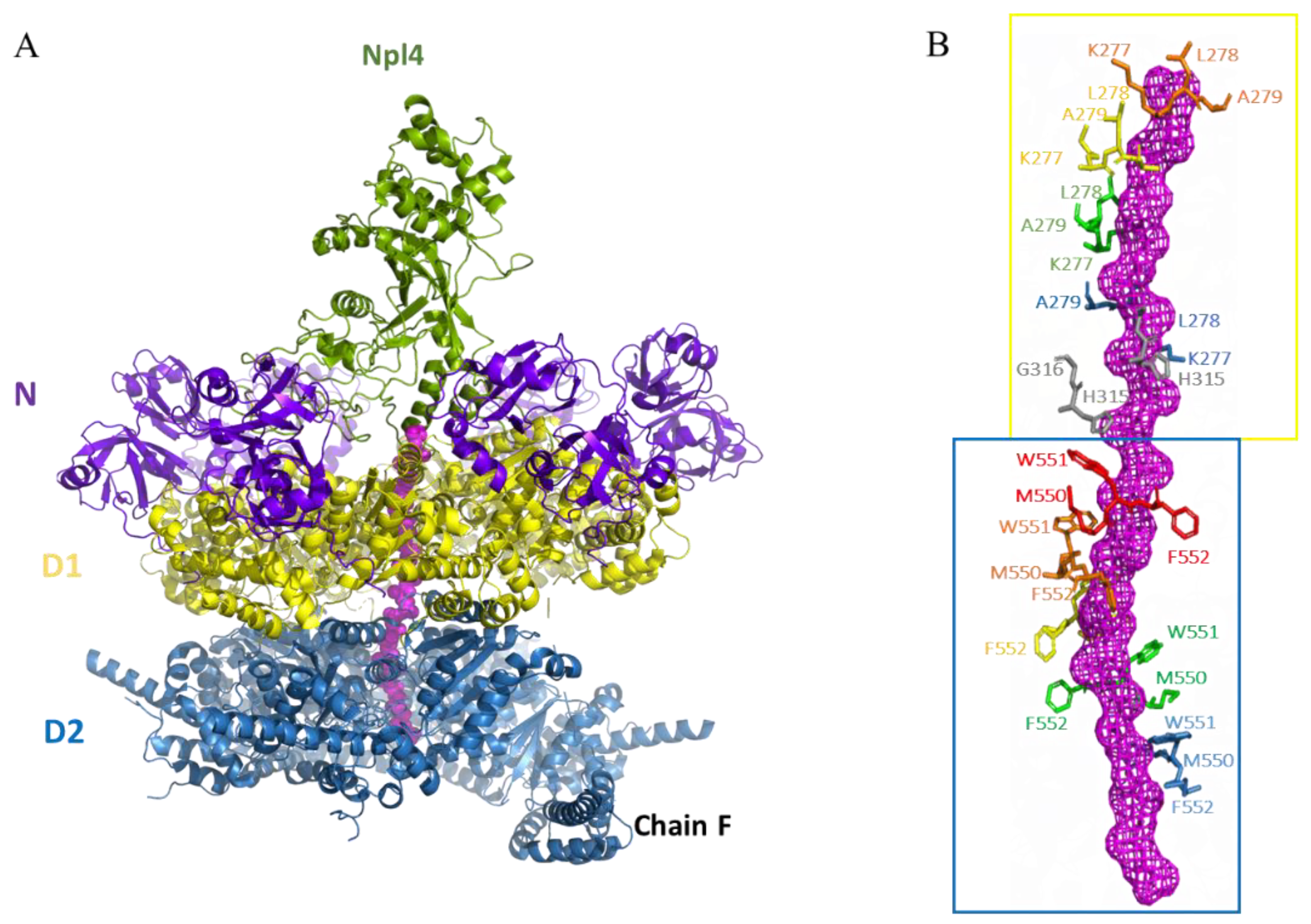
3. Interaction between p97 and Ufd1/Npl4(UN)
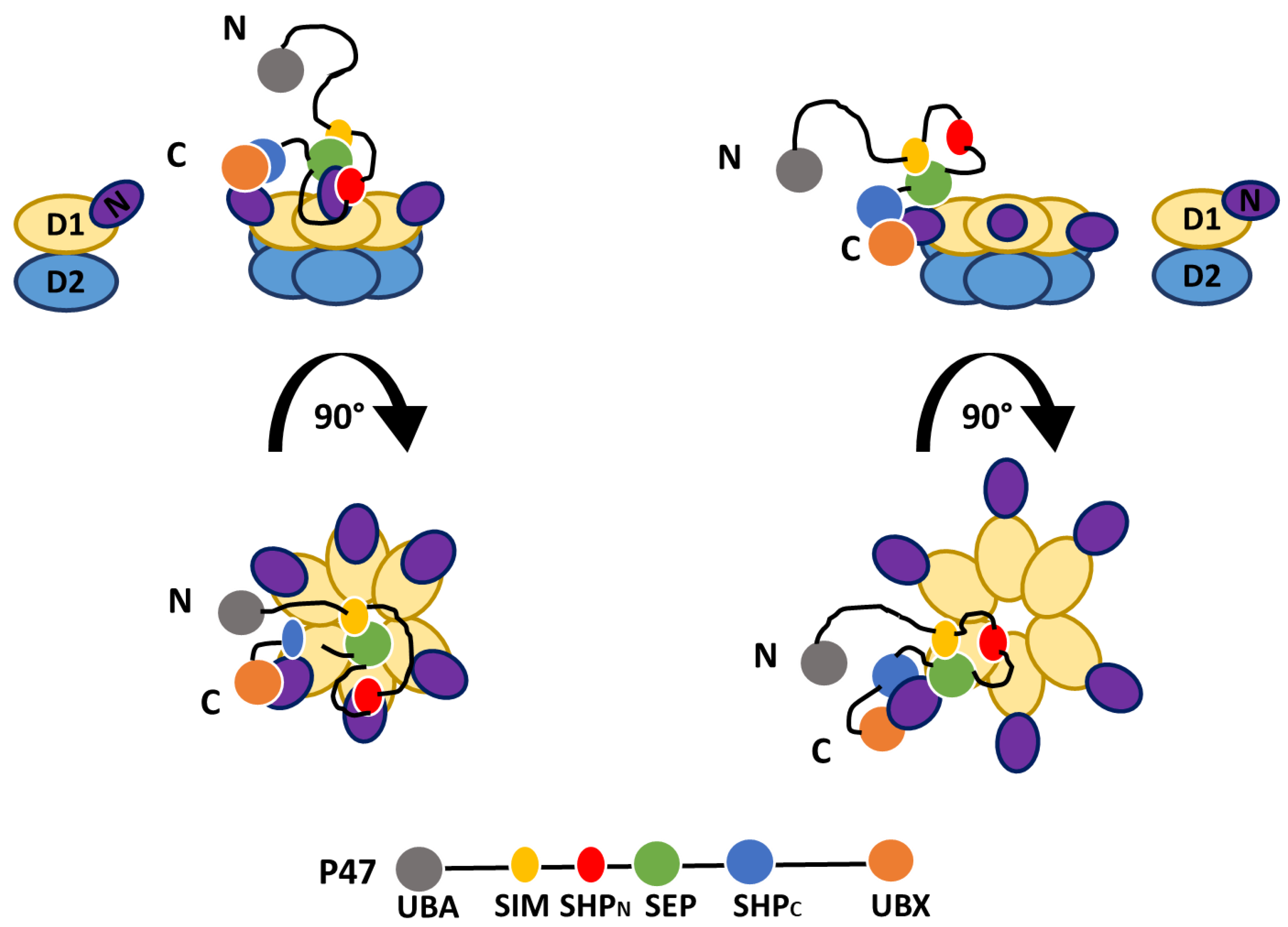
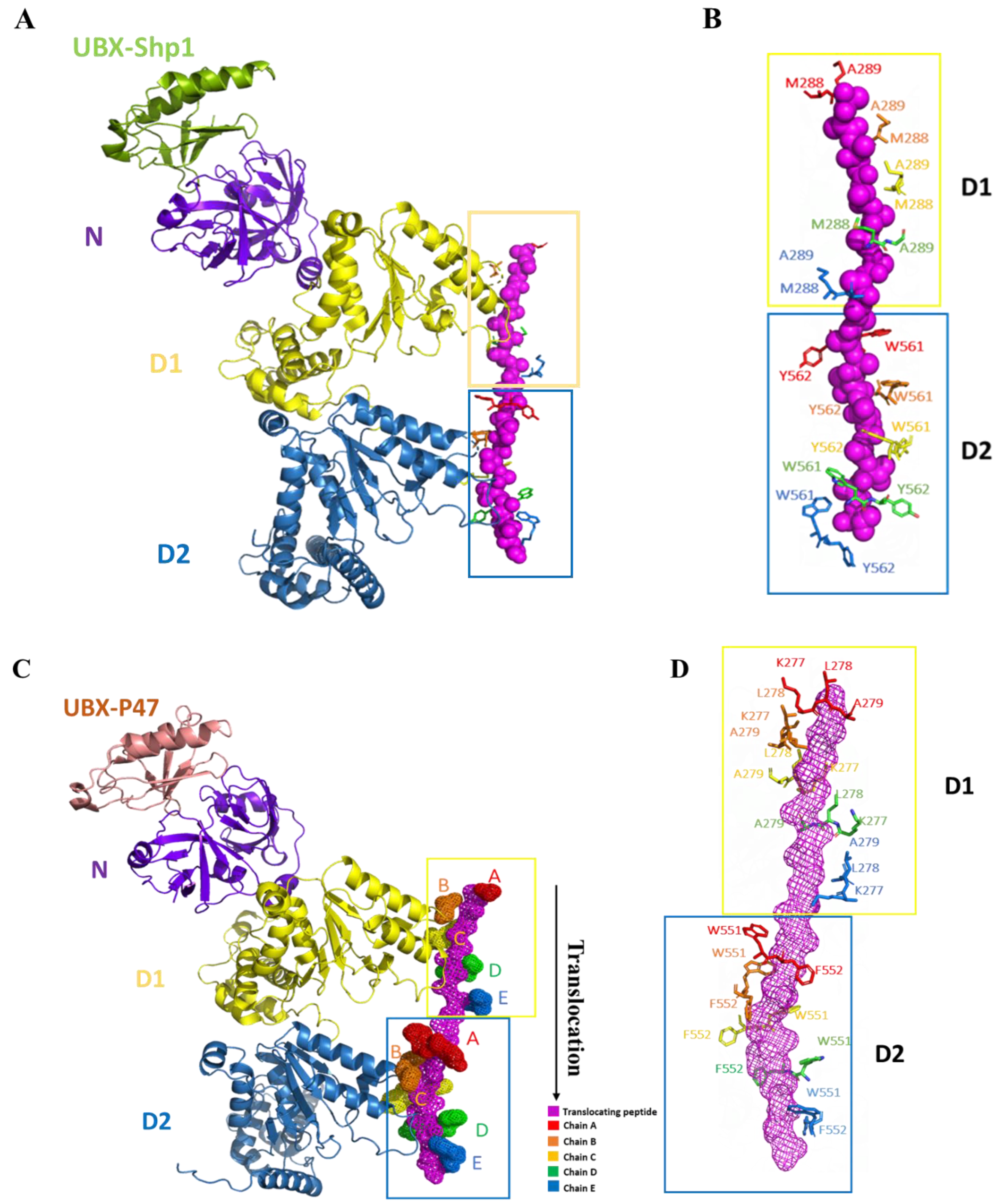
4. Interaction between p97 and p47
5. Interaction between p97 and p37
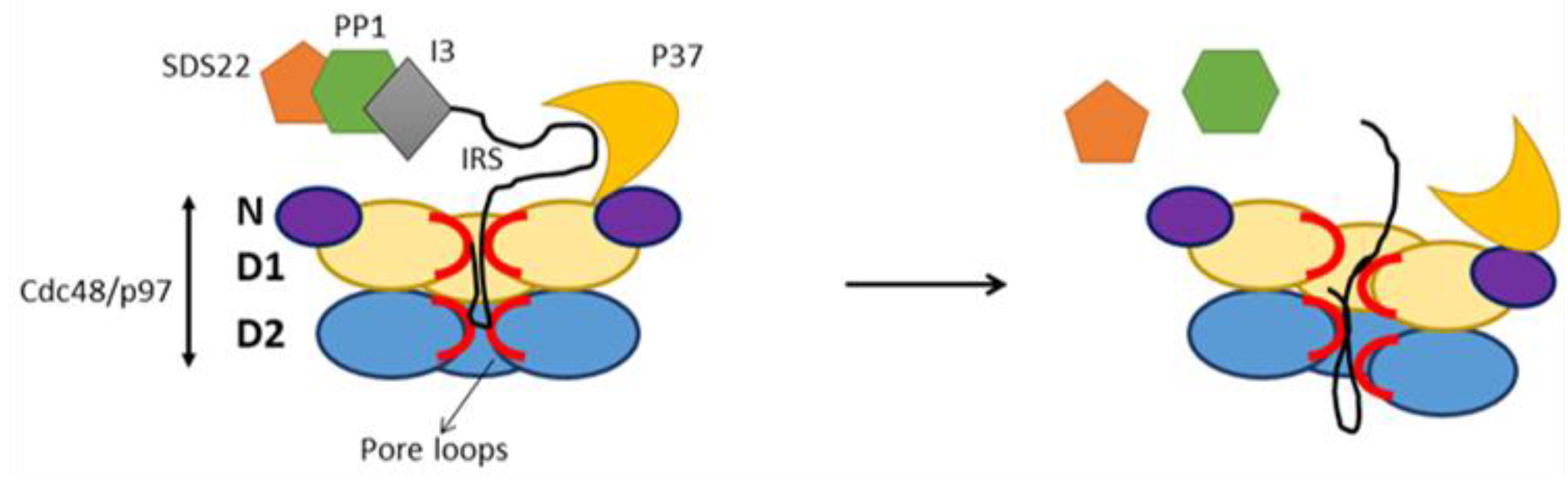
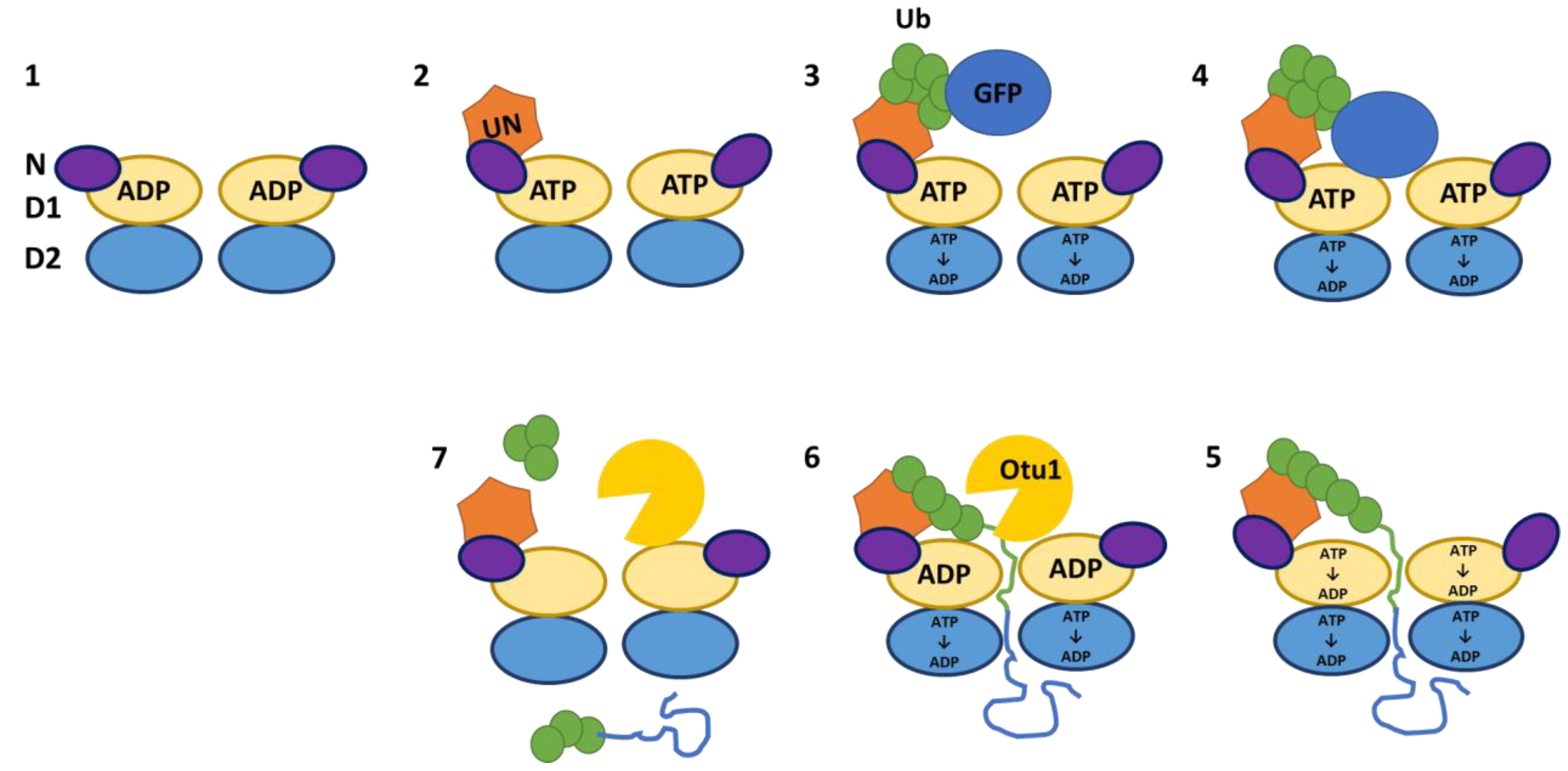
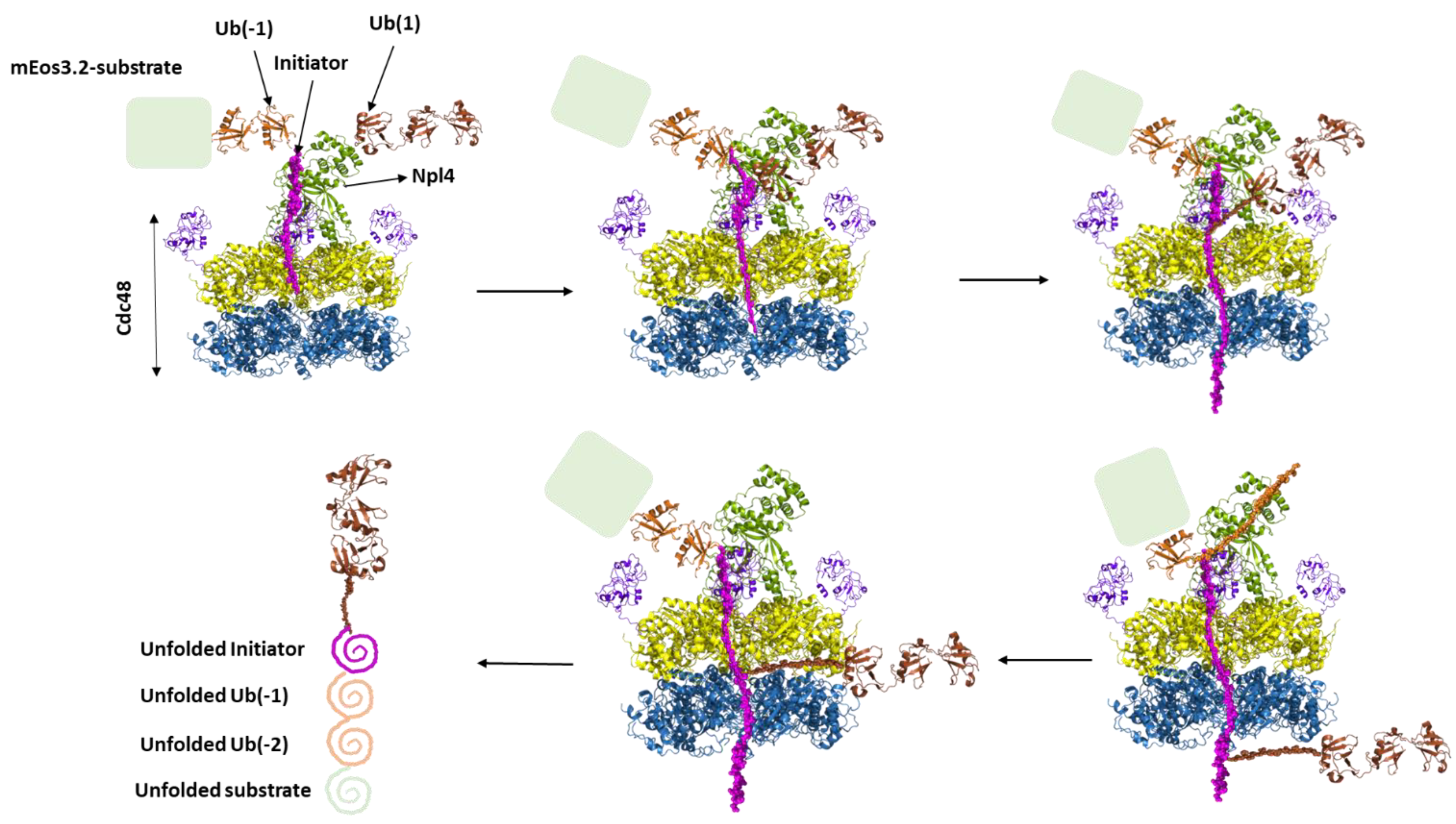
6. Substrate processing by p97 in complex with cofactor
7. Conclusion and Future Directions
Funding
Acknowledgments
References
- Chen, B., Retzlaff, M., Roos, T. & Frydman, J. (2011) Cellular strategies of protein quality control, Cold Spring Harbor perspectives in biology. 3, a004374. [CrossRef]
