Submitted:
13 September 2024
Posted:
16 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Design
2.2. Study Population
2.3. Definitions
2.4. Study Variables
2.5. Analysis Plan and Statistical Analysis
3. Results
3.1. Whole Population
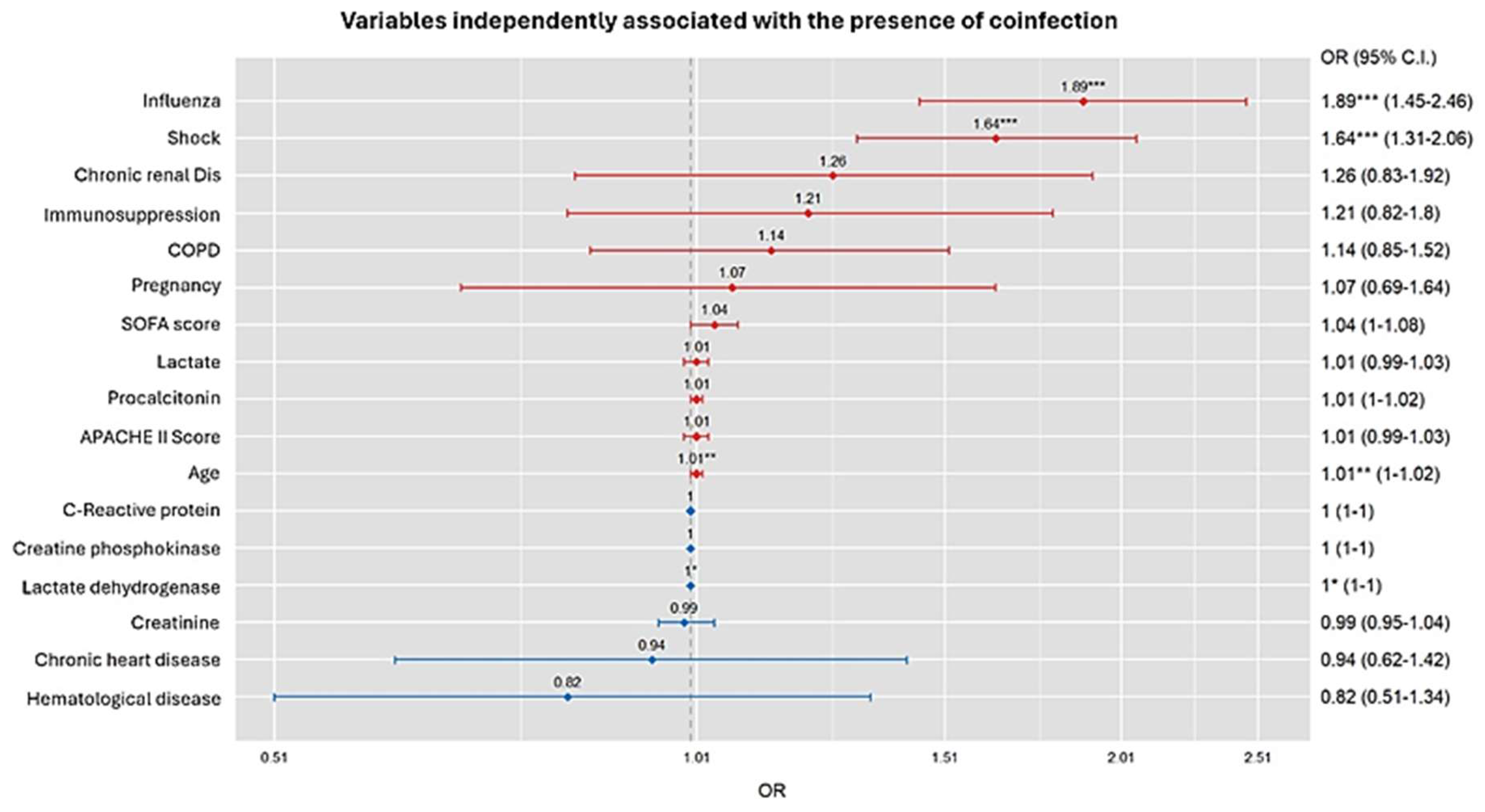
3.2. Factors Associated with COI in the Whole Population According to General Linear Model (GLM)
3.3. GLM Model Validation
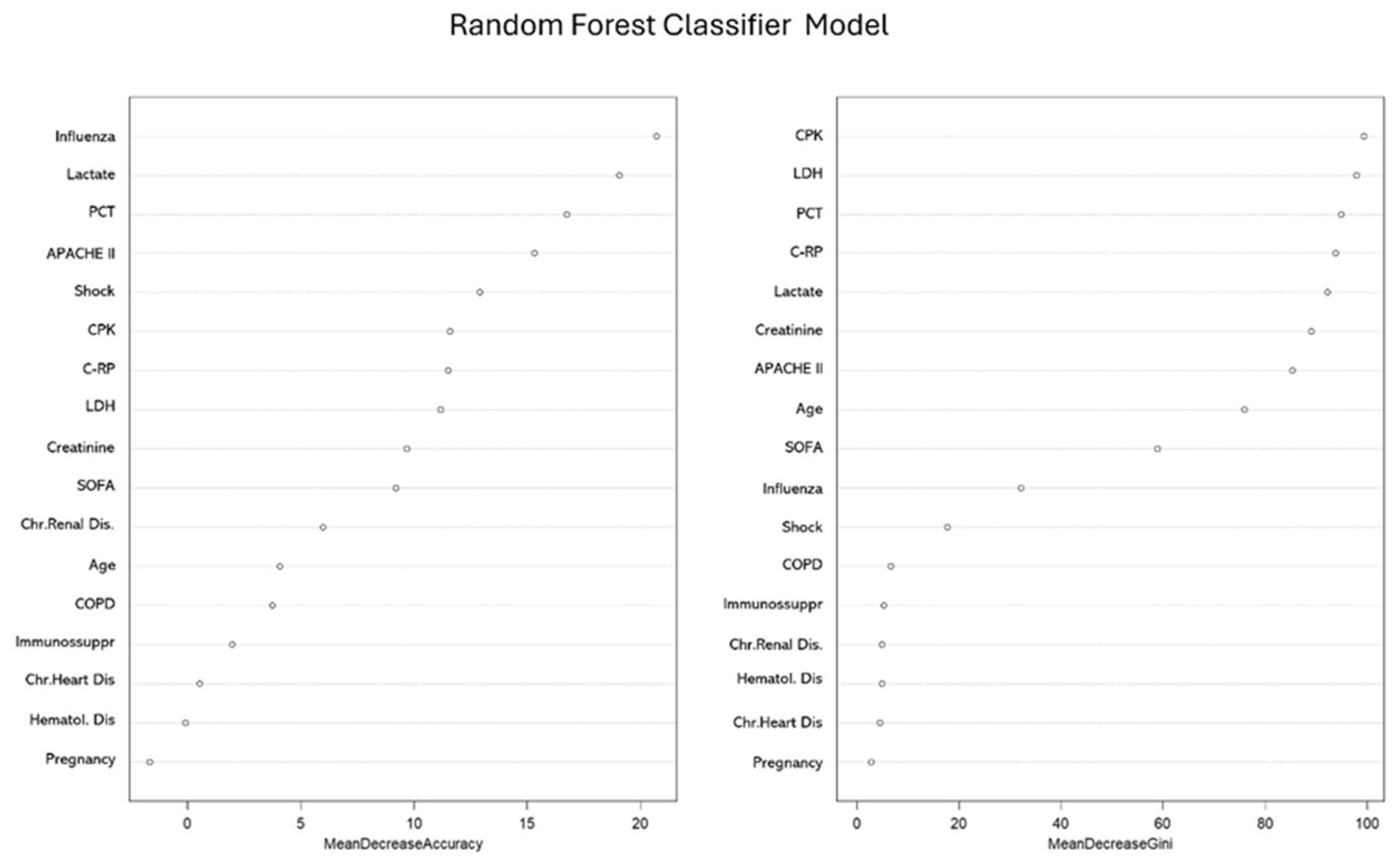
3.4. Factors Associated with COI in the Whole Population According to No-Linear Model (Random Forest)
3.5. Factors Associated with COI in the Influenza Cohort According to general linear model (GLM)
3.6. Factors Associated with COI in the Influenza Cohort According to No-Linear Model (Random Forest)
3.7. Factors Associated with COI in the COVID-19 Cohort According to General Linear Model (GLM)
3.8. Factors Associated with COI in the COVID-19 Cohort According to No-Linear Model (Random Forest)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simonsen,L.; Spreeuwenberg,P.; Lustig,R.; Taylor,R.J.; Fleming, D.M.; Kroneman, M.; Van Kerkhove, M.D. et al. Global Mortality Estimates for the 2009 Influenza Pandemic from the GLaMOR Project: A Modeling Study. PLoS Med. 2013; 10(11): e1001558. [CrossRef]
- Viboud,C.; Simonsen,L. Global mortality of 2009 pandemic influenza A H1N1.Lancet Infect Dis 2012; 9:651-653, . [CrossRef]
- Mathieu, E.; Ritchie, H.; Rodés-Guirao, L.; Appel, C.; Giattino,C.; Hasell,J.; et al. “Coronavirus Pandemic (COVID-19)”. 2020 Published online at OurWorldInData.org. Retrieved from: ‘https://ourworldindata.org/coronavirus’ [Online Resource.
- World Health Organization 2023 data.who.int, WHO Coronavirus (COVID-19) dashboard > Deaths [Dashboard]. https://data.who.int/dashboards/covid19/deaths.
- Martin-Loeches, I.; J Schultz, M.J.; Vincent, J.L.; Alvarez-Lerma, F.; Bos, L.D.; Solé-Violán, J.; Torres, A.; Rodriguez, A. Increased incidence of co-infection in critically ill patients with influenza. Intensive Care Med 2017; 43:48–58. [CrossRef]
