Submitted:
16 September 2024
Posted:
17 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials
2.1. Animals for Cell Culture
- Cortical astrocytes were obtained from the brains of newborn pups (postnatal day (P)1-P4) from either Wistar rats or high-anxiety behavior (HAB) rats (from the laboratory of Prof. Inga Neumann, University of Regensburg) or Wistar Kyoto rats (WKY). Procedures follow institutional guidelines of the University of Regensburg, Germany, as well as the European Community Council Directive (86/609/EEC). The Animal Welfare Office at the Animal Care and Use Facility of the University of Regensburg approves all experiments. In accordance with the German Animal Welfare Act (Tierschutzgesetz, Articles 4 and 7), no additional approval for the post-mortem removal of brain tissue is necessary.
2.2. Media
-
Dissection Solution (DS)
- Hank’s Balanced Salt Solution (HBSS) 500ml e.g., Thermo Scientific 14025092
- Pen/Strep e.g., Thermo Scientific 10378016
- 1M HEPES e.g., Thermo Scientific 15630080
- 200mM L-Glutamine e.g., Thermo Scientific™ 25030081
-
Neuronal Growth Medium (NGM)
- Neurobasal™ Plus Medium 500ml e.g., Gibco 21103049
- B27 Supplement e.g., Gibco 17504044
- 200mM L-Glutamine or GlutaMAX™ Supplement e.g., Thermo Scientific™ 35050061
- Antibiotics/Antimitotic e.g., Sigma A5955
-
Astrocyte Full Medium
- DMEM, Dulbecco’s Modified Eagle + 4.5 g/L D-Glucose, L-glutamine e.g., Gibco 11965092
- Heat Inactivated Fetal Calf Serum (FCS) e.g., Gibco 26010074
- Sodium-Pyruvate 100mM, (100x) e.g., Sigma-Aldrich 11360070
- Antibiotics/Antimitotic
- MEM Non-Essential Amino Acids (NEAA), (100x), 100 mL e.g., Gibco 11140050
- 1M HEPES
-
Low medium
- DMEM, Dulbecco’s Modified Eagle + 4.5 g/L D-Glucose, L-glutamine
- Sodium-Pyruvate 100mM, (100x)
- Antibiotics/Antimitotic
- MEM Non-Essential Amino Acids (NEAA), (100x)
- 1M HEPES
- 24-well plate coated w/ PDL & Laminin e.g., Corning - Costar®
- Trypsin-EDTA solution e.g., Sigma T4049
- Millipore H2O
- Culture One
- Dissection tools
- Petri Dishes
- T75 flasks
- Neubauer Chamber
- Trypan Blue
2.3. Preparation of Slides for Immunofluorescence-Immunocytochemistry (IF-IHC)
- Round-shaped glass (coverslips) coated with poly-D-lysine (PDL) to plate cells
- Phosphate-buffered saline (PBS, 10X): 137 mM NaCl, 27 mM KCl, 14 mM Na2HPO4, and 43 mM KH2PO4. Bring total volume to 1L with deionized water
- Adjust pH to 7.5. Sterilize by autoclaving.
- 4% Paraformaldehyde (PFA) in PBS 1X, pH 7.5
- PBS 1X
- 24-well plates
- Pre-cleaned microscope slides (e.g., Superfrost/plus, Fisher Scientific)
- Pipettes and other general laboratory equipment like dissection kit
- Horizontal shaker
2.4. Immunofluorescent Staining
- 24-well plates
- Washing buffer: PBS 1X
- Normal goat serum (NGS)
- Blocking/permeabilization buffer: PBS 1X, 0.1% Triton X-100 and 2% normal goat serum
- Antibody buffer solution: PBS 1X, 0.1% Triton X-100 and 2% normal goat serum
-
Primary antibodies:
- Mouse monoclonal anti-Glial Fibrillary Acidic Protein (GFAP) antibody (1:400, e.g., Sigma–Aldrich Chemicals G3893) to label astrocytes
- Mouse monoclonal anti-S-100 (β-Subunit) antibody (1:1000, e.g., Sigma–Aldrich Chemicals S2532) to label astrocytes
- Rabbit polyclonal anti-Lysosome Associated Membrane Protein 1 (LAMP1) antibody (1:100, e.g., Abcam ab24170) to label lysosomes
- Chicken polyclonal anti-Synaptophysin1 antibody (1:500, e.g., SySy 101006) to label presynaptic puncta
-
Secondary antibodies:
- Cy3-conjugated anti-mouse IgG antibody (1:400, e.g., Thermo Scientific A10521)
- Biotin-conjugated anti-rabbit IgG antibody (1:500 e.g., Jackson lab 11-065-003)
- Alexa Fluor™ 488-conjugated goat anti-chicken IgG antibody, (1:500 e.g., Invitrogen A11035)
-
Tertiary antibodies:
- Alexa Fluor™ 647-conjugated Streptavidin (1:1000 e.g., Invitrogen S21374)
- 4′,6-diamidino-2-phenylindole (DAPI, 1:1000 e.g., Sigma D9542) to label nuclei
- Mounting medium (e.g., DAKO S3023)
- Round-shaped glass plate to mount slices on slides
- Pre-cleaned microscope slides (e.g., Superfrost/plus, Fisher Scientific).