- Chou, T.-F., Brown, S. J., Minond, D., Nordin, B. E., Li, K., Jones, A. C., Chase, P., Porubsky, P. R., Stoltz, B. M. & Schoenen, F. J. (2011) Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways, Proceedings of the National Academy of Sciences. 108, 4834-4839. [CrossRef]
- van den Boom, J. & Meyer, H. (2018) VCP/p97-mediated unfolding as a principle in protein homeostasis and signaling, Molecular cell. 69, 182-194. [CrossRef]
- Olszewski, M. M., Williams, C., Dong, K. C. & Martin, A. (2019) The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome, Communications biology. 2, 1-8. [CrossRef]
- Rouiller, I., DeLaBarre, B., May, A. P., Weis, W. I., Brunger, A. T., Milligan, R. A. & Wilson-Kubalek, E. M. (2002) Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle, Nature structural biology. 9, 950-957. [CrossRef]
- Brunger, A. T. & DeLaBarre, B. (2003) NSF and p97/VCP: similar at first, different at last, FEBS letters. 555, 126-133.
- Huyton, T., Pye, V. E., Briggs, L. C., Flynn, T. C., Beuron, F., Kondo, H., Ma, J., Zhang, X. & Freemont, P. S. (2003) The crystal structure of murine p97/VCP at 3.6 Å, Journal of structural biology. 144, 337-348.
- Wendler, P., Ciniawsky, S., Kock, M. & Kube, S. (2012) Structure and function of the AAA+ nucleotide binding pocket, Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 1823, 2-14. [CrossRef]
- Hänzelmann, P. & Schindelin, H. (2016) Structural basis of ATP hydrolysis and intersubunit signaling in the AAA+ ATPase p97, Structure. 24, 127-139. [CrossRef]
- Buchberger, A., Schindelin, H. & Hänzelmann, P. (2015) Control of p97 function by cofactor binding, FEBS letters. 589, 2578-2589. [CrossRef]
- DeLaBarre, B. & Brunger, A. T. (2005) Nucleotide dependent motion and mechanism of action of p97/VCP, Journal of molecular biology. 347, 437-452.
- Abaan, O. D., Hendriks, W., Üren, A., Toretsky, J. A. & Erkizan, H. V. (2013) Valosin containing protein (VCP/p97) is a novel substrate for the protein tyrosine phosphatase PTPL1, Experimental cell research. 319, 1-11.
- Bodnar, N. O. & Rapoport, T. A. (2017) Molecular mechanism of substrate processing by the Cdc48 ATPase complex, Cell. 169, 722-735. e9. [CrossRef]
- Bodnar, N. O., Kim, K. H., Ji, Z., Wales, T. E., Svetlov, V., Nudler, E., Engen, J. R., Walz, T. & Rapoport, T. A. (2018) Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1–Npl4, Nature structural & molecular biology. 25, 616-622.