- Rodríguez, A.H.; Avilés-Jurado, F.X.; Díaz, E.; Schuetz, P.; Trefler, S.I.; Solé-Violán, J. et al. Procalcitonin (PCT) levels for ruling-out bacterial coinfection in ICU patients with influenza: A CHAID decision-tree analysis. J Infect. 2016 ;72(2):143-51. [CrossRef]
- Carbonell, R.; Urgelés, S.; Rodríguez, A.; Bodí, M.; Martín-Loeches, I.; Solé-Violán, J.; et al. Mortality comparison between the first and second/third waves among 3,795 critical COVID-19 patients with pneumonia admitted to the ICU: A multicentre retrospective cohort study. Lancet Reg Health Eur. 2021 ;11:100243. [CrossRef]
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Westwood, D.; MacFadden D.R.; et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clinical Microbiology and Infection 2020;26: 1622e1629. [CrossRef]
- Rodríguez, A.; Moreno, G.; Bodi, M.; Martín-Loeches, I. Antibiotics in development for multiresistant gram-negative bacilli. Med Intensiva (Engl Ed). 2022;46(11):630-640. [CrossRef]
- Kariyawasam, R.M.; Julien, D.A.; Jelinski, D.C.; Larose, S.L.; Rennert-May, E.; Conly, J.M.; et al. Antimicrobial resistance (AMR) in COVID-19 patients: a systematic review and meta-analysis (November 2019–June 2021). Antimicrobial Resistance & Infection Control 2022; 11:45 . [CrossRef]
- Rawson, T.M.; Moore, L.S.P.; Zhu, N.; Ranganathan, N.; Skolimowska, K.; Gilchrist, M. et al. Bacterial and Fungal Coinfection in Individuals With Coronavirus: A Rapid Review To Support COVID-19 Antimicrobial Prescribing. Clin Infect Dis 2020;71(9):2459-2468. [CrossRef]
- Varshney, K.; Pillay, P.; Mustafa, A.D.; Shen, D.; · Jenna Renee Adalbert, J.R.; Mahmood, M.Q. A systematic review of the clinical characteristics of influenza-COVID-19 co-infection. Clinical and Experimental Medicine 2023; 23:3265–3275. [CrossRef]