- Confocal microscope
3. Methods
3.1. Preparation of Astrocyte-Neuron Co-Culture
3.1.1. Preparation of Rat Primary Astrocytes (All Steps Are Executed at Room Temperature (RT) Except when Stated Otherwise)
- Primary astrocytes are isolated from your region of interest from pup brains between P1 and P4
- Decapitate pups, open the skull, carefully extract the brains and remove meninges
- Isolate your region of interest by opening the two hemispheres under a light microscope and cut it out in Dissection Solution (we keep male and female brains separated)
- Mince the tissue in small pieces
- Transfer the minced tissues into a 15 mL falcon in dissociation buffer
- Carefully discard buffer
- Wash tissues with 2 mL Trypsin-EDTA
- Incubate tissue with 1,5 mL Trypsin-EDTA for approximately 20 min at 37°C in the waterbath (invert the falcon tube every 5 min)
- Discard trypsin
- Wash 3 times with 3-4 mL with Astrocyte Full Medium
- Add 1,5 mL of Astrocyte Full Medium and homogenize the tissue with a fire-polished pasteur pipette until suspension is homogenous
- Centrifuge at 1200rpm for 3 min
- Discard the supernatant
- Resuspend the cells in Astrocyte Full Medium (~10 mL)
- Plate cells on poly-D-lysine (PDL) coated T75 cm2 flask at 37°C and 5% CO2 in oxygen-reduced conditions (5% O2)
- Let cells grow in culture for approximately 7-10 days in vitro (changing the medium every ~4 days)
- After reaching approximately 90% confluency, shake flasks for 6 hours on a rotating shaker at circa 300 rpm in order to remove non-astrocytic cells from the culture
- Discard medium
- Wash cells with warm PBS
- Pipette 3 mL of warm Trypsin-EDTA and incubate for 4 min at 37°C
- Add 7 mL of Astrocyte Full Medium in the flask to neutralize trypsin
- Collect cells in a 15 mL falcon tube by pipetting up and down several times
- Centrifuge cells at 1200 rpm for 3 mins
- Discard supernatant
- Resuspend cells in 10 mL freezing medium and aliquot them (approximately 1x106 cells/vial)
-
Gradually freeze cells:
- -20°C for 24h
- -80°C for 24h
- Liquid nitrogen for long-term storage
3.1.2. Culture and Maintenance of Rat Primary Astrocytes
- Cell aliquots containing approximately 1x106 cells (stored in liquid nitrogen) are quickly thawed in the water bath and plated on a PDL coated T75 flask in Astrocyte Full Medium at 37°C and 5% CO2 in oxygen-reduced conditions (5% O2).
- Change medium every 3-4 days
3.1.3. Splitting Cells for Experiments/Culture Maintenance
- Once confluent (~80-90%) discard medium and wash once with warm PBS
- Add 3 mL of Trypsin-EDTA in the flask and incubate for 4 min at 37°C
- Add 7 mL of Astrocyte Full Medium and collect cells in a 15 mL falcon
- Centrifuge at 1200 rpm for 3 min
- Discard supernatant and resuspend cells in ~10 mL of Astrocyte medium
- Primary astrocyte cells are used until passage 3 (see Note 1)
3.2. Preparation of Rat Primary Neurons
- Use late embryonic stage (E18) rat fetuses, euthanize dam with CO2, remove uterus, free individual fetuses from the embryonic sack and collect them in T75 flask with 50ml DS
- Place fetuses into a Petri dish and decapitate them, open the skull, carefully extract the brains and remove meninges.