- Cooney, I., Han, H., Stewart, M. G., Carson, R. H., Hansen, D. T., Iwasa, J. H., Price, J. C., Hill, C. P. & Shen, P. S. (2019) Structure of the Cdc48 segregase in the act of unfolding an authentic substrate, Science. 365, 502-505. [CrossRef]
- Twomey, E. C., Ji, Z., Wales, T. E., Bodnar, N. O., Ficarro, S. B., Marto, J. A., Engen, J. R. & Rapoport, T. A. (2019) Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding, Science. 365. [CrossRef]
- Banerjee, S., Bartesaghi, A., Merk, A., Rao, P., Bulfer, S. L., Yan, Y., Green, N., Mroczkowski, B., Neitz, R. J. & Wipf, P. (2016) 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition, Science. 351, 871-875. [CrossRef]
- Schuller, J. M., Beck, F., Lössl, P., Heck, A. J. & Förster, F. (2016) Nucleotide-dependent conformational changes of the AAA+ ATPase p97 revisited, FEBS letters. 590, 595-604.
- Tang, W. K., Li, D., Li, C. c., Esser, L., Dai, R., Guo, L. & Xia, D. (2010) A novel ATP-dependent conformation in p97 N–D1 fragment revealed by crystal structures of disease-related mutants, The EMBO journal. 29, 2217-2229.
- Briggs, L. C., Baldwin, G. S., Miyata, N., Kondo, H., Zhang, X. & Freemont, P. S. (2008) Analysis of nucleotide binding to P97 reveals the properties of a tandem AAA hexameric ATPase, Journal of Biological Chemistry. 283, 13745-13752. [CrossRef]
- Chou, T.-F., Bulfer, S. L., Weihl, C. C., Li, K., Lis, L. G., Walters, M. A., Schoenen, F. J., Lin, H. J., Deshaies, R. J. & Arkin, M. R. (2014) Specific inhibition of p97/VCP ATPase and kinetic analysis demonstrate interaction between D1 and D2 ATPase domains, J Mol Biol. 426, 2886-2899.
- Song, C., Wang, Q. & Li, C.-C. H. (2003) ATPase activity of p97-valosin-containing protein (VCP): D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity, Journal of Biological Chemistry. 278, 3648-3655.
- Ye, Y., Tang, W. K., Zhang, T. & Xia, D. (2017) A mighty “protein extractor” of the cell: structure and function of the p97/CDC48 ATPase, Frontiers in molecular biosciences. 4, 39.
- Erzberger, J. P. & Berger, J. M. (2006) Evolutionary relationships and structural mechanisms of AAA+ proteins, Annu Rev Biophys Biomol Struct. 35, 93-114.
- Bruderer, R. M., Brasseur, C. & Meyer, H. H. (2004) The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism, Journal of Biological Chemistry. 279, 49609-49616.
- Ernst, R., Mueller, B., Ploegh, H. L. & Schlieker, C. (2009) The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER, Molecular cell. 36, 28-38. [CrossRef]
- Uchiyama, K., Jokitalo, E., Kano, F., Murata, M., Zhang, X., Canas, B., Newman, R., Rabouille, C., Pappin, D. & Freemont, P. (2002) VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo, The Journal of cell biology. 159, 855-866.
- Hänzelmann, P. & Schindelin, H. (2017) The interplay of cofactor interactions and post-translational modifications in the regulation of the AAA+ ATPase p97, Frontiers in molecular biosciences. 4, 21. [CrossRef]
- Hänzelmann, P. & Schindelin, H. (2016) Characterization of an additional binding surface on the p97 N-terminal domain involved in bipartite cofactor interactions, Structure. 24, 140-147. [CrossRef]
- Alexandru, G., Graumann, J., Smith, G. T., Kolawa, N. J., Fang, R. & Deshaies, R. J. (2008) UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1α turnover, Cell. 134, 804-816. [CrossRef]
- Schuberth, C. & Buchberger, A. (2008) UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97, Cellular and molecular life sciences. 65, 2360-2371.
- Kloppsteck, P., Ewens, C. A., Förster, A., Zhang, X. & Freemont, P. S. (2012) Regulation of p97 in the ubiquitin–proteasome system by the UBX protein-family, Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 1823, 125-129. [CrossRef]
- Liu, C., Liu, W., Ye, Y. & Li, W. (2017) Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains, Nature communications. 8, 1-15.
- Parker, C. E., Mocanu, V., Warren, M. R., Greer, S. F. & Borchers, C. H. (2005) Mass spectrometric determination of protein ubiquitination in Ubiquitin-Proteasome Protocols pp. 153-173, Springer.
- Kirkpatrick, D. S., Hathaway, N. A., Hanna, J., Elsasser, S., Rush, J., Finley, D., King, R. W. & Gygi, S. P. (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology, Nature cell biology. 8, 700-710. [CrossRef]
- Ye, Y., Meyer, H. H. & Rapoport, T. A. (2003) Function of the p97–Ufd1–Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains, The Journal of cell biology. 162, 71-84.
- Blythe, E. E., Olson, K. C., Chau, V. & Deshaies, R. J. (2017) Ubiquitin-and ATP-dependent unfoldase activity of P97/VCP• NPLOC4• UFD1L is enhanced by a mutation that causes multisystem proteinopathy, Proceedings of the National Academy of Sciences. 114, E4380-E4388.
- Bodnar, N. & Rapoport, T. (2017) Toward an understanding of the Cdc48/p97 ATPase, F1000Research. 6.
- Lee, C., Prakash, S. & Matouschek, A. (2002) Concurrent translocation of multiple polypeptide chains through the proteasomal degradation channel, Journal of Biological Chemistry. 277, 34760-34765. [CrossRef]
- Burton, R. E., Siddiqui, S. M., Kim, Y.-I., Baker, T. A. & Sauer, R. T. (2001) Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine, The EMBO journal. 20, 3092-3100. [CrossRef]
- Han, H. & Hill, C. P. (2019) Structure and mechanism of the ESCRT pathway AAA+ ATPase Vps4, Biochemical Society Transactions. 47, 37-45. [CrossRef]
- Ripstein, Z. A., Huang, R., Augustyniak, R., Kay, L. E. & Rubinstein, J. L. (2017) Structure of a AAA+ unfoldase in the process of unfolding substrate, Elife. 6, e25754.