- Delhommeau, G.; Buetti, N.; Neuville, M.; Siami, S.; Cohen, Y.; Laurent, V. et al. Bacterial Pulmonary Co-Infections on ICU Admission: Comparison in Patients with SARS-CoV-2 and Influenza Acute Respiratory Failure: A Multicentre Cohort Study. Biomedicines 2022, 10,2646. [CrossRef]
- Patton, M.J.; Orihuela, C.J.; Harrod, K.S.; Bhuiyan, M.A.N.; Paari Dominic, P.; Kevil, C.K. et al. COVID-19 bacteremic co-infection is a major risk factor for mortality, ICU admission, and mechanical ventilation. Critical Care 2023; 27:34 . [CrossRef]
- Carbonell, R.; Moreno, G.; Martín-Loeches, I.; Bodí, M.; Rodríguez, A. The Role of Biomarkers in Influenza and COVID-19 Community-Acquired Pneumonia in Adults. Antibiotics (Basel) 2023 ;12 (1):161. [CrossRef]
- Carbonell, R.; Moreno, G.; Martín-Loeches, I.; Gomez-Bertomeu, F.; Sarvisé, C.;, Gómez, J. et al. Prognostic Value of Procalcitonin and C-Reactive Protein in 1608 Critically Ill Patients with Severe Influenza Pneumonia. Antibiotics (Basel). 2021 ;10(4):350. [CrossRef]
- Carbonell, R.; Urgelés, S.; Salgado, M.; Rodríguez, A.; Reyes, L.F.; Fuentes, Y.V. et al. Negative predictive value of procalcitonin to rule out bacterial respiratory co-infection in critical covid-19 patients. J Infect. 2022 ;85(4):374-381. [CrossRef]
- Hu, F.H.; Jia, Y.J.; Zhao, D.Y.; Fu, X.L.; Zhang, W.Q. et al. Clinical outcomes of the severe acute respiratory syndrome coronavirus 2 Omicron and Delta variant: systematic review and meta-analysis of 33 studies covering 6 037 144 coronavirus disease 2019-positive patients. Clinical Microbiology and Infection 2023;29: 835e844. [CrossRef]
- Murakami, Y.; Nozaki, Y.; Morosawa, M.; Toyama, M.; Ogashiwa, H.; Ueda, T. et al. Difference in the impact of coinfections and secondary infections on antibiotic use in patients hospitalized with COVID-19 between the Omicron-dominant period and the pre-Omicron period. Journal of Infection and Chemotherapy 2024;30: 853–859. [CrossRef]