- Dissect the two hemispheres and place brains in a new petri dish with DS
- Isolate the cortex and mince the tissue
- Transfer the minced tissue to a 15ml falcon with DS
- Carefully discard DS
- Wash tissues with 2 ml Trypsin-EDTA
- Incubate tissue with 1,5ml Trypsin-EDTA for approximately 20 min at 37°C in the waterbath (invert the falcon tube every 5 min)
- Discard Trypsin
- Wash 3 times with 3-4 ml with Neuron Growth Medium
- Add 1,5ml of Neuron Growth Medium and homogenize the tissue with a fire-polished Pasteur pipette until suspension is homogenous (circa 20X)
- Centrifuge at 900 rpm for 5 min
- Discard supernatants. Resuspend in 1ml of Neuron Growth Medium
- Counting cells in Neubauer chamber
- Cell plating: 45.000 cells/well for single cell culture, 90.000 cells/well for co-culture
- Renew half of the Neuron Growth Medium once per week
- Let neurons grow 2 to 3 weeks, depending on the maturational stage required
3.2.1. Fixation
- Remove medium
- Wash 3 times for 5 minutes with PBS 1X
- Incubate with PBS 1X with PFA 4% for 40 min
- Wash 3 times for 10 min with PBS 1X
- Store at 4°C
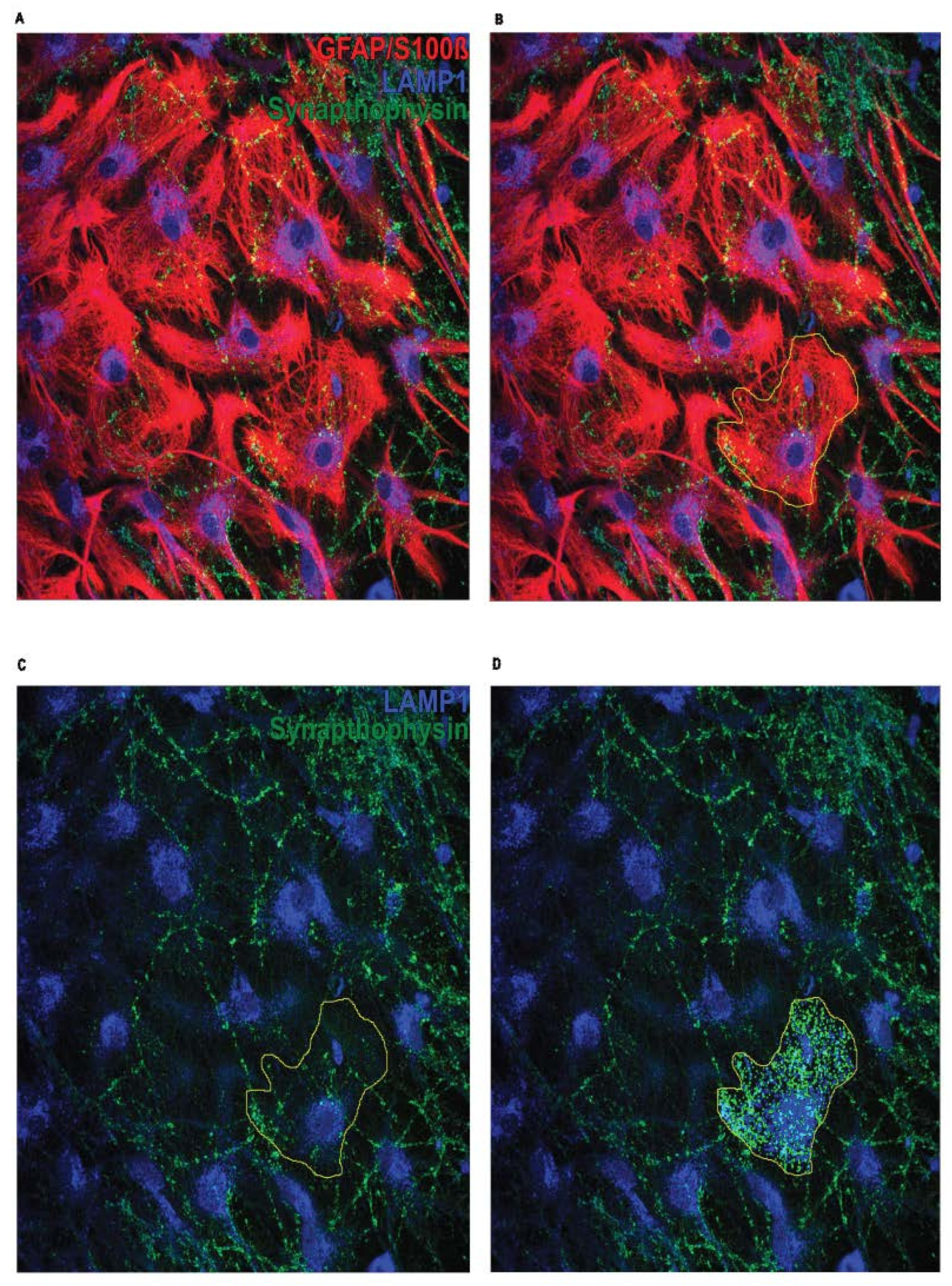
3.3. Immunofluorescent-Immunocytochemistry-GFAP/S100ß/LAMP1/Synaptophysin
- Take out the coverslips and place them in a fresh 24 well plate.