- Puchades, C., Rampello, A. J., Shin, M., Giuliano, C. J., Wiseman, R. L., Glynn, S. E. & Lander, G. C. (2017) Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing, Science. 358, eaao0464. [CrossRef]
- Zhao, M., Wu, S., Zhou, Q., Vivona, S., Cipriano, D. J., Cheng, Y. & Brunger, A. T. (2015) Mechanistic insights into the recycling machine of the SNARE complex, Nature. 518, 61-67. [CrossRef]
- Tang, W. K. & Xia, D. (2013) Altered intersubunit communication is the molecular basis for functional defects of pathogenic p97 mutants, Journal of Biological Chemistry. 288, 36624-36635. [CrossRef]
- Yu, G., Bai, Y., Li, K., Amarasinghe, O., Jiang, W. & Zhang, Z.-Y. (2021) Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation, Iscience. 24, 103310. [CrossRef]
- Hoq, M. R., Vago, F. S., Li, K., Kovaliov, M., Nicholas, R. J., Huryn, D. M., Wipf, P., Jiang, W. & Thompson, D. H. (2021) Affinity Capture of p97 with Small-Molecule Ligand Bait Reveals a 3.6 Å Double-Hexamer Cryoelectron Microscopy Structure, ACS nano. 15, 8376-8385.
- Mori-Konya, C., Kato, N., Maeda, R., Yasuda, K., Higashimae, N., Noguchi, M., Koike, M., Kimura, Y., Ohizumi, H. & Hori, S. (2009) p97/valosin-containing protein (VCP) is highly modulated by phosphorylation and acetylation, Genes to Cells. 14, 483-497.
- Tang, W. K., Odzorig, T., Jin, W. & Xia, D. (2019) Structural basis of p97 inhibition by the site-selective anticancer compound CB-5083, Molecular pharmacology. 95, 286-293. [CrossRef]
- Nandi, P., Li, S., Columbres, R. C. A., Wang, F., Williams, D. R., Poh, Y.-P., Chou, T.-F. & Chiu, P.-L. (2021) Structural and functional analysis of disease-linked p97 ATPase mutant complexes, International journal of molecular sciences. 22, 8079. [CrossRef]
- Gao, H., Li, F., Ji, Z., Shi, Z., Li, Y. & Yu, H. (2022) Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state, Cell Discovery. 8, 1-13.
- Liu, S., Ye, X., Liu, W., Liu, L., Li, D., Lin, Q. & Wang, T. (2022) Cryo-EM structure of dodecamer human p97 in complex with NMS-873 reveals S765-G779 peptide plays critical role for D2 ring oligomerization, Biochemical and Biophysical Research Communications.
- Sato, Y., Tsuchiya, H., Yamagata, A., Okatsu, K., Tanaka, K., Saeki, Y. & Fukai, S. (2019) Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4, Nature communications. 10, 1-13.
- Heo, J.-M., Livnat-Levanon, N., Taylor, E. B., Jones, K. T., Dephoure, N., Ring, J., Xie, J., Brodsky, J. L., Madeo, F. & Gygi, S. P. (2010) A stress-responsive system for mitochondrial protein degradation, Molecular cell. 40, 465-480. [CrossRef]
- Wu, X., Li, L. & Jiang, H. (2016) Doa1 targets ubiquitinated substrates for mitochondria-associated degradation, Journal of Cell Biology. 213, 49-63.
- Moreno, S. P., Bailey, R., Campion, N., Herron, S. & Gambus, A. (2014) Polyubiquitylation drives replisome disassembly at the termination of DNA replication, Science. 346, 477-481. [CrossRef]
- Maric, M., Mukherjee, P., Tatham, M. H., Hay, R. & Labib, K. (2017) Ufd1-Npl4 recruit Cdc48 for disassembly of ubiquitylated CMG helicase at the end of chromosome replication, Cell reports. 18, 3033-3042. [CrossRef]
- Maric, M., Maculins, T., De Piccoli, G. & Labib, K. (2014) Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication, Science. 346. [CrossRef]
- Ye, Y., Tang, W. K., Zhang, T. & Xia, D. (2017) A Mighty “Protein Extractor” of the Cell: Structure and Function of the p97/CDC48 ATPase, Frontiers in Molecular Biosciences. 4.
- Meyer, H. H., Wang, Y. & Warren, G. (2002) Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1–Npl4, The EMBO Journal. 21, 5645-5652.
- Meyer, H.-J. & Rape, M. (2014) Enhanced protein degradation by branched ubiquitin chains, Cell. 157, 910-921.
- Park, S., Isaacson, R., Kim, H. T., Silver, P. A. & Wagner, G. (2005) Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites, Structure. 13, 995-1005. [CrossRef]
- Rumpf, S. & Jentsch, S. (2006) Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone, Molecular cell. 21, 261-269. [CrossRef]
- Hetzer, M., Meyer, H. H., Walther, T. C., Bilbao-Cortes, D., Warren, G. & Mattaj, I. W. (2001) Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly, Nature cell biology. 3, 1086-1091. [CrossRef]
- Isaacson, R. L., Pye, V. E., Simpson, P., Meyer, H. H., Zhang, X., Freemont, P. S. & Matthews, S. (2007) Detailed structural insights into the p97-Npl4-Ufd1 interface, Journal of Biological Chemistry. 282, 21361-21369. [CrossRef]
- Blythe, E. E., Gates, S. N., Deshaies, R. J. & Martin, A. (2019) Multisystem proteinopathy mutations in VCP/p97 increase NPLOC4· UFD1L binding and substrate processing, Structure. 27, 1820-1829. e4. [CrossRef]
- Kimura, Y., Fukushi, J., Hori, S., Matsuda, N., Okatsu, K., Kakiyama, Y., Kawawaki, J., Kakizuka, A. & Tanaka, K. (2013) Different dynamic movements of wild-type and pathogenic VCP s and their cofactors to damaged mitochondria in a P arkin-mediated mitochondrial quality control system, Genes to Cells. 18, 1131-1143.
- Pan, M., Zheng, Q., Yu, Y., Ai, H., Xie, Y., Zeng, X., Wang, C., Liu, L. & Zhao, M. (2020) Seesaw Conformations of Npl4 in the Human p97 Complex and the Inhibitory Mechanism of a Disulfiram Derivative, bioRxiv.