- Graham, M.S.; Sudre, C.H.; May, A.; Antonelli, M.; Murray, B.; Varsavsky, T. et al.Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health 2021; 6: e335–45. [CrossRef]
- Martin-Loeches, I.; Lemiale, V.; Geoghegan, P.; McMahon, M.A.; Pickkers, P; Soares, M. et al. Influenza and associated co-infections in critically ill immunosuppressed patients. Critical Care 2019; 23:152. [CrossRef]
- Corey, L.; Beyrer, C.; Cohen, M.S.; Michael, N.L.; Bedford, T.; Rolland, M. SARS-CoV-2 Variants in Patients with Immunosuppression. N Engl J Med 2021;385(6):562-566. [CrossRef]
- Evans, R.A.; Dube, S.; Lu, Y.; Yates, M.; Arnetorp, S.; Barnes, E. et al. Impact of COVID-19 on immunocompromised populations during the Omicron era: insights from the observational population-based INFORM study. The Lancet Regional Health – Europe 2023;35: 100747. [CrossRef]
- Oliva, I.; Ferré, C.; Daniel, X.; Cartanyà, M.; Villavicencio, C.; Salgado, M. et al. Risk factors and outcome of acute kidney injury in critically ill patients with SARS-CoV-2 pneumonia: a multicenter study. Med Intensiva (Engl Ed). 2024: S2173-5727(24)00176-0. [CrossRef]
- Rodríguez, A.; Ruiz-Botella, M.; Martín-Loeches, I.; Jiménez-Herrera, M.; Solé-Violán, J.; Gómez, J. et al. Deploying unsupervised clustering analysis to derive clinical phenotypes and risk factors associated with mortality risk in 2022 critically ill patients with COVID-19 in Spain. Crit Care. 2021 Feb 15;25(1):63. [CrossRef]
- Uyeki TM, Bernstein HH, Bradley JS, Englund JA Jr, Fry TM,Fry AM, et al. Clinical practice guidelines by the Infectious Diseases Society of America: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal influenza. Clin Infect Dis. 2019;68:e1-47. [CrossRef]
- WHO. Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected: interim guidance, 13 March 2020. https://iris.who.int/bitstream/handle/10665/331446/WHO-2019-nCoV-clinical-2020.4-eng.pdf?sequence=1&isAllowed=y.