- Wash carefully 2 times for 10 min with PBS 1X (all incubation and washing steps require the plate to be on the shaker at around 100rpm, see Note 2)
- Add blocking/permeabilization solution: PBS 1X with 0.1% Triton X-100/2% NGS and incubate for 1 hour (see Note 3)
- Wash carefully 3 times for 10 mix with PBS1X
- Incubate in primary antibody solution: PBS 1X with 0.1% Triton X-100 and 2% NGS O/N at 4°C protected from light. Make sure to cover fully the wells, the recommended minimum amount is 150µl.
- The next day wash carefully 3 times with PBS1X and keep protected it from light
- Incubate in secondary antibody solution: PBS 1X with 2% NGS for 1,5 hours protected from light.
- Wash carefully 3 times with PBS1X and keep protected from light
- Incubate the tertiary staining solution: PBS 1X with 2% NGS for 1,5 hours protected from light.
- Wash carefully 3 times with PBS1X and keep protected from light
- Put a single drop of mounting medium on slides for each coverslip and place the coverslip downwards on slides (see Notes 4).
3.4. Confocal Microscopy
- Choose the suitable lasers to detect the fluorochromes and adjust settings to ensure optimal fluorescent saturation.
- Capture a minimum of 6 to 7 images per coverslip, each consisting of 14 Z-stack slices with a thickness of 0.30 μm, using an Olympus confocal microscope equipped with a 40x oil immersion objective (see Note 5 and Note 6)
- Save the acquired images in the Olympus Image file format.
3.5. Analysis of Astrocyte-Mediated Phagocytosis in Astrocyte-Neuron Co-Cultures
- Open Image J
-
Drag&Drop to Image JColor Mode: Default
- Change to 8-bit: Image → Type → 8Bit; for every channel
- Open ROI manager: Analyse → Tool → ROI manager
-
Merge Channels → Image → Color →Merge Channel
- Preset is C1 (red): (C2) please change it according to colors on the images
- Preset is C2 (green): (C1) please change it according to colors on the images
- Preset is C3 (blue): (C3) please change it according to colors on the images
- Find a cell that is not overlapping with others
- Image → Stacks → Z Projekt → Projekttype: Max Intensity (for every astrocyte use 5-6 slices with the highest intesity)
- Image → Color → Channel tool → choose astrocyte channel
 Freehand selections: mark the astrocyte → Add (to ROI manager)
Freehand selections: mark the astrocyte → Add (to ROI manager)- Image → Color → Split Channels → remove astrocyte channel
- Image → Color → Merge Channels
- Plugins → ComDet v.0.5.5 → Detect particles
- Select pixels and intensity for every channel and keep them identical for every image
-
Suggested starting parameters (see Note 7 and Note 8):
- distance between pixels: 2
- particle size: 5
- threshold: 3
- Summary opens
- Copy to excel file
- Also save ROI → rename: e.g., NAB Pic1 Astro1)
3.6. Statistical Analysis
- For each coverslip: calculate the mean value for “Synaptophysin”, “LAMP1”, and colocalizing spots from the average of 10 astrocytes
- Use unpaired t-Test for the comparison between two groups, and One-way ANOVA when comparing more than two groups.