- Schuberth, C. & Buchberger, A. (2005) Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation, Nature cell biology. 7, 999-1006. [CrossRef]
- Hänzelmann, P., Buchberger, A. & Schindelin, H. (2011) Hierarchical binding of cofactors to the AAA ATPase p97, Structure. 19, 833-843. [CrossRef]
- Lee, J.-J., Park, J. K., Jeong, J., Jeon, H., Yoon, J.-B., Kim, E. E. & Lee, K.-J. (2013) Complex of Fas-associated factor 1 (FAF1) with valosin-containing protein (VCP)-Npl4-Ufd1 and polyubiquitinated proteins promotes endoplasmic reticulum-associated degradation (ERAD), Journal of Biological Chemistry. 288, 6998-7011. [CrossRef]
- Kim, S. J., Cho, J., Song, E. J., Kim, S. J., Kim, H. M., Lee, K. E., Suh, S. W. & Kim, E. E. (2014) Structural basis for ovarian tumor domain-containing protein 1 (OTU1) binding to p97/valosin-containing protein (VCP), Journal of Biological Chemistry. 289, 12264-12274.
- Pye, V. E., Beuron, F., Keetch, C. A., McKeown, C., Robinson, C. V., Meyer, H. H., Zhang, X. & Freemont, P. S. (2007) Structural insights into the p97-Ufd1-Npl4 complex, Proceedings of the National Academy of Sciences. 104, 467-472.
- Ramadan, K. (2012) p97/VCP-and Lys48-linked polyubiquitination form a new signaling pathway in DNA damage response, Cell cycle. 11, 1062-1069.
- Majera, D., Skrott, Z., Chroma, K., Merchut-Maya, J. M., Mistrik, M. & Bartek, J. (2020) Targeting the NPL4 adaptor of p97/VCP segregase by disulfiram as an emerging cancer vulnerability evokes replication stress and DNA damage while silencing the ATR pathway, Cells. 9, 469.
- Pan, M., Zheng, Q., Yu, Y., Ai, H., Xie, Y., Zeng, X., Wang, C., Liu, L. & Zhao, M. (2021) Seesaw conformations of Npl4 in the human p97 complex and the inhibitory mechanism of a disulfiram derivative, Nature communications. 12, 1-12.
- Liu, S., Yang, H., Zhao, J., Zhang, Y.-H., Song, A.-X. & Hu, H.-Y. (2013) NEDD8 ultimate buster-1 long (NUB1L) protein promotes transfer of NEDD8 to proteasome for degradation through the P97UFD1/NPL4 complex, Journal of Biological Chemistry. 288, 31339-31349.
- Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Žídek, A. & Potapenko, A. (2021) Highly accurate protein structure prediction with AlphaFold, Nature. 596, 583-589.
- Kondo, H., Rabouille, C., Newman, R., Levine, T. P., Pappin, D., Freemont, P. & Warren, G. (1997) p47 is a cofactor for p97-mediated membrane fusion, Nature. 388, 75-78.
- Zhang, X., Gui, L., Zhang, X., Bulfer, S. L., Sanghez, V., Wong, D. E., Lee, Y., Lehmann, L., Lee, J. S. & Shih, P.-Y. (2015) Altered cofactor regulation with disease-associated p97/VCP mutations, Proceedings of the National Academy of Sciences. 112, E1705-E1714.
- Bulfer, S. L., Chou, T.-F. & Arkin, M. R. (2016) P97 disease mutations modulate nucleotide-induced conformation to alter protein–protein interactions, ACS chemical biology. 11, 2112-2116.
- Conicella, A. E., Huang, R., Ripstein, Z. A., Nguyen, A., Wang, E., Löhr, T., Schuck, P., Vendruscolo, M., Rubinstein, J. L. & Kay, L. E. (2020) An intrinsically disordered motif regulates the interaction between the p47 adaptor and the p97 AAA+ ATPase, Proceedings of the National Academy of Sciences. 117, 26226-26236.
- Beuron, F., Dreveny, I., Yuan, X., Pye, V. E., Mckeown, C., Briggs, L. C., Cliff, M. J., Kaneko, Y., Wallis, R. & Isaacson, R. L. (2006) Conformational changes in the AAA ATPase p97–p47 adaptor complex, The EMBO journal. 25, 1967-1976.
- Uchiyama, K., Totsukawa, G., Puhka, M., Kaneko, Y., Jokitalo, E., Dreveny, I., Beuron, F., Zhang, X., Freemont, P. & Kondo, H. (2006) p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis, Developmental cell. 11, 803-816.
- Mirzadeh, A., Kobakhidze, G., Vuillemot, R., Jonic, S. & Rouiller, I. (2022) In silico prediction, Characterization, Docking studies and Molecular dynamics simulation of human p97 in complex with p37 cofactor.
- Dreveny, I., Kondo, H., Uchiyama, K., Shaw, A., Zhang, X. & Freemont, P. S. (2004) Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47, The EMBO journal. 23, 1030-1039.
- Soukenik, M., Diehl, A., Leidert, M., Sievert, V., Büssow, K., Leitner, D., Labudde, D., Ball, L. J., Lechner, A. & Nägler, D. K. (2004) The SEP domain of p47 acts as a reversible competitive inhibitor of cathepsin L, FEBS letters. 576, 358-362.
- Weith, M., Seiler, J., van den Boom, J., Kracht, M., Hülsmann, J., Primorac, I., del Pino Garcia, J., Kaschani, F., Kaiser, M. & Musacchio, A. (2018) Ubiquitin-independent disassembly by a p97 AAA-ATPase complex drives PP1 holoenzyme formation, Molecular cell. 72, 766-777. e6.
- van den Boom, J., Kueck, A. F., Kravic, B., Müschenborn, H., Giesing, M., Pan, D., Kaschani, F., Kaiser, M., Musacchio, A. & Meyer, H. (2021) Targeted substrate loop insertion by VCP/p97 during PP1 complex disassembly, Nature Structural & Molecular Biology. 28, 964-971.
- Buchberger, A. (2022) Unfolding by Cdc48/p97: different strokes for different folks, Trends in Cell Biology.
- Van den Boom, J., Marini, G., Meyer, H. & Saibil, H. R. (2023) Structural basis of ubiquitin-independent PP1 complex disassembly by p97, The EMBO Journal. 42, e113110.
- Twomey, E. C., Ji, Z., Wales, T. E., Bodnar, N. O., Ficarro, S. B., Marto, J. A., Engen, J. R. & Rapoport, T. A. (2019) Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding, Science. 365, eaax1033.