- Verweij, PE.; Rijnders, B.J.A.; Brüggemann, R.J.M.; Azoulay, E.; Bassetti, M.; Blot, S.; et al. Review of influenza-associated PA in ICU patients and proposal for a case definition: an expert opinion. Intensive Care Med. 2020; 46:1524-35. [CrossRef]
- Claverias, L.; Daniel, X.; Martín-Loeches, I.; Vidal-Cortez, P.; Gómez-Bertomeu, F.; Trefler, S. et al. Impact of Aspergillus spp. isolation in the first 24 hours of admission in critically ill patients with severe influenza virus pneumonia. Med Intensiva 2022; 46: 426-435. [CrossRef]
- Lunardon, N.; Menardi, G.; Torelli, N. ROSE: A Package for Binary Imbalanced Learning. The R Journal 214. https://journal.r-project.org/archive/2014-1/menardi-lunardon-torelli.pdf.
- Menardi, G.;Torelli, N. Training and assessing classification rules with imbalanced data. Data Mining and Knowledge Discovery, 28(1): 92–122, 2014). [CrossRef]
- Ramsey, J.B. Tests for Specification Errors in Classical Linear Least-Squares Regression Analysis. Journal of the Royal Statistical Society. Series B (Methodological) 1969;21(2):350-371. https://www.jstor.org/stable/2984219).
- Lai, C.C.; Chen, S.Y.; Ko, W.C; Hsueh, P.R. Increased antimicrobial resistance during the COVID-19 pandemic. International Journal of Antimicrobial Agents 2021(57): 106324. [CrossRef]
- Su, L.; Yu, T.; Zhang, C.; Huo, P.; Zhao, Z. A prediction model for secondary invasive fungal infection among severe SARS-CoV-2 positive patients in ICU. Front. Cell. Infect. Microbiol. 2024 ,14. [CrossRef]
- van Grootveld, R.; van der Beek, M.T.; Janssen, N.A.F.; Ergün, M.; van Dijk, K.; Carina Bethlehem, C. et al. Incidence, risk factors and pre-emptive screening for COVID-19 associated pulmonary aspergillosis in an era of immunomodulant therapy. J Crit Care. 2023; 76:154272. [CrossRef]
- Wang,M.; Li, W.; Wang, H.; Song, P. Development and validation of machine learning-based models for predicting healthcare-associated bacterial/fungal infections among COVID-19 inpatients: a retrospective cohort study. Antimicrobial Resistance & Infection Control 2024; 13:42 . [CrossRef]
- López-Herrero, R.; L Sánchez-de Prada, L.; Tamayo-Velasco, A.; Lorenzo-López, M.; E Gómez-Pesquera, E.; Sánchez-Quirós, B. et al. Epidemiology of bacterial co-infections and risk factors in COVID-19-hospitalized patients in Spain: a nationwide study. European Journal of Public Health, Volume 33, Issue 4, August 2023, Pages 675–681, . [CrossRef]