Acknowledgements
| 1 | Use astrocytes ideally between passages 1 and 3 and avoid excessive passaging to preserve their morphological and functional properties |
| 2 | All the incubation and washing steps require the plate to be on a shaker on less than 110 rpm |
| 3 | During the change of solutions, slices should never get dried. |
| 4 | Be careful not to form bubbles in the mounting medium as they might interfere with imaging at the confocal microscope. In case bubbles form, you can gently pull them out with the help of a pipette tip pressed on the coverslip. |
| 5 | Choose the appropriate image frame size, considering the subsequent image analysis. Ensure that one or more astrocytes are captured in the image, avoiding isolation or excessive overlap with dendrites. |
| 6 | Utilize 5 to 6 Z-stack slices where the targeted astrocyte is located. Due to potential variations in image acquisition influenced by experimental procedures related to immunostaining, the condition of the co-culture, and the status of the fluorescent fluorophore, it is advisable to conduct a pilot immunostaining using coverslips from various co-cultures to establish the most suitable settings for image analysis. |
| 7 | Particle size and threshold is the same for both channels |
| 8 | Parameters need adjustment based on the pictures and staining |
References
- Khakh BS, Sofroniew MV (2015) Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci 18:942–952. [CrossRef]
- Sofroniew MV (2020) Astrocyte Reactivity: Subtypes, States, and Functions in CNS Innate Immunity. Trends in Immunology 41:758–770. [CrossRef]
- Abbott NJ, Rönnbäck L, Hansson E (2006) Astrocyte–endothelial interactions at the blood–brain barrier. Nat Rev Neurosci 7:41–53. [CrossRef]
- Allen NJ (2014) Astrocyte Regulation of Synaptic Behavior. Annu Rev Cell Dev Biol 30:439–463. [CrossRef]
- Mahmoud S, Gharagozloo M, Simard C, Gris D (2019) Astrocytes Maintain Glutamate Homeostasis in the CNS by Controlling the Balance between Glutamate Uptake and Release. Cells 8:184. [CrossRef]
- Oberheim NA, Goldman SA, Nedergaard M (2012) Heterogeneity of Astrocytic Form and Function. In: Milner R (ed) Astrocytes: Methods and Protocols. Humana Press, Totowa, NJ, pp 23–45.
- Arnaud AM, Brister TS, Duckworth K, et al. (2022) Impact of Major Depressive Disorder on Comorbidities: A Systematic Literature Review. J Clin Psychiatry 83:. [CrossRef]
- Cobb JA, O’Neill K, Milner J, et al. (2016) Density of GFAP-immunoreactive astrocytes is decreased in left hippocampi in major depressive disorder. Neuroscience 316:209–220. [CrossRef]
- Rajkowska G, Hughes J, Stockmeier CA, et al. (2013) COVERAGE OF BLOOD VESSELS BY ASTROCYTIC ENDFEET IS REDUCED IN MAJOR DEPRESSIVE DISORDER. Biol Psychiatry 73:613–621. [CrossRef]
- Rajkowska G, Stockmeier CA Astrocyte Pathology in Major Depressive Disorder: Insights from Human Postmortem Brain Tissue. Current Drug Targets 14:1225–1236.
- Lee J-H, Kim J, Noh S, et al. (2021) Astrocytes phagocytose adult hippocampal synapses for circuit homeostasis. Nature 590:612–617. [CrossRef]
- Selective induction of astrocytic gliosis generates deficits in neuronal inhibition | Nature Neuroscience. https://www.nature.com/articles/nn.2535. Accessed 27 Sep 2022.
- Antoine MW, Langberg T, Schnepel P, Feldman DE (2019) Increased Excitation-Inhibition Ratio Stabilizes Synapse and Circuit Excitability in Four Autism Mouse Models. Neuron 101:648-661.e4. [CrossRef]
- Dejanovic B, Wu T, Tsai M-C, et al. (2022) Complement C1q-dependent excitatory and inhibitory synapse elimination by astrocytes and microglia in Alzheimer’s disease mouse models. Nat Aging 2:837–850. [CrossRef]
- Park J, Choi Y, Jung E, et al. (2021) Microglial MERTK eliminates phosphatidylserine-displaying inhibitory post-synapses. The EMBO Journal 40:e107121. [CrossRef]
- Lee E, Chung W-S (2019) Glial Control of Synapse Number in Healthy and Diseased Brain. Frontiers in Cellular Neuroscience 13:.
- Neumann ID, Wegener G, Homberg JR, et al. (2011) Animal models of depression and anxiety: What do they tell us about human condition? Progress in Neuro-Psychopharmacology and Biological Psychiatry 35:1357–1375. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