- Alam, S. L., Sun, J., Payne, M., Welch, B. D., Blake, B. K., Davis, D. R., Meyer, H. H., Emr, S. D. & Sundquist, W. I. (2004) Ubiquitin interactions of NZF zinc fingers, The EMBO journal. 23, 1411-1421.
- Pan, M., Yu, Y., Ai, H., Zheng, Q., Xie, Y., Liu, L. & Zhao, M. (2021) Mechanistic insight into substrate processing and allosteric inhibition of human p97, bioRxiv.
- Ji, Z., Li, H., Peterle, D., Paulo, J. A., Ficarro, S. B., Wales, T. E., Marto, J. A., Gygi, S. P., Engen, J. R. & Rapoport, T. A. (2021) Translocation of polyubiquitinated protein substrates by the hexameric Cdc48 ATPase, Molecular Cell.
- Irbäck, A., Mitternacht, S. & Mohanty, S. (2005) Dissecting the mechanical unfolding of ubiquitin, Proceedings of the National Academy of Sciences. 102, 13427-13432.
- Han, H., Fulcher, J. M., Dandey, V. P., Iwasa, J. H., Sundquist, W. I., Kay, M. S., Shen, P. S. & Hill, C. P. (2019) Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases, Elife. 8, e44071.
- Avellaneda, M. J., Franke, K. B., Sunderlikova, V., Bukau, B., Mogk, A. & Tans, S. J. (2020) Processive extrusion of polypeptide loops by a Hsp100 disaggregase, Nature. 578, 317-320.
- Esaki, M., Islam, M. T., Tani, N. & Ogura, T. (2017) Deviation of the typical AAA substrate-threading pore prevents fatal protein degradation in yeast Cdc48, Scientific reports. 7, 1-11.
- Monroe, N., Han, H., Shen, P. S., Sundquist, W. I. & Hill, C. P. (2017) Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase, Elife. 6, e24487.
- Puchades, C., Rampello, A. J., Shin, M., Giuliano, C. J., Wiseman, R. L., Glynn, S. E. & Lander, G. C. (2017) Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing, Science. 358.
- Andres, H., Goodall, E. A., Gates, S. N., Lander, G. C. & Martin, A. (2018) Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis–driven translocation, Science. 362.
- Huang, R., Ripstein, Z. A., Rubinstein, J. L. & Kay, L. E. (2019) Cooperative subunit dynamics modulate p97 function, Proceedings of the National Academy of Sciences. 116, 158-167.
- Han, H., Monroe, N., Sundquist, W. I., Shen, P. S. & Hill, C. P. (2017) The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets, Elife. 6, e31324.
- de la Peña, A. H., Goodall, E. A., Gates, S. N., Lander, G. C. & Martin, A. (2018) Substrate-engaged 26 S proteasome structures reveal mechanisms for ATP-hydrolysis–driven translocation, Science. 362, eaav0725.
- Dong, Y., Zhang, S., Wu, Z., Li, X., Wang, W. L., Zhu, Y., Stoilova-McPhie, S., Lu, Y., Finley, D. & Mao, Y. (2019) Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome, Nature. 565, 49-55.
- Gates, S. N., Yokom, A. L., Lin, J., Jackrel, M. E., Rizo, A. N., Kendsersky, N. M., Buell, C. E., Sweeny, E. A., Mack, K. L. & Chuang, E. (2017) Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104, Science. 357, 273-279.
- Deville, C., Carroni, M., Franke, K. B., Topf, M., Bukau, B., Mogk, A. & Saibil, H. R. (2017) Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase, Science advances. 3, e1701726.
- Yu, H., Lupoli, T. J., Kovach, A., Meng, X., Zhao, G., Nathan, C. F. & Li, H. (2018) ATP hydrolysis-coupled peptide translocation mechanism of Mycobacterium tuberculosis ClpB, Proceedings of the National Academy of Sciences. 115, E9560-E9569.
- White, K. I., Zhao, M., Choi, U. B., Pfuetzner, R. A. & Brunger, A. T. (2018) Structural principles of SNARE complex recognition by the AAA+ protein NSF, Elife. 7, e38888.
- Xu, Y., Han, H., Cooney, I., Guo, Y., Moran, N. G., Zuniga, N. R., Price, J. C., Hill, C. P. & Shen, P. S. (2022) Active conformation of the p97-p47 unfoldase complex, Nature communications. 13, 1-8.
- Arie, M., Matzov, D., Karmona, R., Szenkier, N., Stanhill, A. & Navon, A. (2024) A non-symmetrical p97 conformation initiates a multistep recruitment of Ufd1/Npl4, iScience. 27.
- Oppenheim, T., Radzinski, M., Braitbard, M., Brielle, E. S., Yogev, O., Goldberger, E., Yesharim, Y., Ravid, T., Schneidman-Duhovny, D. & Reichmann, D. (2023) The Cdc48 N-terminal domain has a molecular switch that mediates the Npl4-Ufd1-Cdc48 complex formation, Structure. 31, 764-779. e8. [CrossRef]
| Reference | N-Domain in up conformation | N-Domain in coplanar conformation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| D1 Domain Occupancy | D2 Domain Occupancy | Cryo-EM Map Resolution (A°) | EMDB ID | PDB ID | D1 Domain Occupancy | D2 Domain Occupancy | Cryo-EM Map Resolution (A°) | EMDB ID | PDB ID | |
| Gao et al. (2022) | ATP | ATP | 3.3 | EMD-31894 | 7VCS | ATP | ADP | 3.1 | EMD-31896 | 7VCU |
| G. Yu et al. (2021) | Empty | Empty | 3.9 | EMD-22675 | 7K56 | - | - | - | - | - |
| G. Yu et al. (2021) | Empty | Empty | 3.7 | EMD-22675 (at the presence of apyrase) | 7K57 (at the presence of apyrase) | - | - | - | - | - |
| P. Nandi (2021) | Empty | Empty | 3.3 | EMD-23191 | 7L5W | - | - | - | - | - |
| R. Hoq et al. (2021) | Empty | Empty | 3.6 | NA | NA | - | - | - | - | - |
| S. Liu, X. Ye, W. Liu et al. (2022) | - | - | - | - | - | ADP | NMS-873 | 3 | EMD-32827 | 7WUB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