- Raymond Bak Hei Chu, Shi Zhao, Jack Zhenhe Zhang, King Chung Kenny Chan, Pauline Yeung Ng, Carol Chan, Ka Man Fong, Shek Yin Au, Alwin Wai Tak Yeung et al. Comparison of COVID-19 with influenza A in the ICU: a territory-wide, retrospective, propensity matched cohort on mortality and length of stay. BMJ Open 2023; 13: e067101. [CrossRef]
- Chu, R.B.H.; Zhao, S.; Zhang, J.Z.; Chan, K.C.K.; Ng, P.Y.; Carol Chan, C. et al. Comparison of COVID-19 with influenza A in the ICU: a territory-wide, retrospective, propensity matched cohort on mortality and length of stay. BMJ Open 2023; 13: e067101. [CrossRef]
- Giannella, M.; Rinaldi, M.; Tesini, G.; Gallo, M.; Cipriani, V.; Vatamanu, O. et al. Predictive model for bacterial co-infection in patients hospitalized for COVID-19: a multicenter observational cohort study. Infection (2022) 50:1243–1253 . [CrossRef]
- Masiá, M.; Padilla, S.; Antequera, P.; Ramos, J.M.; Ruiz, M.; Gutiérrez, F. Predictors of Pneumococcal Co-infection for Patients with Pandemic (H1N1) 2009. Emerg Infect Dis. 2011;17(8):1475-1478. [CrossRef]
| Variables# | Whole population (n=8902) | Influenza cohort (n=3702) | COVID-19 cohort(n=5200) | ||||||
| Total | No-COI (n=7639) | COI (n= 1263) | Total | No-COI(n=2897) | COI(n=805) | Total | No-COI(n=4742) | COI(n=458) | |
| General characteristics | |||||||||
| Age, years | 60(49-70) | 60 (49-69) | 62(51-72)*** | 55 (43-67) | 54(42-66) | 59(47-72)*** | 63(54-71) | 63(54-71) | 65(57-72)*** |
| Male sex | 5855 (66.1) | 5012 (65-6) | 843(66.7) | 2203 (59.5) | 1698(58.6) | 505(62.7)* | 3652(70.2) | 3314(69.9) | 338(73.8) |
| APACHE II score | 14 (10-19) | 14 (10-18) | 17(12-22)*** | 15 (11-21) | 15(11-20) | 19(14-24)*** | 13(10-17) | 13(10-17) | 14(11-18)*** |
| SOFA score | 5 (3-7) | 5 (3-7) | 6(4-9) | 5 (4-8) | 5(4-8) | 7(4-10)*** | 4(3-7) | 4(3-7) | 5(4-8)*** |
| Gap-ICU, days | 1 (1-3) | 1 (1-3) | 1(0-2)*** | 3 (1-6) | 1.0(1.0-2.0) | 1.0(1.0-2.0) | 2(0-4) | 2(0-4) | 1(0-3)* |
| COVID | 5200 | 4742(61.1) | 458(36.3)*** | ----- | ----- | ----- | ----- | ----- | ----- |
| Influenza | 3702 | 2897(37.9) | 805(63.7)*** | ----- | ----- | ----- | ----- | ----- | ----- |
| Laboratory | |||||||||
| WBC x103 | 8.6 (5.7-12.5) | 8.6 (5.8-12.3) | 8.9 (5.8-13.7) | 8.1 (4.6-12.4) | 8.0(4.7-12.0) | 8.8(3.6-14.3) | 8.9(6.4-12.5) | 8.9(6.4-12.4) | 9.0(6.1-13.1) |
| LDH U/L | 542 (403-687) | 537 (401-686) | 566(415-697)* | 599 (457-750) | 600(456-754) | 594(449-737) | 501(380-632) | 500(379-632) | 514(390-631) |
| C-RP mg/mL | 19.6(9.8-34.7) | 18.3 (9.3-32.8) | 27.0(14.3-64.2)*** | 36 (19-84) | 34(17-20) | 42(23-100)*** | 14(7-22) | 14(7.2-22.5) | 14(8.6-24.6)*** |
| PCT ng/mL | 0.8(0.20-5.67) | 0.73 (0.19-4.14) | 3.4(0.56-19.3)*** | 7.2 (1.9-22.0) | 6.2(1.7-20.7) | 12.2(3.2-26.4)*** | 0.26(0.11-0.72) | 0.25(0.11-0.70) | 0.30(0.14-0.8)*** |
| Creatinine mg/dL | 0.89(0.70-1.23) | 0.87(0.69-1.17) | 1.0(0.75-1.57)*** | 1.0 (0.70-1.46) | 0.9(0.7-1.4) | 1.2(0.8-1.9)*** | 0.8(0.7-1.0) | 0.8(0.6-1.0) | 0.8(0.7-1.1)*** |
| CPK | 216 (100-420) | 208(98-410) | 272(119-490)*** | 331 (145-585) | 327(146-578) | 344(138-629) | 170(83-321) | 169(83-320) | 193(95-335)* |
| Lactate mmol/L | 2.0 (1.4-3.3) | 1.9(1.3-3.1) | 2.8(1.8-4.2)*** | 3.2 (2.8-4.7) | 3.2(2.3-4.7) | 3.6(2.7-5.0)*** | 1.5(1.1-2.1) | 1.5(1.1-2.0) | 1.6(1.2-2.2)*** |
| Comobidities | |||||||||
| COPD | 1281(14.3) | 1022(13.4) | 259(20.5)*** | 908(24.5) | 696(24.0) | 212(26.3) | 373(7.2) | 326(6.8) | 47(10.3)* |
| Asthma | 698 (7.8) | 595(7.8) | 103(8.1) | 367(10.0) | 296(10.2) | 71(8.8) | 331(6.4) | 299(6.3) | 32(6.9) |
| Chr. Heart Dis | 623 (7.0) | 501(6.6) | 122(9.6)*** | 447(12.0) | 347(12.0) | 100(12.4) | 176(3.4) | 154(3.2) | 22(4.8) |
| Chr.Renal Dis. | 595 (6.7) | 486(6.4) | 109(8.6)*** | 314(8.5) | 235(8.1) | 79(9.8) | 281(5.4) | 251(5.3) | 30(6.5) |
| Hematologic Dis. | 436 (4.9) | 343 (4.5) | 93 (7.4)*** | 272(7.3) | 202(6.7) | 70(8.7) | 164(3.1) | 141(2.9) | 23(5.0)* |
| Pregnancy | 480 (5.4) | 364(4.7) | 116(9.2)*** | 460(12.4) | 344(11.9) | 116(14.4) | 20(0.38) | 20(0.4) | 0(0.0) |
| Obesity | 3046 (34.2) | 2677(35.0) | 369 (29.2)*** | 1178(31.8) | 985(34.0) | 193(24.0)*** | 1868(35.9) | 1692(35.7) | 176(38.4) |
| IS | 711 (7.9) | 564(7.4) | 147(11.6)*** | 419(11.3) | 305(10.5) | 114(14.2)** | 292(5.6) | 1(0.02) | 1(0.22) |
| Treatments and procedures | |||||||||
| EAT | 7410 (83.2) | 6228(81.5) | 1182(93.6)*** | 3240(87.5) | 2452(84.6) | 788(97.9)*** | 4170(80.2) | 3776(79.6) | 394(86.9)*** |
| Corticosteriods | 5275 (59.3) | 4530(59.3) | 745(59.0) | 1438(38.8) | 1048(36.2) | 390(48.4)*** | 3837(73.8) | 3482(73.4) | 355(77.5) |
| IMV | 5998 (67.4) | 3512(46.0) | 740(58.6)*** | 2072(56.0) | 1566(54.1) | 506(62.9)*** | 3926(75.5) | 3510(74.0) | 416(90.8) |
| AKI | 1435 (16.1) | 1081(14.2) | 354(28.0)*** | 904(24.4) | 608(21.0) | 296(36.8)*** | 531(10.2) | 473(9.9) | 58(12.7) |
| Prone IMV | 4064 (45.6) | 3469(45.4) | 595(47.1) | 1101(29.7) | 837(28.9) | 264(32.8)* | 2963(57.0) | 2632(55.5) | 331(72.3)*** |
| Shock | 3549 (39.9) | 2827(37.0) | 722(57.2)*** | 1899 (51.3) | 1363(47.0) | 536(66.6)*** | 1650(31.7) | 1464(30.9) | 186(40.6)*** |
| Outcomes | |||||||||
| LOS ICU, days | 13 (6-23) | 12(6-23) | 14(6-27)*** | 10 (4-18) | 10.0(4-18) | 10.0(5-19) | 15(8-27) | 14(7-26) | 23(13-37)*** |
| IMV days | 12(6-23) | 12(6-23) | 13(7-25) | 8(3-17) | 8(3-16) | 10(4-18)*** | 15(8-27) | 15(8-27) | 19(11-33)*** |
| ICU mortality | 2294 (25.8) | 1872 (24.5) | 422(33.4)*** | 796 (21.5) | 552(19.1) | 244(30.3)*** | 1498(28.8) | 1320(27.8) | 178(38.9)*** |
| Microorganism | COI Whole population n= 1263 |
Influenza cohort n=805 |
COVID-19 cohort n=458 |
| Streptococcus pneumoniae, n (%) | 433 (32.3) | 367 (44.8) | 66(12.4) |
| Staphylococcus aures Methicillin-sensitive, n (%) | 172 (12.8) | 99(12.1) | 73(13.8) |
| Pseudomonas aeruginosa, n (%) | 143 (10.6) | 56(6.9) | 87(16.4) |
| Aspergillus spp. n (%) | 78 (5.8) | 42(5.2) | 36(6.8) |
| Escherichia coli, n (%) | 69 (5.1) | 23(2.8) | 46(8.7) |
| Klebsiella spp. ,n (%) | 66 (4.8) | 19(2.3) | 47(8.8) |
| Haemophilus influenzae, n (%) | 61 (4.5) | 38(4.7) | 23(4.3) |
| Staphylococcus aures Methicillin-resistant, n (%) | 56 (4.2) | 34(4.1) | 22(4.1) |
| Streptococcus pyogenes, n (%) | 45 (3.3) | 45(5.6) | 0(0.0) |
| Enterobacter spp., n (%) | 30 (2.3) | 4(0.5) | 26(4.9) |
| Serratia spp, n (%) | 23 (1.6) | 8(1.0) | 15(2.8) |
| Staphylococcus hominis | 22 (1.6) | 6(0.7) | 16(3.0) |
| Stenotrophomonas maltophilia, n (%) | 21 (1.5) | 6(0.7) | 15(2.8) |
| Moraxella catarrhalis, n (%) | 15 (1.1) | 12(1.4) | 3(0.6) |
| Acinetobacter baumannii, n (%) | 14 (1.0) | 14(1.7) | 0(0.0) |
| Chlamydia spp, n (%) | 10 (0.7) | 5(0.6) | 5(0.9) |
| Mycoplasma spp. n (%) | 10 (0.7) | 5(0.6) | 5(0.9) |
| Staphylococcus haemolyticus, n (%) | 10 (0.7) | 0(0.0) | 10(1.8) |
| Streptococcus agalactiae, n (%) | 5 (0.4) | 0(0.0) | 5(0.9) |
| Coxiella burnetii, n (%) | 5 (0.4) | 0(0.0) | 5(0.9) |
| Pneumocystis jirovecii, n (%) | 5 (0.4) | 5(0.6) | 0(0.0) |
| Morganella morganii, n (%) | 4 (0.3) | 2(0.2) | 2(0.4) |
| Proteus spp, n (%) | 4 (0.3) | 0(0.0) | 4(0.8) |
| Corynebacterium spp., n (%) | 4 (0.3) | 0(0.0) | 4(0.8) |
| Citrobacter spp., n (%) | 4 (0.3) | 0(0.0) | 4(0.8) |
| Others , n (%) | 40(3.0) | 29(3.5) | 11(2.0) |
| Total, n (%) | 1342(100) | 819(100) | 530(100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





